Back to Journals » OncoTargets and Therapy » Volume 10
RNA-binding protein HuR promotes bladder cancer progression by competitively binding to the long noncoding HOTAIR with miR-1
Authors Yu DP, Zhang C, Gui JQ
Received 18 January 2017
Accepted for publication 13 March 2017
Published 17 May 2017 Volume 2017:10 Pages 2609—2619
DOI https://doi.org/10.2147/OTT.S132728
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ingrid Espinoza
Dapeng Yu,1,* Chao Zhang,2,* Junqing Gui2
1Department of Urinary Surgery, the Jining No 1 People’s Hospital, Jining, 2Department of Urinary Surgery, the Qujing No 1 People’s Hospital, Qujing, People’s Republic of China
*These authors contributed equally to this work
Abstract: The elevated expressions of RNA-binding protein HuR and long noncoding HOX transcript antisense RNA (HOTAIR) are observed in numerous cancers. And HuR often exerts its promotive effects on tumorigenesis via binding to AU-rich elements in target transcripts and thus regulating the expression of target transcripts. However, the roles and related mechanisms of HuR/HOTAIR in bladder cancer progression have never been formally tested. Here, we found that the expression level of HuR was higher in clinical bladder cancer samples than in normal adjacent samples, mirroring that of HOTAIR, and their expression showed strong correlation. Knockdown of HuR/HOTAIR in bladder cancer inhibited cell proliferation, migration, invasion, and promoted cell apoptosis. Notably, HuR interacted and stabilized HOTAIR mRNA and knockdown of HuR decreased HOTAIR expression. Additionally, HOTAIR was identified as a potential target of miR-1 in bladder cancer cells. Interestingly, overexpression of HOTAIR enhanced HuR expression and increased cytoplasmic accumulation of HuR, thus enhancing HOTAIR expression in turn. But mutation of miR-1 binding site in HOTAIR canceled the effects of HOTAIR on HuR expression. Overall, we identified a regulatory loop between HOTAIR and HuR during the progression of bladder cancer, which could be exploited to curb bladder cancer progression.
Keywords: HuR, HOTAIR, bladder cancer, miR-1
Introduction
Bladder cancer is the most common genitourinary tumor, and it is a leading cause of cancer-associated deaths in developed countries.1 However, few achievements have been made in identifying new targets for bladder cancer. Therefore, it is vital to understand the mechanisms underlying the progression of bladder cancer, and thus help us find potential novel biomarkers or agents that combat bladder cancer.
Noncoding RNAs (ncRNA) have emerged as a new player in gene regulations that contribute to carcinogenesis, including long noncoding RNAs (lncRNAs)2 and miRNAs.3 The changes in miRNAs or lncRNAs expression levels have been observed in many cancers, and modulated various pathways involved in survival, proliferation, invasion, and apoptosis.4 One of the lncRNAs, HOX transcript antisense RNA (HOTAIR), has been shown to be correlated with disease progression in patients with prostate cancer,5 breast cancer,6 and head and neck squamous cell carcinoma,7 and also in bladder cancer, although HOTAIR has been proved to be related to poor survival of bladder cancer and contained in Bladder Cancer Patient Urinary Exosomes.8 However, the mechanisms regulating the expression of HOTAIR in bladder cancer are unclear.
HuR, as one of the well-known RNA binding proteins implicated in tumor development, is necessary for the mRNA stability via binding to AU-rich RNA motifs in 3′-untranslated region.9 Additionally, HuR enhanced mRNA stability and thus induced lncRNAs expression by binding to AU-rich RNA motifs.10 Although HuR is predominantly located in the nucleus, its nucleo-cytoplasmic translocation has been implicated in tumor development.11 Moreover, HuR inhibition held antimetastatic, proapoptotic, or antiangiogenic effects, demonstrating the strong potential of HuR as a therapeutic target in cancer.12 Recently, a feed-forward regulatory loop between HuR and HOTAIR has been shown in the progression and metastasis of head and neck squamous cell carcinoma,7 and we asked whether this relationship holds true in human bladder cancer.
