Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Resistance to PARP Inhibitors After First-Line Platinum-Based Chemotherapy in a Patient with Advanced Ovarian Cancer with a Pathogenic Somatic BRCA1 Mutation
Authors Zhong L, Yin R, Song L
Received 16 November 2022
Accepted for publication 6 March 2023
Published 14 March 2023 Volume 2023:16 Pages 195—200
DOI https://doi.org/10.2147/PGPM.S397827
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Lan Zhong,1,2 Rutie Yin,1,2 Liang Song1,2
1Department of Obstetrics and Gynecology, West China Second University Hospital, Sichuan University, Chengdu, Sichuan, 610041, People’s Republic of China; 2Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Sichuan, 610041, People’s Republic of China
Correspondence: Liang Song, Department of Obstetrics and Gynecology, West China Second University Hospital of Sichuan University, No. 20 The Third Section of South Renmin Road, Chengdu, Sichuan, 610041, People’s Republic of China, Tel +86-19828966121, Email [email protected]
Abstract: PARP inhibitors (PARPi) are the maintenance therapy after first line platinum-based chemotherapy for patients with advanced epithelial ovarian cancer (EOC) with germline and pathogenic somatic BRCA1/2 mutations. However, as with chemotherapy, patients can develop resistance to PARPi. The selective pressure generated by heterogeneous somatic BRCA mutations may give rise to chemotherapy or PARPi resistant tumors. Here, we present the case of a patient harboring a pathogenic p.Glu143* (c.427G>T) somatic BRCA1 mutation conferring resistance to olaparib following cytoreductive surgery and platinum-based chemotherapy. We ordered a plasma ctDNA analysis (tissue biopsy of recurrent lesions was contraindicated due to their anatomical location) to figure out the possible resistance mechanism. Analysis of ctDNA did not detect the pathogenic somatic BRCA1 p.Glu143* (c.427G>T) mutation seen before. The tumor cells harboring the pathogenic BRCA1 mutation were probably eliminated by the platinum-based chemotherapy, leaving only those without BRCA mutations to proliferate.
Keywords: PARPi resistance, BRCA1 mutation, ovarian carcinoma, circulating tumor DNA, sequencing
Introduction
Ovarian cancer is a leading cause of death from genital tract malignancies in women, epithelial high-grade serous ovarian cancer (HGSOC) is the most frequent subtype. Standard therapy for patients with newly diagnosed advanced ovarian cancer consists of cytoreductive surgery and platinum-based chemotherapy, but only 20–40% of patients are alive 5 years after diagnosis.1 Approximately 50% of HGSOC tumors are characterized by deficient homologous recombination (HR); HR is an important cellular pathway to repair DNA double strand breaks. Inactivation of the BRCA1 or BRCA2 genes, due to germline or somatic mutations or to BRCA1 promoter hypermethylation, lead to deficient HR.2 BRCA1 and BRCA2 somatic pathogenic mutations have been found in different types of cancer, most notably in HGSOCs. Inhibition of the poly-ADP-ribose polymerase (PARP) is lethal in cells with HR deficiency, and the use of PARP inhibitors (PARPi) has significantly improved the outcome of patients with HGSOC and BRCA1/2 mutations.3 Several PARPi agents have been approved for patients with advanced FIGO III–IV stages completing a platinum-based chemotherapy. The SOLO-1 trial demonstrated a remarkable improvement in progression-free survival (PFS) with olaparib (compared with placebo) as maintenance therapy for patients with newly diagnosed advanced ovarian cancer and a germline or somatic BRCA1/2 mutations who achieved complete remission (CR)/partial remission (PR) following primary treatment with surgery and platinum-based first-line therapy.4 However, intrinsic or acquired resistance to PARPi inevitably occurs. A key resistance mechanism involves the restoration of the HR repair pathway through BRCA reversion mutations.5 Moreover, in tumors with somatic BRCA mutations, resistance-causing events may be selected for in a Darwinian fashion.6 The heterogeneity of somatic BRCA mutations may provide an evolutionary basis for development of resistance in epithelial ovarian cancers (EOCs) under the selection pressure afforded by chemotherapy or PARPi treatments. Clones without BRCA mutations may have a selective advantage under the treatment pressure and ultimately emerge as the predominant clones.
In here, we report the case of a patient with newly diagnosed advanced HGSOC and PARPi resistance after treatment with cytoreductive surgery and platinum-based first-line therapy, who had presented a pathogenic somatic BRCA1 mutation.
