Back to Journals » Infection and Drug Resistance » Volume 12
Resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible Escherichia coli in Taiwan, 2012-2015
Authors Chang YT, Siu LK, Wang JT , Wu TL, Chen YH, Chuang YC, Lin JC, Lu PL
Received 10 March 2019
Accepted for publication 20 May 2019
Published 16 July 2019 Volume 2019:12 Pages 2113—2123
DOI https://doi.org/10.2147/IDR.S208231
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Ya-Ting Chang,1 L Kristopher Siu,2 Jann-Tay Wang,3 Tsu-Lan Wu,4 Yu-Hui Chen,5,6 Yin-Ching Chuang,7,8 Jung-Chung Lin,9 Po-Liang Lu1,10
1Department of Internal Medicine, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan; 2National Institutes of Infectious Diseases and Vaccinology, National Health Research Institutes, Miaoli, Taiwan; 3Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; 4Department of Clinical Pathology, Linkou Chang Gung Memorial Hospital, Taoyuan, Taiwan; 5Infection Control Center, Chi Mei Medical Center, Tainan, Taiwan; 6Department of Nursing, College of Medicine and Life Science, Chung Hwa University of Medical Technology, Tainan, Taiwan; 7Department of Internal Medicine and Medical Research, Chi Mei Medical Center, Tainan, Taiwan; 8Department of Internal Medicine, Chi Mei Medical Center, Tainan, Taiwan; 9Division of Infectious Diseases and Tropical Medicine, Department of Internal Medicine, Tri-Service General Hospital, National Defense Medical Center, Taipei, Taiwan; 10College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
Purpose: This study aimed to investigate the resistance mechanisms and molecular epidemiology of carbapenem-nonsusceptible Escherichia coli (CnsEC) in Taiwan.
Patients and methods: From 2012 to 2015, 237 E. coli isolates with minimum inhibitory concentrations of imipenem or meropenem >1 μg/mL were collected in a nationwide surveillance and subjected to polymerase chain reaction (PCR) for carbapenemase, AmpC-type β-lactamase, and extended spectrum β-lactamase (ESBL) genes. We evaluated outer membrane proteins (OmpF and OmpC) loss and conducted multilocus sequence typing and pulsed-field gel electrophoresis (PFGE). Isolates that were resistant to all carbapenems were designated as pan-carbapenem-resistant E. coli (pCREC) in this study.
Results: The predominant resistance mechanism of CnsEC in Taiwan was the CMY-2 β-lactamase in combination with OmpF and OmpC loss. Sequence type 131 was the most prevalent type (29.2%). Among 237 CnsEC isolates, 106 (44.7%) isolates were pCREC and 18 (7.59%) produced carbapenemase. The prevalence of carbapenemases increased from 6% in 2012 to 11.36% in 2015. Various carbapenemases including KPC-2, IMP-8, NDM-1, NDM-5, VIM-1, OXA-48, and OXA-181 were identified, with NDM-1 being the most common (38.9%) carbapenemase. Comparison between pCREC and non-pCREC among the non-carbapenemase-producing CnsEC isolates revealed SHV, CMY, co-carriage of SHV and CTX-M and concurrent loss of both OmpF and OmpC were more commonly detected in the pCREC group. PFGE revealed no nationwide clonal spread of carbapenemase-producing E. coli.
Conclusion: NDM-1 was the most common carbapenemase and combination of CMY-2 and concurrent OmpF and OmpC porin loss was the most prevalent resistance mechanism in CnsEC in Taiwan.