In this study, we found that the expression levels of HuR and HOTAIR were upregulated and positively correlated in bladder cancer tissues and cells. In addition, we showed that knockdown of HuR or HOTAIR inhibited bladder cancer cell proliferation, migration, invasion, and increased cell apoptosis. Furthermore, HuR competitively bound to HOTAIR with miR-1 enhanced HOTAIR mRNA stability and promoted HOTAIR expression in bladder cancer. Conversely, HOTAIR induced the nucleo-cytoplasmic translocation of HuR. Meanwhile, the level of miR-1 was negatively correlated with HuR and HOTAIR expression levels in bladder cancer. Taken together, these results contribute significantly to our understanding of the role of the feed-forward regulatory loop between HuR and HOTAIR in bladder cancer and provide critical insights as to how HuR or HOTAIR may be targeted therapeutically to prevent bladder cancer.
Materials and methods
Clinical samples and cell culture
A total of 43 bladder transitional cell carcinoma (TCC) and 36 normal adjacent bladder transitional cell (NBTC) specimens were obtained from the Qujing No 1 People’s Hospital from May 2014 to October 2016. The Ethics Committees of Qujing No 1 People’s Hospital approved this study, and written informed consent from all patients was obtained before operation. Human TCC J82 and T24 cell lines and human bladder epithelial immortalized SV-HUC-1 cell line, and HEK293T cells were purchased from the Chinese Academy of Sciences Cell Bank, and were cultured in Dulbecco’s Minimum Essential Medium, which contained 10% fetal bovine serum (Life Technologies, USA), and culture condition was 5% CO2 at 37°C.
Quantitative real-time polymerase chain reaction
Total RNA was extracted by using Trizol reagent (Invitrogen, USA) following the manufacturer’s standard procedure, and then reverse transcribed into cDNA using RevertAid™ First Strand cDNA Synthesis Kit (ThermoFisher Scientific, Grand Island, NY, USA) and All-in-One™ miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia) according to the manufacturer’s instructions. The mRNA and miRNA expression levels were determined following the protocols of SYBR mix™ kit (Biomics, China) on an ABI Prism 7500 Detection System (Applied Biosystems, Inc., USA). β-actin was used as an internal reference for mRNAs and U6 was an internal reference for miRNAs. The relative gene expression levels were analyzed via 2−ΔΔct method.
Construction of HuR and HOTAIR stable knockdown cells
The shRNA sequences were purchased from Santa Cruz, and inserted into pLKO.1. HuR coding sequences and HOTAIR sequences were inserted into pLVX-IRES-ZsGreen1, named as Lenti-HuR and Lenti-HOTAIR. The lentivirus was produced in HEK293T cells by co-transfection with pCMV-dR8.2 and pMD2.G constructs. After 72 h, supernatants were collected and centrifuged to remove cell debris. The J82 and T24 cells were infected with the virus in the presence of 2 μg/mL Polybrene. The virus was removed after 24 h, and the infected cells were selected with puromycin (Sigma, 1.5 μg/mL for J82 cells and 2 μg/mL for T24 cells) for 2 weeks. Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analyses were used to verify the stable knockdown cells.
Cell migration and invasion assays
Transwell migration and invasion assays were performed by 24-well MILLIcell Hanging Cell Culture inserts 8 mm PET (MILLIPORE) precoated with or without Matrigel matrix gel (BD Biosciences). The detailed procedure was referred to the previous study.7 A total of 8×104 cells/200 μL in serum-free medium were added to the upper chamber. Twenty percent FBS in 800 μL medium was used as a chemoattractant in the bottom chamber. Cells were allowed to migrate or invade for 24 h. Cells on the upper side of the membrane were wiped off, and cells on the lower side of the membrane were fixed and stained with crystal violet solution. Five random fields from each of the triplicate migration assays were counted by using phase contrast microscopy. Quantification was done by measuring with Microplate Reader (optical density 570 nm) after being destained with glacial acetic acid. All migration and invasion assays were done in triplicate for at least three independent experiments.
