Back to Journals » Infection and Drug Resistance » Volume 15
Risk Factors for Carbapenem Resistant Gram Negative Bacteria (CR-GNB) Carriage Upon Admission to the Gastroenterology Department in a Tertiary First Class Hospital of China: Development and Assessment of a New Predictive Nomogram
Authors Zhang H , Hu S, Xu D, Shen H, Jin H, Yang J, Zhang X
Received 11 November 2022
Accepted for publication 14 December 2022
Published 28 December 2022 Volume 2022:15 Pages 7761—7775
DOI https://doi.org/10.2147/IDR.S396596
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Hongchen Zhang,1– 3,* Shanshan Hu,1– 3,* Dongchao Xu,1– 3 Hongzhang Shen,1– 3 Hangbin Jin,1– 3 Jianfeng Yang,1– 3 Xiaofeng Zhang1– 3
1The Department of Gastroenterology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Zhejiang, People’s Republic of China; 2Key Laboratory of Integrated Traditional Chinese and Western Medicine for Biliary and Pancreatic Diseases of Zhejiang Province, Zhejiang, People’s Republic of China; 3Hangzhou Institute of Digestive Disease, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaofeng Zhang, Department of Gastroenterology, Hangzhou First People’s Hospital, NO. 261 HuanSha Road, Hangzhou, 310006, People’s Republic of China, Tel +86-13588296257, Fax +86-571-56005600, Email [email protected]
Background: With the increasing number of critically ill patients in the gastroenterology department (GED), infections associated with Carbapenem resistant gram-negative bacteria (CR-GNB) are of great concern in GED. As the turn-around time (TAT) for a positive screening culture result is slow, contact precaution and pre-emptive isolation, cohorting methods should be undertaken immediately on admission for high-risk patients. Accurate prediction tools for CR-GNB colonization in GED can help determine target populations upon admission. And thus, clinicians and nurses can implement preventive measures more timely and effectively.
Objective: The purpose of the current study was to develop and internally validate a CR-GNB carrier risk predictive nomogram for a Chinese population in GED.
Methods: Based on a training dataset of 400 GED patients collected between January 2020 and December 2021, we developed a model to predict CR-GNB carrier risk. A rectal swab was used to evaluate the patients’ CR-GNB colonization status microbiologically. We optimized features selection using the least absolute shrinkage and selection operator regression model (LASSO). In order to develop a predicting model, multivariable logistic regression analysis was then undertaken. Various aspects of the predicting model were evaluated, including discrimination, calibration, and clinical utility. We assessed internal validation using bootstrapping.
Results: The prediction nomogram includes the following predictors: Transfer from another hospital (Odds ratio [OR] 3.48), High Eastern Cooperative Oncology Group (ECOG) performance status (OR 2.61), Longterm in healthcare facility (OR 10.94), ICU admission history (OR 9.03), Blood stream infection history (OR 3.31), Liver cirrhosis (OR 4.05) and Carbapenem usage history within 3 month (OR 2.71). The model demonstrated good discrimination and good calibration.
Conclusion: With an estimate of individual risk using the nomogram developed in this study, clinicians and nurses can take more timely infection preventive measures on isolation, cohorting and medical interventions.
Keywords: carbapenem resistant gram-negative bacteria, carrier risk, gastroenterology department, predictive nomogram, screen culture
Introduction
Carbapenem resistant gram-negative bacteria (CR-GNB), namely, carbapenem-resistant Enterobacteriaceae (CRE) (for example, Klebsiella pneumoniae, Escherichia coli), Acinetobacter baumannii (CRAB) and Pseudomonas aeruginosa (CRPA), is an international and national health issue, because they are emerging causes of healthcare-associated infections that are of great concern to the public health.1 As a result of high levels of antimicrobial resistance (AMR), these bacteria are difficult to treat and are also associated with high mortality.2 Importantly, due to mobile genetic elements, they may be able to transmit resistance widely.3 Over 2.8 million cases and 35,000 deaths per year are reported by the disease control and prevention (CDC) as a result of infections caused by resistant bacteria.4 From 2005 to 2021, the proportion of carbapenem-resistant Klebsiella pneumoniae (CRKP) in China has increased from 3.0% to 24.4%, and that of CRAB from 39.0% to 72.3%.5 In China, CR-GNB infection rates differed significantly among provinces, with a high incidence in Zhejiang Province (CRKP 42.2%, CRPA 28.7%, and CRAB 64.7%). This study was conducted at a tertiary, first-class hospital in the province of Zhejiang, China.