Keywords: multidrug resistance, carbapenemase, Enterobacteriaceae, epidemiology
Introduction
Escherichia coli is one of the most common human pathogen, the major etiology of community-acquired urinary tract infection and a major nosocomial Gram negative bacteria.1,2 In the late 1990s, extended spectrum β-lactamase (ESBL)-producing E. coli infections emerged.3 Since the worldwide propagation of ESBL-producing Enterobacteriaceae, carbapenems have been the prevailing treatment of such infections. Carbapenem resistance in Enterobacteriaceae was relatively uncommon before 2000. Nevertheless, the prevalence of carbapenem-resistant Enterobacteriaceae (CRE) rose markedly in the following decade.4 The United States Centers for Disease Control and Prevention (CDC) listed CRE as an urgent threat that requires intensive monitoring and prevention (https://www.cdc.gov/drugresistance/biggest_threats.html). There are two primary mechanisms of carbapenem resistance: (1) carbapenemase production and (2) a combination of β-lactamases, ESBLs, or AmpC cephalosporinases with structural mutations such as outer membrane protein (OMP) deficiency or efflux pump overproduction.5 Carbapenemases are generally considered more hazardous, because the genes are carried mostly on plasmids, which can transmit between Enterobacteriaceae species and facilitate spread.6
Most surveillance reports have shown lower prevalence rates of carbapenam resistance and carbapenemase-producing Enterobacteriaceae (CPE) for E. coli, when compared with the rates of Klebsiella pneumoniae.7–9 Seeing the global dissemination of CTX-M β-lactamase-producing E. coli clones in community-onset infections,3,10 the carbapenem resistant E. coli should be a serious public health concern and its spread may potentially change the status quo. CTM-X 15 is the most common type of ESBL among E. coli and has been associated with E. coli sequence type 131 (ST131). ST131 correlates with extraintestinal infections, fluoroquinolone resistance, and community-onset infections.11 The carbapenemase genes of blaNDM, blaVIM, blaKPC, blaIMP, and blaOXA-48 have all been identified from ST131.11 A global epidemic caused by carbapenemase-producing ST131 E. coli in nosocomial and community-onset infections would be a nightmare. Because less is known about the carbapenem-nonsusceptible E. coli (CnsEC) in Taiwan, we conducted a nationwide surveillance to investigate the characteristics, resistance mechanisms, and molecular typing of CnsEC in Taiwan.
Materials and methods
Bacterial isolates and definitions
A total of 21 hospitals in Taiwan, including 12 tertiary medical centers and 9 regional hospitals, participated in the surveillance program from January 2012 to September 2015. The study was approved by the institutional review boards (IRBs) of all participating hospitals, including Chang Gung Memorial Hospital (IRB No.: 1003399B), Taipei Veterans General Hospital (IRB No.: 2011-11-001IC), National Taiwan University Hospital (IRB No.: 201110043RB), Tri-Service General Hospital (IRB No.: 100-05-205), Kaohsiung Medical University Chung-Ho Memorial Hospital (IRB No.: KMUH-IRB-20110328), Chi- Mei Medical Center (IRB No.: 10012-001), China Medical University Hospital (IRB No.: DMR100-IRB-214), Kaohsiung Armed Forces General Hospital (IRB No.: 100–076), National Cheng Kung University Hospital (IRB No.: A-ER-101–304), Tzu Chi Hospital (IRB No.: ACT-IRB100-14), Chung Shan Medical University Hospital (IRB No.: CS12187) and Taichung Veterans General Hospital (IRB No.: SG14157). The IRBs waived the requirement for informed consents from participants because all bacterial isolates were obtained from clinical samples as part of standard care.
A total of 237 nonduplicate imipenem- or meropenem-nonsusceptible E. coli isolates (minimum inhibitory concentrations [MICs] of imipenem or meropenem >1 μg/mL) were collected. Isolates with imipenem or meropenem MICs of >1 μg/mL were defined as CnsEC. Isolates that were resistant to all carbapenems, including imipenem, meropenem, doripenem (MICs ≥4 μg/mL) and ertapenem (MICs ≥2 μg/mL), were designated as pan-carbapenem-resistant E. coli (pCREC) in this study. The preliminary identification of E. coli and testing of carbapenem susceptibility were performed at the participating hospitals as routine laboratory procedures. All isolates were sent to a reference laboratory at the National Health Research Institutes in Taiwan. Species identification was confirmed with a VITEK 2 automated system (bioMerieux, Marcy l’Etoile, France) and the Bruker Biotyper MALDI-TOF MS system (Bruker Daltonik GmbH, Leipzig, Germany).
