Back to Journals » OncoTargets and Therapy » Volume 13
RING Finger Protein 38 Mediates LIM Domain Binding 1 Degradation and Regulates Cell Growth in Colorectal Cancer
Authors Huang Z, Yang P, Ge H, Yang C, Cai Y, Chen Z, Tian W, Wang H
Received 16 October 2019
Accepted for publication 26 December 2019
Published 14 January 2020 Volume 2020:13 Pages 371—379
DOI https://doi.org/10.2147/OTT.S234828
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Ziming Huang, 1 Peng Yang, 2 Hengfa Ge, 1 Chenchen Yang, 1 Yong Cai, 1 Zhen Chen, 1 Wenze Tian, 3 Haixiao Wang 4
1Department of Emergency Surgery, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, Huai’an, Jiangsu, People’s Republic of China; 2Department of Emergency, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China; 3Department of Cardio-Thoracic Surgery, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, Huai’an, Jiangsu, People’s Republic of China; 4Department of General Surgery, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, Huai’an, Jiangsu, People’s Republic of China
Correspondence: Ziming Huang
Department of Emergency Surgery, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, No. 1 The Yellow River West Road, Huaiyin District, Huai’an 223300, Jiangsu, People’s Republic of China
Tel +86 15896163441
Email [email protected]
Background and Objectives: RING finger protein 38 (RNF38) has been reported to be involved in the tumorigenesis of several tumors, but its role in colorectal cancer (CRC) is still not investigated. In the present study, we aimed to investigate the effect of RNF38 in CRC cells.
Materials and Methods: The public tumor databases GEPIA and Kaplan-Meier Plotter were used to analyze RNF38 expression and patients’ overall survival in CRC. The qRT-PCR was carried out to assess the mRNA levels of RNF38 and LDB1. Western blot and co-immunoprecipitation were used to detect protein expression and ubiquitination. CCK-8 assay was performed to analyze CRC cell growth and viability.
Results: RNF38 was found downregulated in CRC tumor tissues and cell lines, and CRC patients with high RNF38 expression had a longer overall survival than patients with low RNF38 expression. Our further investigations showed that RNF38 interacted with LDB1, and downregulated LDB1 expression by inducing its polyubiquitination. Moreover, overexpression of RNF38 inhibited CRC cell growth but enforced LDB1 could significantly antagonize RNF38-induced cell growth inhibition in CRC cells. Additionally, RNF38/LDB1 axis was involved in the drug sensitivity of 5-FU to CRC cells.
Conclusion: Our studies suggested that RNF38 was functional in CRC cells, and downregulated CRC cell growth by inducing LDB1 polyubiquitination, which indicated that RNF38 could be as a novel target for CRC therapy.
Keywords: RNF38, LDB1, ubiquitination, degradation, colorectal cancer
Introduction
As is known to all, protein ubiquitination is closely associated with the occurrence and development of tumors. And the ubiquitination process of proteins consists of at least five important components, including ubiquitin (Ub), ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), ubiquitin ligase (E3) and deubiquitinating enzyme (DUB).1–3 At present, it is estimated that there are 1–2 E1s, 60–100 E2s, 1000 E3s and nearly 100 DUBs in human cells.4 Among them, the disorder of protein ubiquitination components is closely related to the occurrence and development of cancer.5 At present, the research and development of new drugs targeting E3 ubiquitin ligase have become a research hotspot of scientists.6 Therefore, the study of ubiquitination of proteins in colorectal cancer (CRC) cells is of great significance for the treatment of colorectal cancer caused by the disorder of ubiquitin system.