CR-GNB infected patients in intensive care units and hematological malignancy departments are the main subjects of studies published recently.6,7 Relevant data is scanty regarding the CR-GNB carrier status in gastroenterology department (GED). As technology develops in the field of digestive endoscopy and awareness of early gastrointestinal tumor screenings increases, more patients with malignant or infectious diseases of the biliopancreatic system and major gastrointestinal tract surgery history are received in GED.8 There is usually a poor prognosis and high mortality rate associated with CR-GNB infection in these patients. CR-GNB infections in GED patients have increased dramatically in recent years, which posed a serious health threat to patients.9
A major problem with CR-GNB infections is their high mortality and morbidity rates, as well as their ability to cause outbreaks and contribute to the spread of resistance. Additionally, it has been recognized that colonization with CR-GNB usually precedes or co-occurs with CR-GNB infection.10 Therefore, early recognition of CR-GNB colonization in GED can assist in identifying patients most at risk for subsequent CR-GNB infections. As a result, infection prevention and control (IPC) measures can be introduced earlier to prevent transmission of pathogens to other patients and the hospital environment. Furthermore, a detailed understanding of CR-GNB colonization can be useful not only for infection control but also for antimicrobial stewardship.11
However, it is inevitable that there will be a delay between collecting samples and receiving test results, even with prompt screening. As the turn-around time (TAT) for a positive screening culture result is slow (48–72 h), contact precaution and pre-emptive isolation, cohorting methods should be undertaken immediately on admission for high-risk patient and known CR-GN colonized patients to reduce the transmission risk.12 In light of so many associated risk factors, accurate prediction CR-GNB colonization tools in GED can help determine target populations and make prevention materials more effective. Since nomograms can yield individual probabilities for clinical events by combining clinical characteristics, they are currently one of the most widely used prediction tools.13 In our study, this nomogram was used for the first time in predicting CR-GNB carrier status among gastroenterology patients.
Our study aimed to develop a simple but valid prediction tool for the GED to estimate the risk of rectal colonization with CR-GNB in order to target screening and preventative strategies accordingly upon GED admission.
Methods
Study Design
A retrospective cohort study was conducted in a 120-bed adult gastroenterology department of a government tertiary hospital in China including CR-GNB -infected patients. As the region is known to be an endemic area for cases of CR-GNB, a universal rectal swab performed at admission in GED was conducted as part of the hospital’s infection control policy during this period.14,15 A total of 3335 patients over the age of 18 years were enrolled in the study, who were hospitalized in adult GEDs between January 2020 and December 2021. Infection Control Commission (CCIH) active surveillance protocol was employed for the detection and prevention of CR-GNB in the patients. Based on the guidelines issued by the National Healthcare Commission of China (NHC), as well as the World Health Organization (WHO) and the centers for CDC criteria for antimicrobial resistance, the resolution for the study was drafted. CR-GNB carrier status is detected by screening tests using rectal swabs taken upon admission as part of the active surveillance protocol. Ethics approval of the research protocol was obtained from the institution’s Research Ethics Committee (ZN2022062).