Antimicrobial susceptibility testing
All isolates were tested for MICs of (1) β-lactam agents, including penicillin (piperacillin-tazobactam), cephalosporins (cefazolin, cefoxitin, cefuroxime, cefotaxime, ceftazidime, and cefepime), carbapenems (doripenem, ertapenem, imipenem, and meropenem), and a monobactam (aztreonam); and (2) non-β-lactams, including fluoroquinolones (ciprofloxacin and levofloxacin), aminoglycosides (gentamicin and amikacin), trimethoprim-sulfamethoxazole, colistin, and tigecycline. The MIC of tigecycline was determined through the E-test (AB Biodisk, Solna, Sweden) on Mueller-Hinton media, and the MICs of other agents were determined using the broth microdilution method (Sensititre, Trek Diagnostic Systems, Cleveland, OH, USA). Susceptibilities to colistin and tigecycline were determined based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (http://www.eucast.org/clinical_breakpoints/), and susceptibilities to other agents were determined based on the updated guidelines from the Clinical and Laboratory Standards Institute (CLSI).12
Detection of genes encoding carbapenemases, AmpC, and ESBLs
All verified CnsEC isolates were subjected to polymerase chain reaction (PCR) for the detection of genes encoding carbapenemase (class B families IMP, VIM, NDM, GIM, SPM, and SIM; class A families NMC, IMI, SME, KPC, and GES; class D family OXA-48),13 plasmidic AmpC (CMY, DHA, and ACT),14 and ESBL genes (CTX-M, TEM, and SHV).15
Pulsed-field gel electrophoresis
Total DNA was prepared, and pulsed-field gel electrophoresis (PFGE) was performed as described.16 The restriction enzyme, XbaI (New England Biolabs, Beverly, MA, USA), was used at the temperature suggested by the manufacturer. The Dice coefficient was used to calculate similarities, and the unweighted pair-group method with arithmetic mean was used for cluster analysis with BioNumerics software version 5.10 (Applied Maths, St-Martens-Latem, Belgium).
Isolation and analysis of OMPs
Bacterial OMPs were prepared as described.17 The OMPs were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 7.5% polyacrylamide-6 M urea gels and visualized using Coomassie Blue staining (Bio-Rad). A reference strain, E. coli ATCC25922, was included as a control.
Multilocus sequence typing
Multilocus sequence typing (MLST) with seven housekeeping genes18 —adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate synthetase), and recA (ATP/GTP motif)—was performed on all isolates according to the protocol described on the Enterobase website. (http://mlst.ucc.ie/mlst/dbs/Ecoli/documents/primersColi_html). The allele sequences and STs were verified by the website as well (http://mlst.ucc.ie/mlst/dbs/Ecoli/).
Statistical analyses
SPSS version 17.0 (SPSS, Chicago, IL, USA) was used to perform statistical analyses. Categorical variables were compared using the chi-square test or Fisher’s exact test. A p-value of <0.05 was considered statistically significant.
Results
Of the 237 CnsEC isolates, most were collected from urine (35.0%), followed by abscess, drainage, pus, or wound cultures (22.4%), and blood (11.4%). Among all CnsEC, 18 isolates (7.59%) produced carbapenemase. We identified 44.7% (106 isolates) of the CnsEC to be pCREC, including 17 carbapenemase-producing isolates.