Cell viability assay
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) (KeyGEN BioTECH, China) was used to examine cell viability. Cells with HuR knockdown, HOTAIR knockdown, or the wide-type cells were seeded in 96-well plates at 3,000 cells/well. After 24, 48, and 72 h, the growth rates were determined. Triplicate experiments were performed for each assay.
Cell cycle distribution
Cells (5×105 cells/well) with different treatment were seeded in six-well plates and serum starved for 24 h. Then washed with ice-cold phosphate-buffered saline (PBS), centrifuged, and fixed in ice-cold 75% (v/v) ethanol for overnight at 4°C. Then cells were suspended in propidium iodide (PI) solution (50 μg/mL) (KeyGEN BioTECH, Nanjing, China) with ribonuclease A (RNase A) (0.1 mg/mL) for 30 min in the dark. Cell cycle distribution was assessed using flow cytometer (Becton-Dickinson, San Jose, CA, USA).
Cell apoptosis assay
Flow cytometry analysis was used to determine the cell apoptosis with AnnexinV–FITC (fluorescein isothiocyanate) and PI (ab14085, Abcam). Cells stained with Annexin V–FITC and PI were followed with the flow cytometry analysis (BD Biosciences) according to the manufacturer’s protocol.
Luciferase reporter assays
pMIR-Reporter was used to introduce the sequences of HOTAIR containing the wild-type (wt) (TATGCAGTGGGACCCTGCTGC) and mutant binding sites (mut) (TATGCAGTGGGGATACATGAC) for miR-1. miR-1 mimic and inhibitor were purchased from Biomics (China). β-gal expression was used as an internal control. The HOTAIR promoter sequence was introduced into the pGL3 vector (Promega) for the HOTAIR promoter transcriptional activity assays.
RNA binding protein immunoprecipitation assay
TCC J82 and T24 cells with or without HuR stable knockdown were lysed with 100 U/mL RNase inhibitor (Sigma) and 25 mM Tris–HCl buffer (pH 7.5), and incubated with protein-A Sepharose beads precoated with 3 μg anti-HuR antibody (ab200342, Abcam) or control rabbit IgG for 1.5 h at 4°C. The RNA–protein complexes were pulled down by protein A/G agarose beads and RNA was extracted with Trizol, followed by detecting HOTAIR level with qRT-PCR.
Immunohistochemistry staining
The detailed procedure was referred to in the previous study.13 Antibody against HuR (ab200342) was purchased from Abcam. The horseradish peroxidase polymer-conjugated anti-rabbit secondary antibody (BA1054) was purchased from Boster.
Immunofluorescent assay
Cells were grown on glass bottom plates, fixed with 4% paraformaldehyde in PBS for 30 min, permeabilized in 0.1% Triton X-100 for 30 min, and blocked with 5% bovine serum albumin in PBS for 1.5 h at room temperature. The cells were incubated with anti-HuR antibody overnight at 4°C. Then, the cells washed with PBS for three times and were incubated with FITC-conjugated secondary antibody (ab6717, Abcam) for 1 h, and washed with PBS for three times, and observed under the confocal microscope.
Western blot
Detailed procedure was described elsewhere.2 The antibodies against HuR (ab200342), E-Cadherin (ab40772), vimentin (ab8978), caspase-3 (ab2171), cleaved-caspase-3 (ab32042), cleaved-poly ADP-ribose polymerase (PARP) (ab32064), PARP (ab32138), and β-actin (ab8227) were purchased from Abcam. The antibodies against Bcl-2 (sc-509) and Bax (sc-4239) were purchased from Santa Cruz. Chemiluminescence was detected using an enhanced chemiluminescence kit (Thermo Scientific) followed by visualizing using the ChemiDoc XRS Detection System (Bio-Rad).
Statistical analysis
All data are presented as mean ± SD. For three independent experiments, the Student’s t-test was used to evaluate the differences between the groups. n>3, *P<0.05, or **P<0.01 was considered significant.