Data Collection
Over the course of 2020 and 2021, 3335 records of active CR-GNB surveillance protocol were collected. As a result of review, we excluded records for patients discharged or died before their first rectal swab collection as well as records for patients with duplicated or incomplete records due to collection irregularities. A final review of protocol records revealed that 2020 patients who received CR-GNB screening test between January 2020 and December 2021 were qualified and then included in the study. Data on demographics, clinical outcomes, and microbiological characteristics of each individual record were collected, including the patients’ identification record, age, sex, received ward, Eastern Cooperative Oncology Group (ECOG) score,16 clinical diagnosis upon admission, use of gastrointestinal and other interventions, use of antimicrobials before admission. The medical records provided information such as the patient’s long-term hospitalization history, the history of ICU admission, the history of bloodstream infections, having transferred from another healthcare facility, the history of chemotherapy, the history of long-term steroids (more than 5 days), and the history of antibiotic exposure within 3 months prior to admission.
It is possible that estimates may be biased due to previous differences between carrier and non-carrier characteristics. Propensity score matching (PSM) techniques were used to reduce this bias.17 The propensity to participate in both groups was estimated using a logistic regression model based on a set of observed covariates. Covariates used in the PSM match (1:1) were age, gender, and ward admitted. As a result, 200 carriers and 200 non-carriers were enrolled in the study. According to CDC guidelines, the CR-GNB colonized patients include individuals that were tested positive for CR-GNB cultures and were clinically diagnosed as CR-GNB carriers. Five days was the average TAT for CR-GNB surveillance results. When CR-GNB screening cultures were positive, the microbiology laboratory informed the GED’s medical or nursing staff immediately. Notifications were made in the electronic medical records and prescriptions by the CCIH (Figure 1).
 |
Figure 1 Flow chart for patients’ enrollment. |
Microbiology
In order to screen for CR-GNB, rectal swabs were directly inoculated onto chromogenic agar plates containing carbapenem (CHROMagar, France). Cultures from clinical samples and surveillance samples were screened for identification and susceptibility using an automated system, VITEK2 (BioMe rieux, France). We measured the sensitivity of imipenem and meropenem using Kirby-Bauer’s method. According to M100-ED30 breakpoints established by the Clinical and Laboratory Standards Institute (CLSI), the results were interpreted.
Statistical Analysis
Statistics relating to demographics, disease, and treatment are expressed as counts (%). Using the R programming language (Version 3.1.1; https://www.R-project.org), we performed statistical analysis. Continuous variables are described as median and interquartile range. The Mann–Whitney U-test was used for statistical comparison between two groups. Categorical variables were described as total number and percentage, and the chi-square test or Fisher’s exact test was used for comparison between groups. To select optimal predictive features for risk factors from CR-GNB colonized patients, we used the least absolute shrinkage and selection operator (LASSO) method, suitable for reducing high-dimensional data.18 An analysis of the LASSO regression model was conducted to identify features with nonzero coefficients. After that, the chosen features were incorporated into a multivariable logistic regression model to build a prediction model.19,20 The features were considered as odds ratio (OR) having 95% confidence interval (CI) and as P-value. A two-sided significance level was used for all statistical tests. Statistically significant sociodemographic variables were included in the model, as well as variables pertaining to disease and treatment characteristics. By using the cohort, all potential predictors were applied in order to create a predicting model for CR-GNB colonization risk.21 Using calibration curves, we next evaluated the calibration of the colonization risk nomogram. The presence of a significant test statistic implies that the model is not calibrated perfectly.22 A measurement of Harrell’s C-index was performed to quantify the discrimination performance of the colonization nomogram. To calculate a relatively corrected C-index, a bootstrapping validation (1000 resamples) was performed on the colonization nomogram.23 Based on the numerical results from a decision curve analysis, the clinical usefulness of the colonization nomogram was evaluated at a variety of threshold probabilities by quantifying the net benefits in the GED patients cohort.24 In order to determine the net benefit, we subtracted the proportion of false positives from the proportion of true positives and assessed the relative harm of not intervening versus the negative outcomes of an unnecessary intervention.
Results
Patients’ Characteristics
There were 400 patients in total, and the average age was 66.93±17.33 years (range 19–99 years) and a majority (61.5%) of the population is male. It was discovered that Klebsiella pneumoniae was the most common strain (61%), followed by Escherichia coli (22%), Enterobacter cloacae (7%), Citrobacter freundii (4%), and Pseudomonas aeruginosa (2%).