Antimicrobial susceptibilities
Table 1 shows the resistance rates of CnsEC to antimicrobial agents. With the exception of piperacillin-tazobactam and cefepime, the resistance rates to all noncarbapenem β-lactams were 95–100%. The resistance rates were higher for ertapenem (96.2%), followed by imipenem (74.3%), meropenem(61.6%) and doripenem (47.3%). Figure 1 shows the MIC distribution of carbapenems in CnsEC. The MIC is higher for imipenem than meropenem and doripenem, with 60% of isolates presenting MIC >4 mg/L for imipenem and only 36.3% and 18.1% for meropenem and doripenem, respectively. Amikacin, colistin, and tigecycline were the most effective agents among all antimicrobials. Trends of increasing nonsusceptibility were observed for amikacin (2.69% in 2012–2013 and 8% in 2014–2015) and colistin (1.79% and 4.8% in two consecutive 2-year periods) without statistical significance.
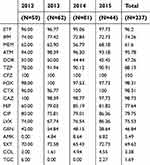 |
Table 1 Antimicrobial resistance rates of carbapenem-nonsusceptible E. coli isolates in Taiwan |
PFGE patterns, MLST profiles, and PCR analyses for carbapenemases
During the study period, eight pulsotypes were found consecutively in three or four of the years. Of the eight pulsotypes, seven corresponded to a specific ST, and four of these belonged to ST131. MLST analysis revealed ST131 to be the most prevalent ST, accounting for 29.2% of all isolates, followed by ST410 (13 isolates, 5.6%) and ST457 (13 isolates, 5.6%). A total of 11, 10, 9, 8, and 6 isolates were designated as ST405, ST2003, ST38, ST68, and ST354, respectively. Moreover, 10 isolates were found to be new STs (4.3%), and the remaining isolates were of diverse STs.
Figure 2 depicts the percentages and distributions of carbapenemases among CnsEC isolates in each year. Increasing prevalence was found with more diverse carbapenemase genes over the 4-year study period. The prevalence of carbapenemases rose from 6% in 2012 to 11.36% in 2015; however, no statistical significance was found. The isolate number of each carbapenemase-producing E. coli and the associated MLST types were as follows: three KPC-2 (two ST131 and one ST410), two IMP-8 (ST410 and a new type), seven NDM-1(two ST10, two ST410, and one of each ST224, ST345, ST4261), one NDM-5 (ST117), three VIM-1(ST349, ST744, and a new ST type), and two OXA-48 (ST68 and ST405) isolates. An ST410 NDM-1-producing isolate also harbored OXA-181. The diverse STs of these carbapenemase-producing E. coli isolates indicate the absence of epidemic clones. The PFGE patterns of carbapenemase-producing isolates are shown in Figure 3. All isolates demonstrated different PFGE patterns.
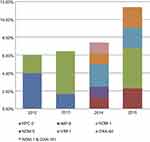 |
Figure 2 Distribution of carbapenemases among carbapenem-nonsusceptible E. coli isolates in Taiwan. |
 |
Figure 3 Dendrogram generated by pulsed-field gel electrophoresis (PFGE) patterns of the 18 isolates of carbapenemase-producing carbapenem-nonsusceptible E. coli using the BioNumerics software. |
AmpC, ESBL, and porin loss
86.5% of CnsEC isolates produced AmpC β-lactamase, and most of them (96.1%) carried CMY-2 gene. CMY-4, CYM-42, DHA-1, and CMY-2/DHA-1 co-carriage were detected in one, three, four, and eight isolates, respectively. CTX-M-type ESBL was detected in 103 isolates (43.5%), whereas only 12 isolates harbored SHV genes (5.1%). Among the 219 CnsEC isolates without carbapenemases, the most common pattern of porin deficiency was the loss of both OmpC and OmpF (157/219, 71.7%), followed by the loss of OmpF alone (42/219, 19.2%). The patterns of coexistence of β-lactamases and Omp loss in non-carbapenemases-producing CnsEC were shown in Table 2. For the 18 isolates of carbapenemase-producing E. coli, there were coexistence of DHA, CMY, SHV and CTX-M genes in 1, 8, 3, 10 isolates, respectively (data not shown). The prevailing type of porin loss in carbapenemase-harboring isolates was the loss of OmpF only (11/18, 61.1%) (Table 2). Furthermore, in the non-carbapenemase-producing subgroup, resistance rates to seven antimicrobial agents were shown to be significantly related to the loss of OmpC or OmpF (Table 3). In order to evaluate the contribution to resistance by each Omp, a categorical comparison was performed between isolated loss of OmpC or OmpF and concurrent loss of both OMPs. Compared to OmpF, the loss of OmpC was more significantly associated with resistance to all carbapenems (Table 3).