RNF38, the ring finger protein 38, belongs to one of the largest E3 ubiquitin ligase family proteins.7 RNF38 protein contains RING-H2 domain, KIL domain and coiled-coil domain at its carboxyl end.7 Among them, RING zinc finger motifs are zinc-binding domains in RNF ubiquitin ligase family proteins, which can promote the transfer of E2-presenting ubiquitin molecules to target proteins, thus promoting the ubiquitination of proteins, and play an important role in many physiological and biochemical processes of cells, such as protein degradation, signal transduction, cell growth and tumorigenesis.8 At present, there are few reports on the function of RNF38 in tumors, involving only the preliminary discovery of hepatocellular carcinoma, lung cancer and gastric cancer, but its function and mechanism in CRC have not been reported. In gastric cancer, RNF38 was highly expressed, and upregulated STAT3 signaling by promoting SHP-1 ubiquitination, thus promoting the growth of gastric cancer cells.9 And in non-small cell lung cancer (NSCLC), it has been reported that RNF38 promoted the occurrence of NSCLC by regulating EMT, and indicated a poor index for NSCLC patients.10 However, the prognostic roles of RNF38 seemed differed by different tumor types according to the data from TCGA samples for mRNA analysis,9 which suggested that RNF38 may exert different effects by interacting with different substrates in tumors. Therefore, in this study, we aimed to investigate the function and mechanism of RNF38 in CRC, which have not been reported yet. For example, we aimed to investigate the expression of RNF38 in CRC cell lines and tumor tissues, and the roles of RNF38 as an E3 ligase in regulating CRC cell growth.
In the present study, we found that RNF38 was downregulated in both of CRC tumor tissues and cell lines, and its low expression was associated with poor prognosis of CRC, which suggested that RNF38 may also act as a tumor suppressor. In addition, we found that LDB1 was a novel substrate of RNF38, and RNF38 induced LDB1 degradation through 26S proteasome. Interestingly, LDB1 had been shown to be elevated in CRC and its high expression was associated with poor prognosis of CRC patients.11 And the present study found that RNF38 was lowly expressed in CRC and RNF38 promoted the ubiquitination-mediated degradation of LDB1, which may explain why LDB1 was overexpressed in CRC to some extent.
Materials and Methods
Cells, Tissues and Chemicals
Colorectal cancer cell lines (HCT116, HT-29, RKO and DLD1), the human normal intestinal epithelial cell line NCM460, and HEK293T cell line were purchased from ATCC (American Type Culture Collection, Manassas, USA). All colorectal cancer cell lines, NCM460 and HEK293T cells were cultured in Dulbecco’s high glucose modified Eagle’s medium with 10% fetal bovine serum, 100 μg/mL of penicillin, and 100 units/mL of streptomycin. The primary colorectal cancer tumor tissues and adjacent normal tissues were collected from the Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. The collection and use of human tissues for this study were approved by the Institutional Review Board of the Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. 5-FU was purchased from Selleck, Houston, USA.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Briefly, twenty pairs of CRC tumor tissues and adjacent normal tissues were treated and grinded in liquid nitrogen. The RNAiso Plus (Takara Bio Group, Japan) was used for RNA isolation according to the manufacturer’s instructions. EasyScript® First-Strand cDNA Synthesis Super Mix (TransGen, Nanjing, China) was used for cDNA synthesis. And then the cDNA template was used for subsequent qRT-PCR analysis, which was carried out as described previously.12 To prepare the reaction mixes, HS-SYBR (Evrogen, Moscow, Russia) was used. The primers used were as follows: RNF38, forward 5ʹ- TTCTTATCGGTTCAATCC-3ʹ and reverse 5ʹ-AACTCGTGGTTACAGGGT-3ʹ; LDB1, forward 5ʹ-GCTGTGCCTGTCCTGGTT-3ʹ and reverse 5ʹ-GCCCACATCCCTATCCAG-3ʹ; GAPDH, forward 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ and reverse 5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ.
Western Blot
Relative cells were lysed for Western blot as described previously.13 The primary antibodies against RNF38, LDB1 and GAPDH were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-Flag and anti-Myc antibodies were purchased from Medical & Biological Laboratories (Tokyo, Japan).
Plasmids Construction and Gene Transfection
The human RNF38 and LDB1 genes were generated by PCR and cloned into pcDNA3.1 vector with a Myc, or Flag tag as previously described.7,14 The primers used were as follows: RNF38, forward 5ʹ-ATGGCTTGTAAGATATCTCC-3ʹ and reverse 5ʹ-TCATTCTGAATCCCGATGC-3ʹ; LDB1, forward 5ʹ-ATGTCAGTGGGCTGTGCCTGTCC-3ʹ and reverse 5ʹ-TTACTGGGAGGCCTGTGACGTGGGG-3ʹ. HA-Ub plasmids were conserved in our laboratory. Plasmids were transiently transfected into HCT116, DLD1 or HEK293T cells by Lipofectamine® 2000 (Invitrogen) according to the manufacturer’s instruction.