Results
The expression levels of HuR and HOTAIR were strongly correlated in bladder cancer tissues
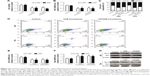
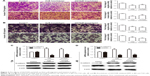
We first investigated whether HuR or HOTAIR expression is linked to bladder cancer. qRT-PCR and immunohistochemistry analyses showed that both HuR and HOTAIR expressions were upregulated in bladder cancer tissues (Figure 1A–D), which is concordant with the previous studies,11,13 and HuR expression was positively correlated to HOTAIR expression (Figure 1E). Additionally, HuR and HOTAIR expression levels were detected in bladder cancer cell lines (J82 and T24 cells) and normal bladder epithelial cell line (SV-HUC-1). As shown in Figure 1F and G, both HuR and HOTAIR expressions were more increased in J82 and T24 cells than in SV-HUC-1 cells. These results indicated that HuR or HOTAIR may hold promotive effects in bladder cancer progression.
Knockdown of HuR or HOTAIR inhibited bladder cancer cell proliferation and apoptosis
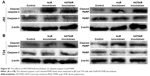
Based on the upregulation of HuR and HOTAIR expression in bladder cancer, we further investigated the effects of HuR and HOTAIR on bladder cancer progression via knockdown of HuR and HOTAIR expression. First, MTT analysis showed that knockdown of HuR or HOTAIR inhibited cell proliferation of bladder cancer (Figure 2A), and ki67, a cell proliferation marker, was blocked in HuR or HOTAIR knockdown cells (Figure 2B). Furthermore, knockdown of HuR or HOTAIR blocked bladder cancer cells in G1 phase of the cell cycle (Figure 2C), and cell apoptosis analysis was performed and indicated that knockdown of HuR or HOTAIR promoted cell apoptosis in J82 (23%±0.4% vs 11%±0.5% and 33%±0.3% vs 11%±0.5%) and T24 (16%±0.5% vs 8%±0.4% and 20%±0.3% vs 8%±0.4%) cells (Figure 2D). And the mRNA and protein levels of antiapoptotic gene (Bcl-2) and proapoptotic gene (Bax) were also measured via qRT-PCR and Western blot assays. Consistent with the above results, the expression levels of Bcl-2 were downregulated, but Bax expression levels were upregulated in HuR or HOTAIR knockdown cells (Figure 2E–G). In addition, the cleaved-caspase-3 and cleaved-PARP levels were evaluated in J82 and T24 cells with HuR/HOTAIR knockdown. HuR/HOTAIR knockdown significantly increased cleaved-caspase-3 and cleaved-PARP levels (Figure S1). Overall, these results suggested that HuR or HOTAIR holds promotive effects on cell proliferation and inhibitory effects on cell apoptosis in bladder cancer.
Knockdown of HuR or HOTAIR inhibited bladder cancer cell migration, invasion, and epithelial–mesenchymal transition process
We further investigated whether HuR or HOTAIR holds promotive effects on bladder cancer cell migration, invasion, and epithelial–mesenchymal transition (EMT) process. Transwell migration and invasion assays showed that knockdown of HuR or HOTAIR significantly inhibited bladder cancer cell migration (Figure 3A) and invasion (Figure 3B). Additionally, the EMT markers (E-cadherin and vimentin) expression were further determined. As shown in Figure 3C and D, E-cadherin’s (the epithelial marker) expression levels were markedly upregulated in HuR or HOTAIR knockdown cells, while vimentin’s (the mesenchymal marker) expression levels were significantly downregulated. These results demonstrated that knockdown of HuR or HOTAIR could significantly inhibit bladder cancer migration, invasion, and EMT process.