 |
Table 2 Outer membrane profiles for carbapenem-nonsusceptible E. coli isolates |
 |
Table 3 Association of outer membrane proteins loss and antimicrobial resistance rates (%) among the 219 carbapenem-nonsusceptible E. coli isolates without carbapenemase |
Comparisons between pan-carbapenem-resistant E. coli (pCREC) and non-pCREC among the non-carbapenemase producing CnsEC isolates
With the exclusion of 18 carbapenemase-producing E. coli isolates, subgroup analyses were performed between pCREC (89 isolates) and non-pCREC (130 isolates) in order to compare the differences of resistance mechanisms and patterns. Regarding the presence of ESBL and AmpC genes, Figure 4A demonstrates that SHV, SHV and CTX-M co-carriage, and CMY were more commonly detected in the pCREC group with statistical significance. Figure 4B depicts the resistance rates of the three most effective antibiotics for CnsEC, namely colistin, tigecycline and amikacin. A trend of higher resistance rates to colistin and amikacin were noticed in pCREC isolates, but with no statistical significance. For the other antimicrobial agents, the resistance rates were similar between pCREC and non-pCREC except for piperacillin-tazobactam (89.9% vs 66.9%, p<0.0001) and cefepime (97.8% vs 80%, p<0.0001). Isolated loss of OmpF was more likely to be found among non-pCREC isolates (28.5% vs 5.6%, p<0.0001) while concurrent loss of both OmpF and OmpC was more commonly detected in pCREC isolates (87.6% vs 60.8%, p<0.0001) (Figure 4C).
Discussion
The predominant mechanism of CnsEC in Taiwan in 2012–2015 was observed to be AmpC β-lactamase CMY-2, in combination with OmpF and OmpC porin loss, similar to a previous report for 2010 and 2012 isolates in Taiwan.19 The most prevalent carbapenemase was the NDM type (44.4%), among which NDM-1 type was the most common (7/8). In Taiwan, the first KPC-2- and NDM-1-harboring E. coli were identified in 2012.19 Herein we report the first NDM-5 and OXA-181 in Enterobacteriaceae in Taiwan in this surveillance study.
There were 106 (44.7%) isolates of CnsEC to be pCREC. Among the 18 carbapenemase-producing E. coli, 17 isolates were pCREC except one that harbored OXA-48 type carbapenemase. The result is compatible with previous reports that OXA-48 producers are more frequent to be susceptible to carbapenems.20 Aside from carbapenemase, the major resistance mechanisms that contributed to CnsEC were the CMY-type AmpC β-lactamase and the concurrent porin loss of OmpF and OmpC (Table 2).