Co-Immunoprecipitation Analysis
Cells were lysed for co-immunoprecipitation (Co-IP) as described previously.15 Briefly, cells were lysed and whole-cell lysates were isolated. Then, whole-cell lysates were incubated with indicated antibodies overnight at 4°C, followed by incubating with protein A/G agarose (Santa Cruz, CA, USA). Finally, the precipitated proteins were boiled and analyzed by Western blot.
Cell Growth and Viability
HCT116 and DLD1 cells were transfected with empty vector, Myc-RNF38 or Flag-LDB1 plasmids for indicated time. Then, transfected cells were incubated with 5-FU or directly prepared for Cell Counting Kit-8 (CCK-8) assay according to the manufacturer’s instructions (Biotool, Houston, USA).
Statistical Analysis
The GraphPad Prism 5 and IBM SPSS 22.0 software were used for statistical analysis. To compare the differences between two groups in the study, the student’s t test was used. To compare more than 2 groups, ANOVA was used. In this study, a p value less than 0.05 was considered statistically significant.
Results
RNF38 Is Downregulated in Colorectal Cancer and Predicts a Positive Index for Patients
To investigate the expression level of RNF38 in colorectal cancer (CRC), the public tumor database GEPIA matched TCGA and GTEx data was firstly used. As shown in Figure 1A, the database revealed that RNF38 was decreased in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ). To further confirm it, twenty pairs of CRC tumor tissues and adjacent normal tissues were collected and prepared for qRT-PCR analysis. As shown in Figure 1B and C, RNF38 was markedly downregulated in CRC tumor tissues compared with the adjacent normal tissues. Moreover, the overall survival of CRC patients with low or high RNF38 expression was analyzed by Kaplan-Meier Plotter, a public online tumor database (Figure 1D). And it showed that CRC patients with high RNF38 expression had a longer overall survival than those with low RNF38 expression (Figure 1D). Thus, above results indicated that RNF38 was closely associated with CRC tumorigenesis.
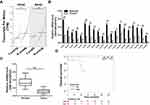 |
Figure 1 RNF38 is downregulated in colorectal cancer and predicts a positive index for patients. (A). The expression level of RNF38 was analyzed by GEPIA online (http://gepia.cancer-pku.cn/). COAD: Colon adenocarcinoma; READ: Rectum adenocarcinoma. (B). Twenty pairs of colorectal cancer tumor tissues and adjacent normal tissues were prepared for qRT-PCR against RNF38. GAPDH was used as an internal control. (C). Statistical analysis for Figure 1B. (D). The overall survival of colorectal cancer patients with low or high RNF38 expression was analyzed by Kaplan-Meier Plotter (http://kmplot.com). *p<0.05, **p<0.01. |
RNF38 Destabilizes LDB1 Protein via Proteasomal Pathway in Colorectal Cancer Cells
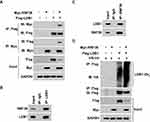
Our further results showed that the expression levels of RNF38 were also decreased in four CRC cell lines compared with a human normal intestinal epithelial cell line (Figure 2A). However, the expression levels of LDB1 were elevated in CRC cell lines compared with the human normal intestinal epithelial cell line (Figure 2A), and RNF38 expression was also significantly negatively associated with LDB1 expression (Figure 2B). To further confirm it, indicating concentrations of Myc-RNF38 plasmids were transfected into CRC cells, and we found that overexpression of RNF38 could markedly downregulate the protein levels of LDB1 in HCT116 and DLD1 cells (Figure 2C), but had no effects on the mRNA levels of LDB1 (Figure 2D), which suggested that RNF38 destabilized LDB1 protein in CRC cells. As RNF38 is a ubiquitin ligase and ubiquitin ligases can induce target protein degradation through the 26S proteasome, then the proteasome inhibitor MG132 was used in the study. As shown in Figure 2E and F, RNF38-induced LDB1 downregulation could be obviously abolished by MG132 treatment, which further indicated that RNF38 destabilized LDB1 protein via proteasomal pathway in colorectal cancer cells.