HuR increased HOTAIR expression through directly binding with HOTAIR in bladder cancer
As HuR is known to interact with HOTAIR in head and neck squamous cell carcinoma,7 it is, thus, plausible that this interaction holds true in human bladder cancer. First, HOTAIR expression levels were downregulated in HuR knockdown cells (Figure 4A). In addition, when HuR was knocked down and the de novo synthesis was blocked with actinomycin D, the decay rate of HOTAIR was faster in HuR knockdown cells than in control cells (t1/2 =5.2±0.4 h and t1/2 =2.2±0.3 h vs t1/2 =5.5±0.3 h and t1/2 =2.1±0.4 h) (Figure 4B and C). In particular, the HOTAIR promoter activity was not changed in HuR knockdown cells (Figure 4D), indicating that HuR could not affect the activity of HOTAIR promoter. Furthermore, RNA binding protein immunoprecipitation assays showed that HuR could interact with HOTAIR directly in bladder cancer cells (Figure 4E). HuR knockdown was then performed with Lenti-HOTAIR infection in bladder cancer cells, as shown in Figure 4F, and Lenti-HOTAIR infection could significantly upregulate HOTAIR level and overexpression of HOTAIR could attenuate the inhibitory effects of HuR knockdown on bladder cancer cell apoptosis and migration, characterized as the modulation of apoptotic (Figure 4G) and EMT (Figure 4H) markers. What is more, HOTAIR knockdown could attenuate or even reverse the promotive effects of HuR overexpression on bladder cancer cell apoptosis and migration, characterized as the modulation of apoptotic (Figure 4I and J) and EMT (Figure 4K and L) markers. Thus, these results indicated that HuR could also bind to and stabilize HOTAIR in bladder cancer, and regulate bladder cancer progression partly dependent on HOTAIR expression.
HOTAIR increased cytoplasmic accumulation and expression of HuR in a miR-1-dependent way
In silico analysis was performed to search for miRNAs that could potentially interact with HOTAIR and showed putative binding sites for miR-1. Transfection with miR-1 mimics reduced HOTAIR level, while transfection with miR-1 inhibitor increased HOTAIR level in bladder cancer cells (Figure 5A). Furthermore, transfection with miR-1 mimics significantly decreased the activity of luciferase constructs carrying the sequences of HOTAIR containing the wt for miR-1, but no repression of luciferase activity was observed when miR-1 binding sites were mutated (Figure 5B), indicating that miR-1 could bind to HOTAIR. Interestingly, upregulation of HOTAIR promoted HuR expression, but mutation of miR-1 binding sites in HOTAIR canceled the effects of HOTAIR on HuR expression in bladder cancer cells, and knockdown of HOTAIR also decreased HuR expression (Figure 5C and D). And knockdown of HOTAIR increased HuR cytoplasmic accumulation in J82 cells (Figure 5E). In addition, miR-1 level was examined in bladder cancer and NBTC tissues. The level of miR-1 was higher in normal tissues than in bladder cancer tissues (Figure 5F) and negatively correlated with HuR and HOTAIR expression levels (Figure 5G and H).
Discussion
LncRNA has been emerging as a new class of potential therapeutic target for cancers. A number of studies have shown that one of its members, HOTAIR, could modulate the progression of various cancers: regulating c-KIT expression through sponging miR-193a in acute myeloid leukemia,14 promoting carcinogenesis and invasion of gastric adenocarcinoma,15 and inducing cell migration and invasion via downregulation of RNA binding motif protein 38 in hepatocellular carcinoma cells.16 However, the roles of HOTAIR in bladder are elusive. Here, we have observed that HOTAIR expression was elevated in bladder cancer tissues and cells, and knockdown of HOTAIR promoted bladder cancer apoptosis, and inhibited cell proliferation, migration, and invasion.