Case Presentation
A 45-year-old woman presented with abdominal distention and dyspnea in January 2021. Physical examination revealed a markedly distended abdomen and a firm mass in its lower portion that was approximately 8 cm in diameter. Computed tomography (CT) revealed cystic masses at Douglas’ pouch, an omental cake, a nodule on the surface of the liver, massive ascites, and thickening of the peritoneum. No neoplastic lesions were detectable by colon fiberscopy or gastroscopy. The carbohydrate antigen (CA)-125 was 1084.5 U/mL (normal range 0–35 U/mL) (Figure 1) and CA-153 was 187.9 U/mL, but other tumor markers, such as CA-199, CA-153, CEA, alpha fetoprotein, and routine blood tests were all within normal ranges. The pelvic cyst with solid components, high CA-125 level, and massive ascites strongly suggested the presence of an advanced ovarian cancer. The patient underwent an exploratory laparotomy and partial omental resection. Intraoperative findings included multiple peritoneal disseminations larger than 2 cm in diameter, a nodule of approximately 1 cm in diameter on the liver surface, and an omental cake. Pathology analysis results identified a high-grade serous adenocarcinoma (Figure 2). The laparoscopic-based score and multiple disciplinary team (MDT) discussions suggested neoadjuvant chemotherapy (NACT) was a better option than upfront debulking surgery for this patient. After NACT with two cycles of paclitaxel and carboplatin every 3 weeks, the patient underwent a transabdominal interval cytoreductive surgery consisting of hysterectomy, bilateral salpingo-oophorectomy, omentectomy, pelvic and para-aortic lymphadenectomy, resection of implanted peritoneal nodules, and optimal debulking (R0). Pathology results revealed a high-grade serous adenocarcinoma of ovarian origin, multiple peritoneal metastases beyond the pelvis, and multiple positive para-aortic lymph nodes; the cancer was staged as IIIC. Following this, the patient received six cycles of adjuvant chemotherapy with carboplatin and paclitaxel every 3 weeks (last dose on August 2021).
 |
Figure 1 H&E staining showed histological features of high-grade serous ovarian cancer. ((A) original magnification 200×; (B) original magnification 400×). |
 |
Figure 2 Serum CA125 levels during the treatment course. Cycle 1–6 indicating chemotherapy cycles; 1st month, 2nd month and 3rd month indicating the time point after treatment of olaparib. |
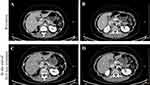
BRCA1/2 genetic testing assay (BGI) results detected variants of uncertain significance (VUS) of the germline BRCA2 mutation (c.9106C>G). Somatic gene analysis revealed pathogenic somatic BRCA1 p.Glu143* (c.427G>T, 19.84%) and somatic VUS BRCA2 p.Gln3036Glu (c.9106C>G, 47.25%) mutations (Table 1). The patient achieved complete regression (CR) according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 guidelines. She was then treated with the PARPi olaparib (300 mg BID) and underwent regular CA-125 monitoring tests (Figure 1). Three months later, her radiologic assessment showed a lymph node recurrence at the renal artery level between the aorta and inferior vena cava (Figure 3).
 |
Table 1 Germline, Somatic and ctDNA Mutation Status for Our Patient |
The cancer of our patient was supposed to be sensitive to PARPi due to the pathogenic somatic BRCA1 mutation. But, the patient did not respond to olaparib. We ordered a circulating tumor DNA (ctDNA) test to find a possible cause for the PARPi resistance because biopsy was not feasible due to the tumor’s anatomical location. Blood was sent for plasma ctDNA analysis and the results identified a VUS BRCA2 mutation c.9106C>G p.Gln3036Glu without the original pathogenic mutation BRCA1 p.Glu143* (c.427G>T) (Table 1). According to NCCN guidelines, we prescribed a first cycle of liposomal doxorubicin (40 mg/m2) and bevacizumab (7.5 mg/kg) for recurrent disease on December 2021. The patient was still being treated at the time of writing this.