RNF38 Interacts with LDB1 and Induces Its Polyubiquitination
To evaluate whether RNF38 interacted with LDB1, the reciprocal Co-IP was performed. As shown in Figure 3A, the reciprocal Co-IP indicated that the exogenous RNF38 interacted with exogenous LDB1 protein in HEK293T cells. And it has been also verified in CRC cells, and the results showed that endogenous RNF38 also interacted with endogenous LDB1 in CRC cells (Figure 3B and C). As RNF38 is a ubiquitin ligase, then we assessed if RNF38 regulated the ubiquitination of LDB1. As shown in Figure 3D, the Co-IP showed that RNF38 induced the polyubiquitination of LDB1. These results further indicated that RNF38 downregulated LDB1 expression by inducing its polyubiquitination.
Overexpression of LDB1 Antagonizes RNF38-Induced Cell Growth Inhibition in Colorectal Cancer Cells
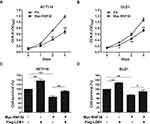
To further investigate the roles of RNF38 in CRC cells, RNF38 was overexpressed in CRC cells. As shown in Figure 4A and B, overexpression of RNF38 significantly suppressed CRC cell growth in HCT116 and DLD1 cells. As stated above, LDB1 was a substrate of RNF38, and RNF38 destabilized LDB1 protein. Then, to further assess whether RNF38 exerted its effect through LDB1, LDB1 was overexpressed in RNF38-overexpressed CRC cells. As shown in Figure 4C and D, LDB1 overexpression could significantly antagonize RNF38-induced cell growth inhibition in CRC cells. For example, in HCT116 cells, about 65% cells were still survived after cells were overexpressed with RNF38, but about 90% cells were survived after cells were co-overexpressed with RNF38 and LDB1 (Figure 4C).
RNF38 and LDB1 Regulates the Drug-Sensitivity of 5-FU in Colorectal Cancer Cells
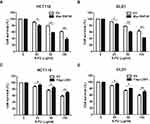
It is known that 5-FU is commonly used in adjuvant chemotherapy for colorectal cancer. In order to further investigate whether RNF38 has a guiding significance in drug chemotherapy, RNF38 was overexpressed in CRC cells under 5-FU treatment. As shown in Figure 5A and B, our results showed that CRC cells overexpressed with RNF38 were more sensitive to 5-FU in both of HCT116 and DLD1 cells. However, CRC cells overexpressed with the substrate of RNF38, LDB1, produce a certain degree of drug resistance to 5-FU in both of HCT116 and DLD1 cells (Figure 5C and D). These results further confirmed that LDB1 was a negative effector of RNF38, and RNF38 could be as a potential target for the treatment or diagnosis of CRC in clinic.
Discussion
In recent years, with the successful application of proteasome inhibitor bortezomib in the treatment of multiple myeloma, ubiquitination of proteins has entered a new research level.16 A series of anti-tumor drugs targeting ubiquitination-related enzymes, including E1, E2, E3 and DUBs, have been developed for cancer treatment.17 For example, PYR-41, an inhibitor of E1, was used in anti-cancer research.18 MI-219, an inhibitor of MDM2, was used in the study of anti-lung cancer.19 Triptolide, a small molecule inhibitor of E3 ubiquitin ligase XIAP, was used in the study of anti-breast cancer and anti-gastric cancer.20 In view of these applications, the study of ubiquitination of proteins in CRC cells is of great significance for the treatment of CRC caused by the disorder of ubiquitin system.