We summarize the prevalence rates and predominant resistance mechanisms of CnsEC reported in recent literatures in Table 4.7–9,21,22 Compared with the global data, the carbapenemase prevalence of CnsEC in Taiwan was relatively low (Table 4). The predominant type of carbapenemase in CnsEC varies in different areas, with NDM being the major type detected in both China and Taiwan.7,22 Several novel β-lactam/β-lactamase inhibitor combinations, such as ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam, have been developed to treat multidrug-resistant organisms but all have limited activity against metallo-β-lactamases (MBL) producers (NDM or VIM).23,24 These new antibiotics are expected to have a potential role for CnsEC treatment in Taiwan due to the low rate of carbapenemase production. However, NDM-1 as the major carbapenemase in carbapenemase-producing E. coli in Taiwan necessitates continuous monitoring of the molecular epidemiology. In addition, it is worth mentioning that different from E. coli, the predominant resistance gene of carbapenemase-producing K. pneumoniae (CPKP) in Taiwan during the same study period was KPC.13,25
 |
Table 4 The prevalence rates and genotypes of carbapenemase-producing E. coli among carbapenem-nonsusceptible E. coli isolates |
The most commonly detected AmpC β-lactamase, ESBL type, and ST for CnsEC in Taiwan were CMY-2 (91.75%), CTX-M-type ESBLs (46.41%), and ST131 (29.18%), respectively. E. coli ST131 is a vehicle for the global dissemination of fluoroquinolone resistance and blaCTX-M-15 among extraintestinal pathogenic E. coli. NDM, KPC, VIM, IMP, OXA-48 have all been identified in ST131 globally.11 In our surveillance, we observed two ST131 strains among the carbapenemase-producing E. coli, and both harbored KPC-2 genes. The high prevalence of either nosocomial or community-onset ESBL-producing E. coli is a well-known issue in Taiwan, as in many Asia-Pacific countries.26,27 The emergence of community-onset CRE infections worldwide, with reported prevalence rates of 0.04–29.5%,28 further complicates the empiric treatment of community-onset Enterobacteriaceae infections. Continuous surveillance of carbapenemase-producing E. coli and epidemic clone such as ST131 is warranted in order to develop instantaneous containment strategies.
Amikacin, colistin, and tigecycline were the most effective antimicrobial agents for CnsEC in Taiwan. One review article reported that the treatment efficacy of carbapenems decreases from 69% for CPKP isolates with a MIC ≤4 mg/liter to 29% for isolates with MICs >8 mg/liter.20 Some cohort studies found that adding high dose meropenem (2g every 8 hrs by extended infusion) to another active drug was associated with lower mortality among patients with bloodstream infection caused by CPE with MIC≤8 mg/liter.29,30 However, all the aforementioned studies contained predominantly KPC-producing K. pneumoniae and whether the “high-dose-extended-infusion meropenem” strategy also applies for other Enterobacteriaceae requires more research. According to our result, only 36.3% CnsEC isolates’ MIC for meropenem is >4 mg/L. High-dose-extended-infusion meropenem for CnsEC might be applicable for some CnsEC infections in Taiwan, especially for infection sources not appropriate for tigecycline combination therapy (urinary tract infection or bloodstream infection) or for isolates with lower meropenem MIC.
Conclusion
The predominant mechanism of carbapenem nonsusceptibility in E. coli isolates in Taiwan was CMY-2-type AmpC β-lactamase in combination with OmpF and OmpC porin loss. The most common type of carbapenemase was NDM. About 45% of the CnsEC were resistant to all carbapenems(pCREC). Besides carbapenemase, the major differences in resistant mechanisms between pCREC and non-pCREC were the significantly higher percentage of SHV and/or CMY and concurrent loss of both OmpF and OmpC in pCREC. Diverse PFGE patterns and ST analysis suggested that no nationally spread clones identified.
Acknowledgments
This surveillance program was sponsored and coordinated by the Taiwan Centers for Disease Control (MOHW 104-CDC-C-114-144406).
Disclosure
Dr Ya-Ting Chang reports non-financial support from Taiwan Centers for Disease Control, during the conduct of the study. The authors report no other conflicts of interest in this work.
References
1. Gaynes R, Edwards JR, National Nosocomial Infections Surveillance S. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–854. doi:10.1086/432803
2. Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect. 2014;20:380–390. doi:10.1111/1469-0691.12646
3. Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8:159–166. doi:10.1016/S1473-3099(08)70041-0
4. Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–67. doi:10.1093/cid/cir202
5. Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new beta-lactamases from gram-negative bacteria. Annu Rev Microbiol. 2011;65:455–478. doi:10.1146/annurev-micro-090110-102911