The patients were categorized into two groups: (1) colonized patients (n = 200), comprised of patients who tested positive in the CR-GNB surveillance testing (including colonized-infected patients) and (2) control patients (n = 200), comprised of patients without CR-GNB colonization (ie, negative surveillance testing). Baseline characteristics are shown in Table 1. Cases and controls were matched in terms of age, gender and received regular floor ward.
 |
Table 1 Demographic and Clinical Characteristics of Patients Colonized with CR-GNB and Matched Controls |
The difference between categorical variables was compared using Chi-square or Fisher’s exact tests. A significant difference was found between CR-GNB-colonized and control groups in terms of past history of long term in healthcare facility (6% and 28%, respectively, p < 0.001), ICU admission history (3% and 19%, respectively, p < 0.001), blood stream infection history within 1 year (6% and 14%, respectively, p = 0.006), transfer from another healthcare facility (16% and 45%, respectively, p < 0.001), ECOG performance status (PS) (p < 0.001), liver cirrhosis (20% and 45%, respectively, p = 0.001), steroid more than 5 days (9% and 18%, respectively, p = 0.005) carbapenem usage (15% and 36%, respectively, p < 0.001) and polyantibiotic usage (33% and 48%, respectively, p = 0.002).
Univariate and Multivariate Logistic Regression Analysis for the Risk Factors of CR-GNB Colonization
To identify the risk factors that exhibited statistical differences between the CR-GNB carrier group and control group, univariate logistic regression analyses were conducted. It was observed that past history of long term in healthcare facility, ICU admission history, blood stream infection history within 1 year, transfer from another healthcare facility, steroid more than 5 days, ECOG performance status (PS), liver cirrhosis, carbapenem usage and polyantibiotic usage were the risk factors affecting the rectal CR-GNB colonization, as shown in Table 2.
 |
Table 2 Univariate and Multivariate Analysis of Risk Factors Related to CR-GNB Colonization |
In addition to univariate logistic regression, multivariate logistic regression was conducted on the risk factors determined by univariate logistic regression. By adjusting confounders, we have identified past history of long term in healthcare facility (odds ratio [OR] 10.94, 95% confidence interval [CI] 4.99–23.95, P < 0.001), ICU admission history (OR 9.03, 95% CI 3.39–24.03, P < 0.001), blood stream infection history within 1 year (OR 3.31, 95% CI 1.28–8.59, P < 0.05), transfer from another healthcare facility (OR 3.48, 95% CI 1.96–6.18, P < 0.001), ECOG performance status (PS) (OR 2.61, 95% CI 1.96–3.48, P < 0.001), liver cirrhosis (OR 4.05, 95% CI 2.14–8.42, P<0.002) and carbapenem usage (OR 2.71, 95% CI 1.49–4.93, P = 0.001) as independent risk factors affecting the rectal CR-GNB colonization, as shown in Table 2.
Feature Selection
As shown in Figure 2A and B, a LASSO regression analysis of 400 patients in the cohort revealed that 10 potential predictors identified from 40 features were with nonzero coefficients (~4:1 ratio). These features included past history of long term in healthcare facility, ICU admission history, blood stream infection history within 1 year, transfer from another healthcare facility, steroid more than 5 days, ECOG performance status (PS), bowel diversion surgery history, liver cirrhosis, carbapenem usage and polyantibiotic usage.
