Back to Journals » OncoTargets and Therapy » Volume 8
rs712 polymorphism within let-7 microRNA-binding site might be involved in the initiation and progression of colorectal cancer in Chinese population
Authors Jiang Q, Peng H, Zhang Y, Tian P, Xi Z, Chen H, Yang Z
Received 3 June 2015
Accepted for publication 15 August 2015
Published 22 October 2015 Volume 2015:8 Pages 3041—3045
DOI https://doi.org/10.2147/OTT.S89746
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr William C. Cho
Qiang-Hua Jiang,1,* Hong-Xin Peng,2,3,* Yi Zhang,4 Peng Tian,4 Zu-Lian Xi,4 Hao Chen4
1Department of Surgery, Dongyang People’s Hospital, Dongyang, Zhejiang, 2Medical College, Southeast University, 3Central Laboratory, Nanjing First Hospital, Nanjing, Jiangsu, 4Hubei Key Laboratory of Biological Targeted Therapy, Yichang Yiling Hospital, Yichang, Hubei, People’s Republic of China
*These authors contributed equally to this work
Abstract: rs712 within 3'-untranslated region of KRAS can affect the specific binding between the mRNA and its targeted microRNAs, leading to the activation of KRAS oncogene. However, the possible association between the locus and susceptibility to colorectal cancer (CRC) remains unclear. We investigated genotypes of the locus in 586 cases and 476 controls to explore the possible association between them. Results of our case–control study showed that genotypes TT (6.5% vs 2.5%, P=0.002, adjusted odds ratio [OR] =2.810, 95% confidence interval [CI] =1.342–5.488) and GT/TT (36.5% vs 30.5%, P=0.038, adjusted OR =1.342, 95% CI =1.030–1.712) and allele T (21.5% vs 6.5%, P=0.004, adjusted OR =1.328, 95% CI =1.105–1.722) of rs712 were significantly associated with an increased risk of CRC, and the significant association was also observed in the recessive model (TT vs GG/GT, 6.5% vs 2.5%, P=0.003, adjusted OR =0.372, 95% CI =0.191–0.725). However, there was no association between genotype GT and risk of CRC (30.0% vs 28.0%, P=0.235, adjusted OR =1.210, 95% CI =0.903–1.548). Furthermore, genotype GT (P=0.003) and allele T (P=0.003) were significantly associated with poor differentiation, and genotypes GT and TT and allele T were significantly associated with tumor-node-metastases stage III (P=0.001 for GT vs GG, P<0.001 for TT vs GG, and P<0.001 for T vs G) and node metastasis (P<0.001 for GT vs GG, P=0.001 for TT vs GG, and P<0.001 for T vs G), respectively. These findings indicated that allele T and genotypes TT and GT/TT of rs712 might be susceptible factors for CRC, and mutated allele and genotypes of the locus might predict a poor clinical outcome in Chinese population.
Keywords: rs712, CRC, polymorphism, susceptibility
Introduction
Colorectal cancer (CRC) is a complex disease initiated by the interaction of environmental and personal genetic factors. According to the latest report of American Cancer Society, ~132,700 persons will be diagnosed as new CRC cases and 49,700 individuals with CRC will die in 2015.1 In 2011, there were 310,244 newly diagnosed CRC patients and 149,722 dead CRC cases in the People’s Republic of China.2 Although there is an increase in the research and development of strategies for its prevention, diagnosis, and therapy, the etiology of CRC cases remains poorly understood, and the clinical outcome of the case is not significantly improved.
It is well known that many causes, such as chromosome instability, loss of heterozygosity, and promoter CpG island methylation, can lead to carcinogenesis and progression of CRC.3 However, these can only account for a small proportion of the cases. Recently, accumulating evidence indicated that single nucleotide polymorphisms (SNPs) of oncogenes and suppressed genes were involved in the onset of CRC.4 A recent meta-analysis reported that rs16892766 polymorphism within the region of 8q23.3 was significantly associated with its susceptibility to CRC.5 Wu et al6 reported that C-1306T polymorphism within MMP-2 is a risk factor for CRC susceptibility, especially in Caucasians. These studies suggested that SNP within key gene might be an important research field to explore the cause of CRC individual.