Methods
DNA Extraction and Sequencing
Blood was collected in two 10 mL Streck tubes (Streck). Cell-free DNA (cfDNA) and Genomic DNA was extracted from plasma and blood cell using QIAmp circulating nucleic acid kit (Qiagen) and QIAamp DNA Blood Midi Kit (Qiagen) according to the manufacturer’s standard protocol. Qubit fluorometer 3.0 (Invitrogen) was used for DNA quantification and 1% Agarose Gel Electrophoresis to determine DNA quality. The DNA was then subjected to targeted capture using the 688 gene panel. The 688 gene panel includes 182 targeted drug-related genes, 68 immunosuppressive checkpoint-related genes, 18 chemotherapeutic drugs-related genes, and 442 genes related to tumorigenesis and development. After targeted capturing, the captured DNA were PCR amplified to achieve a higher density. Then, the post-PCR products were quantified by Qubit 2.0 (Life Tech, Invitrogen, USA) using QubitTM dsDNA HS Assay Kits (Life Tech, Invitrogen, USA). Samples with a concentration of ≥ 4 ng/μL were regarded as being of a sufficient quality for further analysis. The amplified double stranded DNA were then thermally denatured into single strands after pooling, after which cyclic buffer and ligase were added to generate circular DNA by the circularization reaction. DNA circles were used to make DNBs by rolling circle replication. The concentration of DNBs was quantified by Qubit 2.0 using QubitTM ssDNA Assay Kits (Life Tech, Invitrogen, USA) and the DNBs concentration in the range of 8 ng / μL-40 ng/μL was considered an appropriate concentration. The DNBs were loaded onto chips and sequenced on the BGISEQ-2000 sequencing platform (BGI, Shenzhen, China) for targeted sequencing in accordance with the manufacturer’s instructions. To achieve an average sequencing depth of at least 1000X.
Bioinformatic Analyses
Raw paired-end reads were subjected to fastq_tools (v0.1.0) processing to remove sequencing adapters and low-quality reads. High-quality reads were aligned to the reference human genome (build hg19), using the BWA sequence alignment software (version 0.6.2) and and Samtools (v0.1.19). Duplicate PCR reads were marked out using the Picard (MarkDuplicate) package v1.98. Alignment refinement was performed using GATK (v2.3–9). For each sample, variants were called from BAM files using the in-house software developed by BGI. Final list of mutations was annotated using bgicg_anno developed by BGI.
The identified genomics variants include single nucleotide variants (SNV), insertions and deletions (InDels), copy number variants and gene fusions. After variant calling, these variants were named according to HGVS (Human Genome Variation Society; http://www.hgvs.org/). The mutations were interpreted in accordance with the “Genetic Variation Annotation Standards and Guidelines” (2015 Edition) issued by the American College of Medical Genetics (ACMG) for germline mutation and the “Cancer mutation interpretation of guidelines and standards (2017 Edition)” for somatic mutation, respectively.
Discussion
Almost250000women worldwide are annually diagnosed with high-grade serous cancers of the ovary, fallopian tube, or peritoneum. Following the standard therapy of cytoreductive surgery and platinum-based chemotherapy, the average 5-year survival rate is approximately 30%.7 HGSOCs with homologous recombination repair deficiency (HRD) exhibit distinct clinical features, including sensitivity to platinum-based chemotherapies and PARPi.8 The most common HRD mechanisms in HGSOCs are germline or somatic mutations in the BRCA1 and BRCA2 genes, they are detected in 12–15% and 5–8.6% of patients, respectively.9–11 Ovarian cancers with germline BRCA mutations are known to present a high sensitivity to PARPi and platinum-based chemotherapy.12,13 However, the biological effects and full clinical relevance of somatic BRCA mutations remain undetermined. Whether patients with somatic BRCA mutations benefit from PARPi to the same extent as those with germline BRCA mutations is unclear due to small number of patients with somatic BRCA mutations in clinical studies.
Apoptosis and necrosis are the main processes for ctDNA origin, and they can provide a real-time picture of disease status, owing to their short half-life.14 Besides, tissue-based assays have limitations in the clinical setting, it can be challenging for specific anatomic sites. Additionally, biopsies can miss important drivers due to tumor heterogeneity or distant metastatic lesions.15 Liquid biopsy poses a relatively low risk to patients.16 Regarding to these considerations, ctDNA testing is a promising approach that could overcome these shortcomings. An ongoing multicentre, multicohort, phase 2a clinical trial was aimed at assessing the predictive value of ctDNA screening (NCT03182634).