Model Development for Individualized Prediction
A logistic regression analysis to examine the effects of selected features from LASSO regression analysis was done. Nomograms (Figure 3) were developed by incorporating the identified independent predictors. Figure 4 shows the example of how the nomogram can be used.
 |
Figure 3 Nomogram to predict the probability of CR-GNB carrier status for patients in GED upon admission. |
 |
Figure 4 The example of how the predictive nomogram can be used. |
CR-GNB Carrier Risk Nomogram’s Apparent Performance
The prediction nomogram’s C-index was 0.857, and bootstrapping validation demonstrated the model’s good discrimination which was confirmed by the C-index of 0.847 for the cohort (Figure 5). As shown in Figure 6, the calibration curve for the CR-GNB colonization risk nomogram performed well in this cohort for predicting CR-GNB colonization risk. According to the CR-GNB colonization risk nomogram, apparent performance is a good indicator of prediction accuracy.
 |
Figure 5 ROC curves for validating the discrimination power of the nomogram. ((A) training group; (B) bootstrapping group). |
Clinical Use
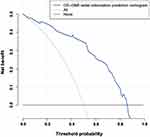
Figure 7 illustrates the decision curve analysis for the CR-GNB colonization nomogram. Based on the decision curve, if the patient’s threshold probability was between 8% and 86%, using this model to predict rectal CR-GNB colonization would result in a greater net benefit for the patient.
Discussion
With the number of critically ill patients with gastrointestinal malignancy and past history of major gastrointestinal tract surgery are increasing in GED, infections associated with CR-GNB are of great concern in GED.25,26 Our study, for the first time, provided nomogram predicting CR-GNB carrier status for patients in GED. By merely utilizing seven easily available variables, we developed and validated a new prediction tool for CR-GNB colonization risk in GED patients. As a result of internal validation in the cohort, good discrimination, calibration power, and a high C-index were observed.
The current study found that CR-GNB screening using rectal swabs had a positivity rate of 9.9% in GED with K. pneumoniae being the most prevalent strain (61%), followed by E. coli (22%) and E. cloacae (7%). Although CR-GNB infection were commonly reported in the ICU, these findings suggested that baseline colonization by CR-GNB was relatively high in GED. Several independent risk factors were identified as being associated with CR-GNB carriage at the time of a GED admission in our study, including transfer from another hospital, high ECOG score, longterm in healthcare facility, ICU admission history within 1 year, blood stream infection history within 1 year, liver cirrhosis and carbapenem usage history within 3 month. In GED, these variables might play an important role in determining risk for CR-GNB rectal colonization.
It was found that transfer from another hospital, long-term in healthcare facility and ICU admission history within 1 year were associated with higher risk for CR-GNB carriage, as in previous studies.27,28 It might due to prolonged exposure to areas with a high prevalence of CR-GNB. This study demonstrated that patients with blood stream infection history within 1 year and carbapenem usage history were much more susceptible to being colonized by CR-GNB, which might signify the important role of selective pressure caused by the wide use of broad-spectrum antibiotic. A performance status equal to or above 3 on the ECOG was independently associated with CR-GNB carriage. In multivariate analyses, steroid and poly-antibiotic use histories were excluded due to their multicollinearity despite significant associations in univariate analyses. The study did not identify other well-known conditions associated with a greater risk of infection in ICUs, such as end-stage renal disease requiring dialysis and gastrointestinal cancer, as risk factors for CR-GNB carrier status.29,30 We speculate that this might be due to the particularity of the patients population of GED.
In the view of uniqueness of the patient population in GED, a range of common digestive disorders were included in the univariate and multivariate analyses. It is interesting to note that liver cirrhosis showed a strong association with CR-GNB colonization among the analyzed comorbidities. It is speculated that a multitude of immunological mechanisms, which is named as cirrhosis associated immune disorder, may be involved in the mechanisms of pathophysiology for susceptibility to CR-GNB colonization among liver cirrhosis patients.31 In addition to immunological mechanisms, liver cirrhosis might also promote CR-GNB colonization by altering the microbiome and degrading intestinal barriers.32
In some previous literature, outbreaks of CR-GNB have been linked to contaminated GI endoscopes.33–35 Our work is the first study to investigate whether commonly performed endoscopy interventions history are associated with CR-GNB colonization upon admission in GED. In this study, it was demonstrated that commonly performed endoscopic interventions did not increase the risk of CR-GNB carriage. As it has been noted that improper cleaning procedure for endoscopes was associated with outbreaks of CR-GNB,36 we speculate that the advances of endoscope disinfection techniques and improvement of awareness of sterile operation have decreased the risk of CR-GNB transmission by GI endoscopic intervention.