In this paper, we aimed to investigate the function and mechanism of RNF38 in CRC. First of all, we found that RNF38 was decreased in colon adenocarcinoma and rectum adenocarcinoma compared with normal tissues analyzed by GEPIA online, which was contrary to previous reports. Recent studies showed that RNF38 was elevated and could be as an oncoprotein in hepatocellular carcinoma and gastric cancer by inducing the ubiquitination-mediated degradation of AHNAK or SHP-1.9,21 In order to verify the expression levels of RNF38 in CRC, CRC tumor tissues and adjacent normal tissues were collected for qRT-PCR, and the results also confirmed that RNF38 was downregulated in CRC. And we also found that CRC patients with high RNF38 expression had a longer overall survival than those with low RNF38 expression. Moreover, we found that overexpression of RNF38 inhibited CRC cell growth by destabilizing LDB1 protein, which suggested that RNF38 exerted anti-tumor activity in CRC cells. At the same time, RNF38 gene is located in 9p12-p13, which belongs to the short arm of chromosome 9, and there are many tumor suppressor genes in the short arm and long arm of chromosome 9, such as CDKN2A, PTCH, DBC1 and TSC1.22
As is known to all, E3 ubiquitin ligases exert their cancer-promoting or anti-cancer function by binding with oncoproteins or tumor suppressors, and its function is closely related to the type of binding proteins.23–25 And in this study, we confirmed that LDB1 was a novel substrate of RNF38, and LDB1 was reported to be elevated in CRC and its high expression was associated with poor prognosis of CRC patients.11 What’s more, it has been reported that RNF12 could induce the ubiquitination-mediated degradation of LDB1 in Xenopus laevis functional tissues, and they also found that other RING finger family proteins RNF6 and RNF38 had similar effects in mediating degradation of LDB1, but it was not further investigated in the follow-up of the article.26 Collectively, above information suggested that RNF38, an E3 ubiquitin ligase, could exert its function as a tumor suppressor by reducing LDB1 expression in CRC. In addition, we showed that CRC cells overexpressed of RNF38 were more sensitive to 5-FU. However, overexpression of LDB1 produced a certain degree of drug resistance to 5-FU in CRC cells. These results further suggested that targeting RNF38-LDB1 axis could be a novel strategy for CRC treatment.
Conclusion
In conclusion, RNF38 was downregulated in CRC and induced LDB1 degradation by mediating its polyubiquitination, which indicated that targeting RNF38-LDB1 axis could be as a novel strategy for CRC therapy in the future.
Ethics Approval and Consent to Participate
This study was approved by the Review Board and Ethical Committee of the Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, and each patient provided written informed consent to donate tissues for this study after clinical procedures.
Acknowledgments
This study was supported by the Natural Science Foundation of Jiangsu Province (BK20170369) and the Natural Science Foundation of China (81802917).
Disclosure
All authors disclosed no relevant conflicts of interest in this work.
References
1. Mansour MA. Ubiquitination: friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi:10.1016/j.biocel.2018.06.001
2. Zhang YK, Tian WZ, Zhang RS, Zhang YJ, Ma HT. Ubiquitin-specific protease 44 inhibits cell growth by suppressing AKT signaling in non-small cell lung cancer. Kaohsiung J Med Sci. 2019;35:535–541. doi:10.1002/kjm2.12096
3. Shen WM, Yin JN, Xu RJ, Xu DF, Zheng SY. Ubiquitin specific peptidase 49 inhibits non-small cell lung cancer cell growth by suppressing PI3K/AKT signaling. Kaohsiung J Med Sci. 2019;35:401–407. doi:10.1002/kjm2.12073
4. Nguyen LK. Dynamics of ubiquitin-mediated signalling: insights from mathematical modelling and experimental studies. Brief Bioinform. 2016;17:479–493. doi:10.1093/bib/bbv052