Our patient had a somatic BRCA1 p.Glu143* (c.427G>T) pathogenic mutation which should have conferred sensitivity to PARPi, but the cancer failed to respond to olaparib. Initially, following cytoreductive surgery and platinum-based chemotherapy, our patient achieved CR, and was then treated with olaparib. Three months later, the radiologic assessment showed para-aortic lymph node recurrence at the renal artery level. We sent blood for plasma ctDNA analysis to the laboratory because a biopsy was contraindicated due to the location of the tumor. The results of the ctDNA analysis identified a somatic VUS BRCA2 c.9106C>G p.Gln3036Glu mutation, but the previously detected pathogenic somatic BRCA1 p.Glu143* (c.427G>T) mutation was undetectable. It is possible that tumor cells harboring the somatic pathogenic BRCA1 mutation were killed by the platinum-based drugs (given that tumor cells with pathogenic BRCA mutations are known to be sensitive to that chemotherapy) and that only those cells without the BRCA mutations survived. The surviving tumor cells without BRCA mutations were probably resistant to the PARPi and proliferated out of control during the three-month treatment. An essential difference between germline and somatic BRCA mutations is determined by the homogeneity or heterogeneity of the mutations in tumor tissues. The heterogeneity of somatic BRCA mutations provides a background for evolutionary pressures during intervening therapies.
Tissue biopsy is the gold standard for recurrent tumor gene testing, but the procedure is invasive and can be challenging depending on the tumor’s anatomical location. Thus, a noninvasive blood-based test (such as a circulating tumor DNA [ctDNA] profiling) may offer significant advantages for the identification of various types of BRCA mutations. Moreover, postsurgical ctDNA analyses can detect minimal residual disease after a tumor surgical resection.17 Somatic BRCA mutation tumor cells may not all be eliminated by platinum-based chemotherapy, and studies need to clarify the status of BRCA mutations after first-line platinum-based chemotherapy at CR/PR to provide evidence for maintenance therapy selection.
Patient Consent Statement
Written informed consent was obtained from the patient for the publication of this case report. The study was approved by the Ethics Committee of West China Second University Hospital of Sichuan University (approval number: 2020076).
Acknowledgments
The study was supported grants from the Key Project of Science and Technology Department of Sichuan province (CN) (19ZDYF), New Seed Fund of West China Second University Hospital, Sichuan University (kz022) and Horizontal Topic of Sichuan University (22H0226, 22H0851).
Disclosure
All authors declare no conflicts of interest in this work.
References
1. Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240–1253. doi:10.1016/S0140-6736(18)32552-2
2. Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi:10.1038/nature03445
3. Chiappa M, Guffanti F, Bertoni F, Colombo I, Damia G. Overcoming PARPi resistance: preclinical and clinical evidence in ovarian cancer. Drug Resist Updat. 2021;55:100744. doi:10.1016/j.drup.2021.100744
4. Massari F, Di Nunno V, Santoni M, et al. Sunitinib alone or after nephrectomy in metastatic renal-cell carcinoma. N Engl J Med. 2018;379:417–427. doi:10.1016/j.euo.2018.08.002
5. McMullen M, Karakasis K, Madariaga A, Oza AM. Overcoming platinum and PARP-inhibitor resistance in ovarian cancer. Cancers. 2020;12(6). doi:10.3390/cancers12061607
6. Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi:10.1038/nature10762
7. Reid BM, Permuth JB, Sellers TA, et al. Epidemiology of ovarian cancer: a review. Cancer Biol Med. 2017;14(1):9–32. doi:10.20892/j.issn.2095-3941.2016.0084
8. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. doi:10.1158/1078-0432.CCR-13-2287
9. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi:10.1038/nature10166
10. Kanchi KL, Johnson KJ, Lu C, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi:10.1038/ncomms4156
11. Ji G, Yao Q, Bao L, et al. Germline and tumor BRCA1/2 mutations in Chinese high grade serous ovarian cancer patients. Ann Transl Med. 2021;9(6):453. doi:10.21037/atm-20-6827
12. Fong PC, Yap TA, Boss DS, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi:10.1200/JCO.2009.26.9589
13. Tan DS, Yap TA, Hutka M, et al. Implications of BRCA1 and BRCA2 mutations for the efficacy of paclitaxel monotherapy in advanced ovarian cancer. Eur J Cancer. 2013;49:1246–1253. doi:10.1016/j.ejca.2012.11.016
14. Aggarwal C, Thompson JC, Black TA, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2019;5:173–180. doi:10.1001/jamaoncol.2018.4305
15. Stewart CM, Kothari PD, Mouliere F, et al. The value of cell-free DNA for molecular pathology. J Pathol. 2018;244:616–627. doi:10.1002/path.5048
16. Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol Hematol. 2020;155:103109. doi:10.1016/j.critrevonc.2020.103109
17. Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5(12):1710–1717. doi:10.1001/jamaoncol.2019.3616
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.