In our study, we for the first time developed a valid CR-GNB colonization risk prediction tool, which assisted clinicians with early identification of patients at high risk of CR-GNB carriage upon admission to GED. As the increase of the number of critically ill patients in GED who might bare higher mortality upon CR-GNB infection, the colonization predictive nomogram will help identify those with high risk to undertake infection control measures, like contact precaution, patients isolation and patient cohorting, for the purpose of preventing intra-facility transmission. Moreover, in case of emergence of a septic status for a patient upon GED admission, knowledge of CR-GNB colonization risk can be relevant to the selection of empirical antimicrobial drugs, such as anti-CR-GNB agents. Furthermore, with the wide application of gastrointestinal endoscopy, a lot of patients in GED need to receive endoscopic treatment immediately upon admission, who were still waiting for the result of screening test. It is necessary to identify patients with high risk for CR-GNB colonization and purposefully strengthen endoscopic disinfection, which is helpful to prevent the intra-facility transmission of CR-GNB.
So, accurate CR-GNB carrier risk assessment will assist physicians with identifying those in high risk and taking timely preemptive precautions, preventing wasting infection preventive resources in low-risk situations, and avoiding delays of anti-CR-GNB agents in treatment when there is a high probability of a CR-GNB infection.
Limitations
We found several limitations in our study as well. First, our acquired data from 2020 and 2021 was a partial representation of people with GEDs. The cohort only performed in a tertiary hospital in Zhejiang province was not representative of all Chinese patients in GED. Additionally, not all potential factors affecting CR-GNB colonization were considered in the risk factor analysis. A number of factors that could contribute to colonization risk were not fully disclosed, including occupations and other conditions. The third issue is that, although we extensively validated the robustness of our nomogram using bootstrap testing, no external validation was conducted. It is unclear whether the results are generalizable to other GED populations in different regions and countries. There is a need for external evaluation in a wider population of patients.
Conclusions
Clinical practitioners can use the nomogram developed in this study to identify patients with CR-GNB colonization upon admission with a relatively high degree of accuracy. With an estimate of individual risk, clinicians and nurses can take more timely infection preventive measures on isolation/cohorting and medical interventions. There is a need for external validation of this nomogram, and further research is needed to establish whether individual interventions based on this nomogram are likely to reduce the risk of CR-GNB transmission within facilities and achieve better treatment outcomes.
Data Sharing Statement
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The study was conducted in accordance with the Declaration of Helsinki. Ethics Committee of Hangzhou First People’s Hospital approved this study (ZN2022062). A waiver of informed consent was granted as all anonymized data was collected retrospectively in accordance with the ethics approval.
Acknowledgments
The department of gastroenterology, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine provided the data used for this project.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This project was supported by National Natural Science Foundation of China (NSFC No. 82000516), Zhejiang Chinese Traditional Medicine Scientific Research Fund Project (2022ZB271), Hangzhou Medical and Health Science and Technology Plan A20200737 and the Construction Fund of Key Medical Disciplines of Hangzhou (OO20190001). The founder of the study has no role in the study design, data collection, data analysis, data interpretation or writing of the article.
Disclosure
No conflicts of interest exist for all authors.