Differential regulation of lncRNAs has been reported in different types of cancer, and given the critical roles of HuR in stabilizing various RNA transcripts, we further investigated whether HuR could bind to HOTAIR in bladder cancer. Previous study had indicated that high expression of HuR was associated with malignant aggressiveness and prognosis in bladder cancer.11 In the present study, we demonstrated that HuR and HOTAIR expression exhibited positive correlation in bladder cancer tissues. Notably, HuR directly interacted with HOTAIR and enhanced HOTAIR mRNA stability, and HuR exerted its promotive effects in bladder cancer in a HOTAIR-dependent manner. Interestingly, HOTAIR could also induce HuR expression dependent on miR-1 expression, and knockdown of HOTAIR decreased cytoplasmic accumulation of HuR in bladder cancer. As competitive endogenous RNAs (ceRNAs) have recently been shown to regulate expression in a miRNA-dependent way2 and HOTAIR has been shown to act as a ceRNA for HuR in other tumors,7 further studies should be performed to prove whether HOTAIR could also act as a ceRNA for HuR in bladder cancer and other cancers. Although HOTAIR had been identified as a target for miR-7 in head and neck squamous cell carcinoma,7 we did not observe any correlation between miR-7 and HOTAIR expression in bladder cancer (data not shown), which suggests that the ceRNA network between HOTAIR and HuR may be mediated by different miRNAs in different cancers. Here, we speculate that the targeting of miR-1 to HuR is necessary for the ceRNA loop between HOTAIR and HuR in bladder cancer. Meanwhile, as inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in bladder cancer,17 our study strongly indicated that the feed-forward regulatory loop between HuR and HOTAIR did exist in bladder cancer, and further studies should be carried out to confirm whether this feed-forward regulatory loop is a universal phenomenon.
Based on our study, we illustrate the roles of the relationship between HuR and HOTAIR in promoting bladder cancer progression. We strongly believe that our studies on the effect of the feed-regulatory loop between HuR and HOTAIR in bladder cancer could bring new lights in bladder tumorigenesis, and further research on this loop could establish it as a potential target or new biomarker for the treatment or diagnosis of bladder cancer.
Acknowledgment
We thank Professor Hu for critical review of this work.
Disclosure
The authors report no conflicts of interest in this work.
References
Martínez-Fernández M, Feber A, Dueñas M, et al. Analysis of the Polycomb-related lncRNAs HOTAIR and ANRIL in bladder cancer. Clin Epigenetics. 2015;7:109. | ||
Zheng L, Li X, Meng X, et al. Competing endogenous RNA networks of CYP4Z1 and pseudogene CYP4Z2P confer tamoxifen resistance in breast cancer. Mol Cell Endocrinol. 2016;427:133–142. | ||
Kim do Y, Park EY, Chang E, et al. A novel miR-34a target, protein kinase D1, stimulates cancer stemness and drug resistance through GSK3/beta-catenin signaling in breast cancer. Oncotarget. 2016;7(12):14791–14802. | ||
Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. | ||
Aiello A, Bacci L, Re A, et al. MALAT1 and HOTAIR long non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci Rep. 2016;6:38414. | ||
Zhang L, Song X, Wang X, et al. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res Treat. 2015;152(1):199–208. | ||
Xu CZ, Jiang C, Wu Q, Liu L, Yan X, Shi R. A feed-forward regulatory loop between HuR and the long noncoding RNA HOTAIR promotes head and neck squamous cell carcinoma progression and metastasis. Cel Physiol Biochem. 2016;40(5):1039–1051. | ||
Berrondo C, Flax J, Kucherov V, et al. Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS One. 2016;11(1):e0147236. | ||
Brennan CM, Steitz JA. HuR and mRNA stability. Cel Mol Life Sci. 2001;58(2):266–277. | ||
Jakstaite A, Maziukiene A, Silkuniene G, Kmieliute K, Gulbinas A, Dambrauskas Z. HuR mediated post-transcriptional regulation as a new potential adjuvant therapeutic target in chemotherapy for pancreatic cancer. World J Gastroenterol. 2015;21(46):13004–13019. | ||
Miyata Y, Watanabe S, Sagara Y, et al. High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer. PLoS One. 2013;8(3):e59095. | ||
Dong R, Yang GD, Luo NA, Qu YQ. HuR: a promising therapeutic target for angiogenesis. Gland Surg. 2014;3(3):203–206. | ||
Yan TH, Lu SW, Huang YQ, et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol. 2014;35(10):10249–10257. | ||
Xing CY, Hu XQ, Xie FY, et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–1987. | ||
Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451(2):171–178. | ||
Ding C, Cheng S, Yang Z, et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15(3):4060–4076. | ||
Guo J, Lv J, Chang S, et al. Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder. Oncotarget. 2016;7(29):45249–45262. |
Supplementary material
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.