Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 10
Repeated short climatic change affects the epidermal differentiation program and leads to matrix remodeling in a human organotypic skin model
Authors Boutrand LB, Thépot A, Muther C, Boher A, Robic J, Guéré C, Vié K, Damour O, Lamartine J
Received 27 August 2016
Accepted for publication 8 November 2016
Published 13 February 2017 Volume 2017:10 Pages 43—50
DOI https://doi.org/10.2147/CCID.S120800
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Laetitia-Barbollat Boutrand,1 Amélie Thépot,2 Charlotte Muther,3 Aurélie Boher,2 Julie Robic,4 Christelle Guéré,4 Katell Vié,4 Odile Damour,5 Jérôme Lamartine1,3
1Departement de Biologie, Université Claude Bernard Lyon I, 2LabSkinCreations, 3CNRS UMR5305, Laboratoire de Biologie Tissulaire et d’Ingénierie Thérapeutique (LBTI), Lyon, 4Laboratoires Clarins, Cergy-Pontoise, 5Banque de Tissus et Cellules, Hospices Civiles de Lyon, Lyon, France
Abstract: Human skin is subject to frequent changes in ambient temperature and humidity and needs to cope with these environmental modifications. To decipher the molecular response of human skin to repeated climatic change, a versatile model of skin equivalent subject to “hot–wet” (40°C, 80% relative humidity [RH]) or “cold–dry” (10°C, 40% RH) climatic stress repeated daily was used. To obtain an exhaustive view of the molecular mechanisms elicited by climatic change, large-scale gene expression DNA microarray analysis was performed and modulated function was determined by bioinformatic annotation. This analysis revealed several functions, including epidermal differentiation and extracellular matrix, impacted by repeated variations in climatic conditions. Some of these molecular changes were confirmed by histological examination and protein expression. Both treatments (hot–wet and cold–dry) reduced the expression of genes encoding collagens, laminin, and proteoglycans, suggesting a profound remodeling of the extracellular matrix. Strong induction of the entire family of late cornified envelope genes after cold–dry exposure, confirmed at protein level, was also observed. These changes correlated with an increase in epidermal differentiation markers such as corneodesmosin and a thickening of the stratum corneum, indicating possible implementation of defense mechanisms against dehydration. This study for the first time reveals the complex pattern of molecular response allowing adaption of human skin to repeated change in its climatic environment.
Keywords: skin, organotypic tissue, climatic changes, transcriptome, collagen
Introduction
The human skin constantly needs to get used to various changes in its environment. Ambient temperature and atmospheric humidity are key climatic parameters that can suddenly change as one moves from indoors to outdoors and vice versa. The range and frequency of these temperature changes have been increased by the spread of air conditioning in many countries, even with temperate climate. Surprisingly, very sparse data are available in the literature concerning the molecular response of human skin to repeated climate change. In this study, a versatile skin equivalent (SE) model1 was used subjected to short daily climatic changes to mimic transient exposure to external winter or summer conditions. To obtain an exhaustive and definitive view of the molecular mechanisms elicited by climate changes, this study analyzed genome-scale gene expression change and looked for functional change by bioinformatic ontology annotation. This analysis revealed several functions, including epidermal differentiation and extracellular matrix, impacted by repeated variations in climatic conditions. Some of these molecular changes were confirmed by histological examination and protein expression. The study revealed, for the first time, the complex pattern of molecular response allowing human skin to adapt to repeated change in its climatic environment.
Experimental procedures
Culture of SEs
Normal human skin samples were obtained from anonymous healthy donors. Residues of plastic surgeries were harvested according to French regulations, including declaration to the Research Ministry (DC n° 2008162) and procurement of written informed consent from the donor. Because the authors used residues of plastic surgeries with informed consent of the patient, they did not need the approval of an ethical review board according to the French law.