KRAS is one of the most important oncogene, which is located at 12p12.1. It is a member of RAS family and encodes a GDP/GTP-binding protein that belongs to a small GTPase superfamily.7 The product is involved in RAF/MEK/MAPK, AKT, and ERK signal transduction pathways and regulates cell proliferation and differentiation in many kinds of cancer cell lines, including CRC cell lines. Moreover, high frequency of KRAS mutation had been examined in many kinds of solid cancer, such as CRC, thyroid cancer, and non-small-cell lung cancer.8–10 In CRC, a frequency of 59% CRC patients were detected as KRAS-mutated cases.11 Furthermore, genetic variation of KRAS is involved in tumorigenesis and progression of CRC as well as response to antiepidermal growth factor receptor monoclonal antibody in metastatic CRC.12–14 rs712 has been reported to be significantly associated with the risk of cancer.12 Blons et al14 reported that KRAS exon 2 mutation was involved in the clinical outcome of CRC and might be an independent prognostic biomarker to predict recurrence in resected stage III distal colon cancer patients undergoing adjuvant therapy. Additionally, cetuximab has been recommended to be used in the treatment for metastatic CRC patients with wild-type KRAS.15
Let-7, the first discovered microRNA containing let-7a–g and I, acted as a tumor suppressor in many kinds of malignancies including CRC.16 It can bind to specific seed regions in mRNA 3′-untranslated region (UTR) of KRAS, and the downregulated expression can affect its function in the processes of CRC carcinogenesis and progression. There are ten let-7 complementary binding sites (LCSs) in 3′-UTR of KRAS mRNA.17 rs712, which is located in LCS1 of KRAS 3′-UTR,17 has been intensively investigated in many kinds of malignancies, such as gastric cancer, oral squamous cell carcinoma, and nasopharyngeal carcinoma.12,18 So, we speculated that the locus of KRAS may also associate with CRC. Thus, we performed this case–control study to investigate the effect of rs712 within KRAS on the susceptibility to CRC in Chinese population.
Materials and methods
A total of 1,062 participants were enrolled in the present study. The case group consisted of 586 clinically and pathologically confirmed tumor-node-metastasis stage I–III CRC patients, and 476 healthy checkup individuals were selected as the controls. All of them were recruited from Yichang Yiling Hospital, Dongyang People’s Hospital, and Nanjing First Hospital between January 2012 and December 2014. Meanwhile, 1 mL peripheral blood from each individual was collected. Clinical data, such as individual’s age, sex, and pathological features, were collected from clinical medical record. Written informed consent was signed by each participant, and this study was approved by the ethics committee of Yichang Yiling Hospital, Hubei, People’s Republic of China.
Genomic DNA was extracted from 200 μL peripheral blood using CWBIO genomic DNA extracted kit (CWBIO, Beijing, People’s Republic of China) according to the manufacturer’s protocol and stored at −80°C for detection. Ultraviolet spectrophotometer (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA) was used to examine the concentration and purity of the extracted DNA. Polymerase chain reaction-restriction fragment length polymorphism was selected to detect rs712 genotype. The restriction enzyme, reaction, and agarose electrophoresis condition were used as reported by Jin et al.19 Meanwhile, 5% samples were randomly selected to DNA sequencing.
Genotype frequencies of two groups were obtained by counting. GenAIEx 6.5 software (Peakall R and Smouse SE, Australian National University) was used to test Hardy–Weinberg equilibrium (HWE) in the control group.20 Two-sided Student’s t-test was selected to compare the differences in the quantitative data, and χ2-test was used to analyze the rs712 genotype or allele frequency differences between the two groups. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were selected to estimate the strength of association between rs712 and risk of CRC. All statistics were performed using SPSS software 17.0 (SPSS Inc., Chicago, IL, USA), and P<0.05 was considered as statistically significant.
Results
A260/A280 value of each sample was in an interval of 1.8–2.1, thus all 586 cases and 476 healthy checkup individuals were included in our study. The baseline characteristics of the two groups were listed in Table 1. As shown in Table 1, there was no significant difference in age, sex, and status of smoking and drinking in the case and control groups. In the case group, 332 and 254 patients were colon and rectum cancer cases, respectively. Patients with well and poor differentiation were 412 and 174, respectively. The frequency distributions of tumor-node-metastasis stage (I–II/III), invasion (T1–2/T3–4), and node metastasis (N0/N1–3) were 69.6% and 30.4%, 47.8% and 52.2%, and 69.6% and 30.4%, respectively.