References
1. Jean SS, Harnod D, Hsueh PR. Global threat of carbapenem-resistant gram-negative bacteria. Front Cell Infect Microbiol. 2022;12:823684. doi:10.3389/fcimb.2022.823684
2. Wilke MH, Preisendörfer B, Seiffert A, Kleppisch M, Schweizer C, Rauchensteiner S. Carbapenem-resistant gram-negative bacteria in Germany: incidence and distribution among specific infections and mortality: an epidemiological analysis using real-world data. Infection. 2022;50(6):1535–1542. doi:10.1007/s15010-022-01843-6
3. Acman M, Wang R, van Dorp L, et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene bla(NDM). Nat Commun. 2022;13(1):1131. doi:10.1038/s41467-022-28819-2
4. Miller C, Gilmore J. Detection of quorum-sensing molecules for pathogenic molecules using cell-based and cell-free biosensors. Antibiotics. 2020;9(5):259. doi:10.3390/antibiotics9050259
5. Zhen X, Stålsby Lundborg C, Sun X, Gu S, Dong H. Clinical and economic burden of carbapenem-resistant infection or colonization caused by Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii: a multicenter study in China. Antibiotics. 2020;9(8):514. doi:10.3390/antibiotics9080514
6. Salomão MC, Freire MP, Boszczowski I, Raymundo SF, Guedes AR, Levin AS. Increased risk for carbapenem-resistant Enterobacteriaceae colonization in intensive care units after hospitalization in emergency department. Emerg Infect Dis. 2020;26(6):1156–1163. doi:10.3201/eid2606.190965
7. Lu L, Xu C, Tang Y, et al. The threat of carbapenem-resistant gram-negative bacteria in patients with hematological malignancies: unignorable respiratory non-fermentative bacteria-derived bloodstream infections. Infect Drug Resist. 2022;15:2901–2914. doi:10.2147/idr.s359833
8. Peery AF, Crockett SD, Murphy CC, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2021. Gastroenterology. 2022;162(2):621–644. doi:10.1053/j.gastro.2021.10.017
9. Wu D, Xiao J, Ding J, et al. Predictors of mortality and drug resistance among carbapenem-resistant Enterobacteriaceae-infected pancreatic necrosis patients. Infect Dis Ther. 2021;10(3):1665–1676. doi:10.1007/s40121-021-00489-5
10. Lima EM, Cid PA, Beck DS, et al. Predictive factors for sepsis by carbapenem resistant Gram-negative bacilli in adult critical patients in Rio de Janeiro: a case-case-control design in a prospective cohort study. Antimicrob Resist Infect Control. 2020;9(1):132. doi:10.1186/s13756-020-00791-w
11. Bedos JP, Daikos G, Dodgson AR, et al. Early identification and optimal management of carbapenem-resistant Gram-negative infection. J Hosp Infect. 2021;108:158–167. doi:10.1016/j.jhin.2020.12.001
12. Ambretti S, Bassetti M, Clerici P, et al. Screening for carriage of carbapenem-resistant Enterobacteriaceae in settings of high endemicity: a position paper from an Italian working group on CRE infections. Antimicrob Resist Infect Control. 2019;8:136. doi:10.1186/s13756-019-0591-6
13. Li J, Xie X, Zhang J, et al. Novel bedside dynamic nomograms to predict the probability of postoperative cognitive dysfunction in elderly patients undergoing noncardiac surgery: a retrospective study. Clin Interv Aging. 2022;17:1331–1342. doi:10.2147/cia.s380234
14. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
15. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2). doi:10.1128/aac.01882-17
16. Sok M, Zavrl M, Greif B, Srpčič M. Objective assessment of WHO/ECOG performance status. Support Care Cancer. 2019;27(10):3793–3798. doi:10.1007/s00520-018-4597-z
17. Austin PC. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res. 2011;46(3):399–424. doi:10.1080/00273171.2011.568786
18. Vasquez MM, Hu C, Roe DJ, Chen Z, Halonen M, Guerra S. Least absolute shrinkage and selection operator type methods for the identification of serum biomarkers of overweight and obesity: simulation and application. BMC Med Res Methodol. 2016;16(1):154. doi:10.1186/s12874-016-0254-8
19. Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–80. doi:10.1016/s1470-2045(14)71116-7
20. Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–1370. doi:10.1200/jco.2007.12.9791
21. Tang N, Chen H, Chen R, Tang W, Zhang H. Predicting mucosal healing in crohn’s disease: a nomogram model developed from a retrospective cohort. J Inflamm Res. 2022;15:5515–5525. doi:10.2147/jir.s378304
22. Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35(9):2052–2056. doi:10.1097/01.ccm.0000275267.64078.b0
23. Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. doi:10.1002/sim.1802
24. Vickers AJ, Cronin AM, Elkin EB, Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi:10.1186/1472-6947-8-53
25. Reintam Blaser A, Preiser JC, Fruhwald S, et al. Gastrointestinal dysfunction in the critically ill: a systematic scoping review and research agenda proposed by the section of metabolism, endocrinology and nutrition of the European Society of Intensive Care Medicine. Crit Care. 2020;24(1):224. doi:10.1186/s13054-020-02889-4
26. Saki M, Amin M, Savari M, Hashemzadeh M, Seyedian SS. Beta-lactamase determinants and molecular typing of carbapenem-resistant classic and hypervirulent Klebsiella pneumoniae clinical isolates from southwest of Iran. Front Microbiol. 2022;13:1029686. doi:10.3389/fmicb.2022.1029686
27. Chuah CH, Gani Y, Sim B, Chidambaram SK. Risk factors of carbapenem-resistant Enterobacteriaceae infection and colonisation: a Malaysian tertiary care hospital based case-control study. J R Coll Physicians Edinb. 2021;51(1):24–30. doi:10.4997/jrcpe.2021.107
28. Ray MJ, Lin MY, Weinstein RA, Trick WE. Spread of carbapenem-resistant Enterobacteriaceae among Illinois healthcare facilities: the role of patient sharing. Clin Infect Dis. 2016;63(7):889–893. doi:10.1093/cid/ciw461
29. Tang SSL, Chee E, Teo JQ, Chlebicki MP, Kwa ALH. Incidence of a subsequent carbapenem-resistant Enterobacteriaceae infection after previous colonisation or infection: a prospective cohort study. Int J Antimicrob Agents. 2021;57(6):106340. doi:10.1016/j.ijantimicag.2021.106340
30. Kumar A, Mohapatra S, Bakhshi S, et al. Rectal carriage of carbapenem-resistant Enterobacteriaceae: a menace to highly vulnerable patients. J Glob Infect Dis. 2018;10(4):218–221. doi:10.4103/jgid.jgid_101_17
31. Albillos A, Martin-Mateos R, Van der Merwe S, Wiest R, Jalan R, Álvarez-Mon M. Cirrhosis-associated immune dysfunction. Nat Rev Gastroenterol Hepatol. 2022;19(2):112–134. doi:10.1038/s41575-021-00520-7
32. Tsiaoussis GI, Assimakopoulos SF, Tsamandas AC, Triantos CK, Thomopoulos KC. Intestinal barrier dysfunction in cirrhosis: current concepts in pathophysiology and clinical implications. World J Hepatol. 2015;7(17):2058–2068. doi:10.4254/wjh.v7.i17.2058
33. Muscarella LF. Risk of transmission of carbapenem-resistant Enterobacteriaceae and related “superbugs” during gastrointestinal endoscopy. World J Gastrointest Endosc. 2014;6(10):457–474. doi:10.4253/wjge.v6.i10.457
34. Bruns MA, Gibbs ER. Multidepartmental response to a duodenoscope used on a CRE patient: a case study. Gastroenterol nurs. 2017;40(1):56–62. doi:10.1097/sga.0000000000000268
35. Akinyemi KO, Al-Khafaji NSK, Al-Alaq FT, et al. Extended-spectrum beta-lactamases encoding genes among salmonella enterica serovar typhi isolates in patients with typhoid fever from four Academic Medical Centers Lagos, Nigeria. Revista de investigacion clinica. 2022;74(3):165–171. doi:10.24875/ric.22000078
36. Assimos DG. Re: carbapenem-resistant Enterobacteriaceae and endoscopy: an evolving threat. J Urol. 2017;197(1):171–172. doi:10.1016/j.juro.2016.10.006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.