5. Veggiani G, Gerpe MCR, Sidhu SS, Zhang W. Emerging drug development technologies targeting ubiquitination for cancer therapeutics. Pharmacol Ther. 2019;199:139–154. doi:10.1016/j.pharmthera.2019.03.003
6. Cheng J, Guo J, North BJ, et al. The emerging role for Cullin 4 family of E3 ligases in tumorigenesis. Biochim Biophys Acta Rev Cancer. 2019;1871:138–159. doi:10.1016/j.bbcan.2018.11.007
7. Eisenberg I, Hochner H, Levi T, et al. Cloning and characterization of a novel human gene RNF38 encoding a conserved putative protein with a RING finger domain. Biochem Biophys Res Commun. 2002;294:1169–1176. doi:10.1016/S0006-291X(02)00584-3
8. Liu L, Wong CC, Gong B, Yu J. Functional significance and therapeutic implication of ring-type E3 ligases in colorectal cancer. Oncogene. 2018;37:148–159. doi:10.1038/onc.2017.313
9. Zhang J, Wu H, Yi B, et al. RING finger protein 38 induces gastric cancer cell growth by decreasing the stability of the protein tyrosine phosphatase SHP-1. FEBS Lett. 2018;592:3092–3100. doi:10.1002/feb2.2018.592.issue-18
10. Xiong D, Zhu SQ, Wu YB, et al. Ring finger protein 38 promote non-small cell lung cancer progression by endowing cell EMT phenotype. J Cancer. 2018;9:841–850. doi:10.7150/jca.23138
11. Garcia SA, Swiersy A, Radhakrishnan P, et al. LDB1 overexpression is a negative prognostic factor in colorectal cancer. Oncotarget. 2016;7:84258–84270. doi:10.18632/oncotarget.12481
12. Artyukhov AS, Dashinimaev EB, Tsvetkov VO, et al. New genes for accurate normalization of qRT-PCR results in study of iPS and iPS-derived cells. Gene. 2017;626:234–240. doi:10.1016/j.gene.2017.05.045
13. Ni D, Xu P, Gallagher S. Immunoblotting and immunodetection. Curr Protoc Cell Biol. 2017;74:6 2 1–6 2 37. doi:10.1002/cpcb.18
14. Semina EV, Altherr MR, Murray JC. Cloning and chromosomal localization of two novel human genes encoding LIM-domain binding factors CLIM1 and CLIM2/LDB1/NLI. Mamm Genome. 1998;9:921–924. doi:10.1007/s003359900899
15. Xu X, Huang A, Cui X, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. 2019;9:4208–4220. doi:10.7150/thno.33803
16. Schlafer D, Shah KS, Panjic EH, Lonial S. Safety of proteasome inhibitors for treatment of multiple myeloma. Expert Opin Drug Saf. 2017;16:167–183. doi:10.1080/14740338.2017.1259310
17. Leestemaker Y, Ovaa H. Tools to investigate the ubiquitin proteasome system. Drug Discov Today Technol. 2017;26:25–31. doi:10.1016/j.ddtec.2017.11.006
18. Yang Y, Kitagaki J, Dai RM, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi:10.1158/0008-5472.CAN-07-0568
19. Azmi AS, Philip PA, Beck FW, et al. MI-219-zinc combination: a new paradigm in MDM2 inhibitor-based therapy. Oncogene. 2011;30:117–126. doi:10.1038/onc.2010.403
20. Cheng X, Shi W, Zhao C, et al. Triptolide sensitizes human breast cancer cells to tumor necrosis factor-α-induced apoptosis by inhibiting activation of the nuclear factor-kappa B pathway. Mol Med Rep. 2016;13:3257–3264. doi:10.3892/mmr.2016.4931
21. Peng R, Zhang PF, Yang X, et al. Overexpression of RNF38 facilitates TGF-beta signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:113. doi:10.1186/s13046-019-1113-3
22. Panzeri E, Conconi D, Antolini L, et al. Chromosomal aberrations in bladder cancer: fresh versus formalin fixed paraffin embedded tissue and targeted FISH versus wide microarray-based CGH analysis. PLoS One. 2011;6:e24237. doi:10.1371/journal.pone.0024237
23. Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010;10:737–747. doi:10.4161/cbt.10.8.13417
24. Li X, Elmira E, Rohondia S, et al. A patent review of the ubiquitin ligase system: 2015–2018. Expert Opin Ther Pat. 2018;28:919–937. doi:10.1080/13543776.2018.1549229
25. Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi:10.1146/annurev-biochem-060815-014922
26. Hiratani I, Yamamoto N, Mochizuki T, Ohmori SY, Taira M. Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development. 2003;130:4161–4175. doi:10.1242/dev.00621
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.