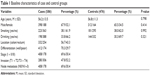  | Table 1 Baseline characteristics of case and control groups |
P-value of HWE in the control group was 0.753, suggesting that the genotype distribution of the locus was fit for HWE. The frequencies of genotypes GG, GT, and TT were 63.5%, 30.0%, and 6.5% in cases and 69.5%, 28.0%, and 2.5% in controls, respectively. There was no difference between the two groups in comparison of GT vs TT (30.0% vs 28.0%, P=0.235, adjusted OR =1.210, 95% CI =0.903–1.548) (Table 2). Furthermore, genotype TT distribution (6.5% vs 2.5%, P=0.002, adjusted OR =2.810, 95% CI =1.342–5.488) in cases was significantly higher than that in controls. Frequencies of allele T (21.5% vs 16.5%, P=1.328, adjusted OR =1.328, 95% CI =1.105–1.722) and allele T carrier (GT/TT) (36.5% vs 30.5%, P=0.038, adjusted OR =1.342, 95% CI =1.030–1.712) were significantly higher in cases compared to controls (Table 2). In addition, distribution of genotype allele G carrier (GG/GT) was significantly lower in cases than controls (93.5% vs 97.5%, adjusted OR =0.372, 95% CI =0.191–0.725) (Table 2).
The association between rs712 and clinical baseline characteristics was described in Table 3. There was no difference between rs712 genotype and allele frequency in colon and rectum cancer subgroups (P=0.853 for GT vs GG, P=0.795 for TT vs GG, and P=0.946 for T vs G). No association was found between rs712 and invasion depth in the case group (P=0.885 for GT vs GG, P=0.460 for TT vs GG, and P=0.530 for T vs G), whereas genotype GT (P=0.003) and allele T (P=0.003) were significantly associated with poor differentiation; genotypes GT and TT and allele T were significantly associated with stage III (P=0.001 for GT vs GG, P<0.001 for TT vs GG, and P<0.001 for T vs G) and node metastasis (P<0.001 for GT vs GG, P=0.001 for TT vs GG, and P<0.001 for T vs G) in the case group, suggesting that rs712 would be associated with CRC progression in Chinese population.
  | Table 3 rs712 within KRAS and clinical pathological feature in cases |
Discussion
KRAS, encoding a member of small GTPase superfamily proteins, is an important oncogene in the initiation of CRC.21 Variations within KRAS have been reported to be significantly associated with susceptibility, drug resistance, and poor prognosis in patients with CRC.12,22,23 Recently, some studies reported the association between SNP of KRAS and susceptibility to CRC. rs61764370 within KRAS 3′-UTR was not related to the risk of ovarian or breast cancer.24 Significant associations were observed between rs712 and susceptibility to papillary thyroid cancer, gastric cancer, and oral squamous cell carcinoma.18,19,25 However, few studies reported the association between rs712 and the risk of CRC, and the possible association between them in Chinese population remains unclear. A case–control designed study including 586 CRC clinically and pathologically confirmed CRC patients and 476 healthy individuals was conducted. Our results showed that there was no significant distribution difference between the two groups in comparison of GT vs TT, suggesting that genotype GT of the locus was not associated with the risk of CRC. However, frequencies of allele T and genotypes TT and GT/TT within rs712 were significantly higher in cases than controls, indicating that mutated allele and genotypes of the locus were significantly related to an increased susceptibility to CRC. Furthermore, frequency of genotype GG/GT within the locus was significantly lower in cases than controls, suggesting that genotype GG/GT was negatively associated with CRC and it might be a protective factor for the onset of CRC. Additionally, genotype GT and allele T within rs712 were significantly associated with poor differentiation in cases; genotypes GT and TT and allele T were significantly associated with an increased risk of stage III and node metastasis, respectively, suggesting that rs712 might involve in the progression of CRC, and genotype TT and allele T of the locus could predict poor prognosis in Chinese population. The following interpretations might be accounted for our findings. rs712, which is located in LCS1 of KRAS 3′-UTR, alternation of allele G to T, can alter the secondary structure of the mRNA, disrupt let-7 binding site, affect the accessibility of let-7, and influence combining affinity between them.26,27 Moreover, lower levels of let-7a, let-7b, and let-7c were detected in the CRC cell line SW480 when it harbored rs61764370, showing that lower level of let-7 might be associated with mutated allele of rs712.28 Consequently, aberrant expression of KRAS and low level of let-7 promote CRC cell proliferation and migration, contributing to carcinogenesis and progression of the disease.28
Conclusion
In summary, rs712 within 3′-UTR of KRAS was involved in the initiation and progression of CRC and mutated allele, and genotypes might be used as susceptible and prognostic biomarkers in Chinese population. With the limitations of this study, larger sample size and well-designed studies and function analysis are warranted to further validate our findings.