6. Iovleva A. Carbapenem-resistant Enterobacteriaceae. Clin Lab Med. 2017;37:303–315.
7. Guh AY, Bulens SN, Mu Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA. 2015;314:1479–1487. doi:10.1001/jama.2015.12480
8. Jeong SH, Kim HS, Kim JS, et al. Prevalence and molecular characteristics of carbapenemase-producing Enterobacteriaceae from five hospitals in Korea. Ann Lab Med. 2016;36:529–535. doi:10.3343/alm.2016.36.6.529
9. Karlowsky JA, Lob SH, Kazmierczak KM, et al. In vitro activity of imipenem against carbapenemase-positive Enterobacteriaceae isolates collected by the SMART Global Surveillance Program from 2008 to 2014. J Clin Microbiol. 2017;55:1638–1649. doi:10.1128/JCM.02316-16
10. Oteo J, Perez-Vazquez M, Campos J. Extended-spectrum beta-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr Opin Infect Dis. 2010;23:320–326.
11. Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–591. doi:10.1128/CMR.00116-14
12. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
13. Chiu SK, Wu TL, Chuang YC, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013;8:e69428. doi:10.1371/journal.pone.0069428
14. Alvarez M, Tran JH, Chow N, Jacoby GA. Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–537. doi:10.1128/aac.48.2.533-537.2004
15. Eckert C, Gautier V, Saladin-Allard M, et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004;48:1249–1255. doi:10.1128/aac.48.4.1249-1255.2004
16. D’Agata EM, Gerrits MM, Tang YW, Samore M, Kusters JG. Comparison of pulsed-field gel electrophoresis and amplified fragment-length polymorphism for epidemiological investigations of common nosocomial pathogens. Infect Control Hosp Epidemiol. 2001;22:550–554. doi:10.1086/501950
17. Freire P, Vieira HL, Furtado AR, et al. Effect of the morphogene bolA on the permeability of the Escherichia coli outer membrane. FEMS Microbiol Lett. 2006;260:06–111. doi:10.1111/j.1574-6968.2006.00307.x
18. Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi:10.1111/j.1365-2958.2006.05172.x
19. Ma L, Siu LK, Lin JC, et al. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis. 2013;13:599. doi:10.1186/1471-2334-13-599
20. Daikos GL, Markogiannakis A. Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect. 2011;17:1135–1141. doi:10.1111/j.1469-0691.2011.03553.x
21. Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17:153–163. doi:10.1016/S1473-3099(16)30257-2
22. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62:e01882–17.
23. Haidar G, Clancy CJ, Chen L, et al. Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:e00642-17. doi:10.1128/AAC.00642-17
24. Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, et al. Treatment of infections caused by Extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31:e00079-17.
25. Chiu SK, Ma L, Chan MC, et al. Carbapenem nonsusceptible Klebsiella pneumoniae in Taiwan: dissemination and increasing resistance of carbapenemase producers during 2012-2015. Sci Rep. 2018;8:8468. doi:10.1038/s41598-018-26691-z
26. Chang YT, Coombs G, Ling T, et al. Epidemiology and trends in the antibiotic susceptibilities of Gram-negative bacilli isolated from patients with intra-abdominal infections in the Asia-Pacific region, 2010-2013. Int J Antimicrob Agents. 2017;49:734–739. doi:10.1016/j.ijantimicag.2017.01.030
27. Jean SS, Coombs G, Ling T, et al. Epidemiology and antimicrobial susceptibility profiles of pathogens causing urinary tract infections in the Asia-Pacific region: results from the Study for Monitoring Antimicrobial Resistance Trends (SMART), 2010-2013. Int J Antimicrob Agents. 2016;47:328–334. doi:10.1016/j.ijantimicag.2016.01.008
28. Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents. 2017;50:127–134. doi:10.1016/j.ijantimicag.2017.03.012
29. Gomez-Simmonds A, Nelson B, Eiras DP, et al. Combination regimens for treatment of carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Antimicrob Agents Chemother. 2016;60:3601–3607. doi:10.1128/AAC.03007-15
30. Gutierrez-Gutierrez B, Salamanca E, de Cueto M, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis. 2017;17:726–734. doi:10.1016/S1473-3099(17)30228-1
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