Acknowledgment
This study was supported by the Hubei Key Laboratory of Biological Targeted Therapy, Yichang Yiling Hospital, Hubei, People’s Republic of China.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. | ||
Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27:2–12. | ||
Cisyk AL, Penner-Goeke S, Lichtensztejn Z, et al. Characterizing the prevalence of chromosome instability in interval colorectal cancer. Neoplasia. 2015;17:306–316. | ||
Kraus S, Nabiochtchikov I, Shapira S, Arber N. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347:15–21. | ||
Liao M, Wang G, Quan B, et al. Analyzing large-scale samples confirms the association between rs16892766 polymorphism and colorectal cancer susceptibility. Sci Rep. 2015;5:7957. | ||
Wu Z, Jiang P, Zulqarnain H, Gao H, Zhang W. Relationship between single-nucleotide polymorphism of matrix metalloproteinase-2 gene and colorectal cancer and gastric cancer susceptibility: a meta-analysis. Onco Targets Ther. 2015;8:861–869. | ||
Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–82. | ||
Su J, Zhang XC, An SJ, et al. Detecting the spectrum of multigene mutations in non-small cell lung cancer by Snapshot assay. Chin J Cancer. 2014;33:346–350. | ||
Oczko-Wojciechowska M, Pfeifer A, Rusinek D, et al. The prevalence of somatic RAS mutations in medullary thyroid cancer – a Polish population study. Endokrynol Pol. 2015;66:121–125. | ||
Smith G, Bounds R, Wolf H, Steele RJ, Carey FA, Wolf CR. Activating K-Ras mutations outwith ‘hotspot’ codons in sporadic colorectal tumours – implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. | ||
Sakai K, Tsurutani J, Yamanaka T, et al. Extended RAS and BRAF mutation analysis using next-generation sequencing. PLoS One. 2015;10:e0121891. | ||
Ying HQ, Wang F, He BS, et al. The involvement of Kras gene 3′-UTR polymorphisms in risk of cancer and influence on patient response to anti-EGFR therapy in metastatic colorectal cancer: a meta-analysis. Onco Targets Ther. 2014;7:1487–1496. | ||
Ishige T, Itoga S, Sato K, et al. High-throughput screening of extended RAS mutations based on high-resolution melting analysis for prediction of anti-EGFR treatment efficacy in colorectal carcinoma. Clin Biochem. 2014;47:340–343. | ||
Blons H, Emile JF, Le Malicot K, et al. Prognostic value of KRAS mutations in stage III colon cancer: post hoc analysis of the PETACC8 phase III trial dataset. Ann Oncol. 2014;25:2378–2385. | ||
Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–1796. | ||
Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. | ||
Saridaki Z, Weidhaas JB, Lenz HJ, et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin Cancer Res. 2014;20:4499–4510. | ||
Wang WY, Chien YC, Wong YK, Lin YL, Lin JC. Effects of KRAS mutation and polymorphism on the risk and prognosis of oral squamous cell carcinoma. Head Neck. 2012;34:663–666. | ||
Jin H, Liang Y, Wang X, et al. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol. 2014;31:221. | ||
Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research – an update. Bioinformatics. 2012;28:2537–2539. | ||
Steven A, Heiduk M, Recktenwald CV, et al. Colorectal carcinogenesis: connecting K-RAS-induced transformation and CREB activity in vitro and in vivo. Mol Cancer Res. 2015;13(8):1248–1262. | ||
Deng Y, Wang L, Tan S, et al. KRAS as a predictor of poor prognosis and benefit from postoperative FOLFOX chemotherapy in patients with stage II and III colorectal cancer. Mol Oncol. 2015;9:1341–1347. | ||
Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. | ||
Ovarian Cancer Association Consortium; Breast Cancer Association Consortium; Consortium of Modifiers of BRCA1 and BRCA2, et al; Australian Ovarian Cancer Study Group; Breast Cancer Family Register, GEMO Study Collaborators; GENICA Network; KConFab Investigators. No clinical utility of KRAS variant rs61764370 for ovarian or breast cancer. Gynecol Oncol. 2015;15:00863-X. | ||
Li ZH, Pan XM, Han BW, et al. A let-7 binding site polymorphism rs712 in the KRAS 3′ UTR is associated with an increased risk of gastric cancer. Tumour Biol. 2013;34:3159–3163. | ||
Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. | ||
Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. | ||
Crowley EH, Arena S, Lamba S, Di Nicolantonio F, Bardelli A. Targeted knock-in of the polymorphism rs61764370 does not affect KRAS expression but reduces let-7 levels. Hum Mutat. 2014;35:208–214. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