SEs were prepared as described previously.1 Briefly, for preparation of dermal equivalent, normal human dermal fibroblasts from young donors (2 years old) were seeded at a density of 25×104 cells/cm² onto a dermal substrate made of chitosan-cross-linked collagen–glycosaminoglycan matrix prepared as previously described. The dermal equivalent was grown for 14 days at 37°C in 5% CO2 atmosphere. For the preparation of SEs, normal human epidermal keratinocytes from young donors were seeded on the dermal equivalent on day 14, at a density of 25×104 cells/cm². After 7 days of submerged culture, the SE was raised at the air–liquid interface and cultured for 19 additional days before climatic treatment.
Climatic treatment of cell cultures and SEs
Preliminary analyses demonstrated that strong heat and changes in relative humidity (RH) (4°C, 40% RH for 60 min, and 42°C, 80% RH for 60 min) were not cytotoxic for monolayer-cultured fibroblasts or keratinocytes. The dermal substrates elevated at the air–liquid interface were cultured in classical conditions at 37°C and then put in the climatic chamber at 4°C or 42°C. Temperature was measured every 5 min without opening the cell culture dish. After 15 min at 4°C, the temperature at the dermal substrate surface was 14°C; after 42°C for 30 min, the surface was at 41.1°C. Taking account of the repeatability of the climatic stress (3 times), it was considered that 15 min at 10°C and 30 min at 42°C were suitable to mimic repeated climatic change. Then, these conditions were used for the treatment of SEs at days 40, 42, and 44: SEs were stressed under 40°C and 80% RH for 30 min or 10°C and 40% RH for 15 min. All samples were harvested, after 45 days of total cell culture, for histology, immunohistochemistry, and DNA array studies. For each cell culture condition and analysis, SEs were produced in triplicate.
RNA extraction and quantitative polymerase chain reaction (qPCR) analysis
SE samples were placed in RNAlater® (Ambion, Foster City, CA, USA) and stocked at 4°C until RNA extraction, which was performed, after initial grinding with plastic pistons, using the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Total RNA was reverse-transcribed into cDNA by the PrimeScript™ RT reagent Kit (TaKaRa, Shiga, Japan) and analyzed by real-time qPCR using SYBR® Premix Ex Taq™ II (TaKaRa) on a Mx3000P real-time PCR system (Stratagene, San Diego, CA, USA). Results were obtained from 3 SEs per culture condition and normalized to the 18S rRNA expression level. Relative quantification was performed using the 2∆∆Cq quantification method.2 The oligonucleotide sequences are given in Table S1.
Microarray analysis
Gene expression profiles of SEs were analyzed using a whole human genome microarray containing 47,231 probes (HumanHT-12 v4 Expression BeadChip; Illumina Inc., San Diego, CA, USA). RNA amplification, microarray hybridization, scanning, as well as data normalization and analysis were performed at the ProfileXpert genome facility (Lyon, France) as previously described.3
Genes significantly modulated were selected with a fold change cutoff of 1.5 and p<0.05 (one-way analysis of variance [ANOVA]). A Venn diagram was used to visualize relationships between the gene lists thus created.
Histological and immunohistological analysis
Harvested SEs were immediately fixed in neutral buffered formalin 4% (Diapath, Martinengo, Italy) for 24 h, embedded in paraffin (for corneodesmosin, cFos, and LCE6 expression analysis) or in OCT compound (for LCE1A expression analysis), and frozen at −20°C. Preparation of SE section and analysis of protein expression by immunohistochemistry were performed exactly as described previously4 using the primary antibodies cFos monoclonal antibody (sc-8047; Santa Cruz, Dallas, TX, USA) and corneodesmosin polyclonal antibody (13184-1-AP; Proteintech, Rosemont, IL, USA). Images were acquired using an Axioskop 2 Plus optical microscope (Zeiss, Oberkochen, Germany).4 Six representative images were obtained for each condition.
For immunofluorescence, 5 μm air-dried cryosections were incubated with primary antibodies LCE1A polyclonal antibody (PA5-24375; Thermo Fisher Scientific, Waltham, MA, USA) or LCE6 polyclonal antibody (HPA046376; Sigma, St Louis, MO, USA). Revelation by secondary antibody and nuclear counterstaining was performed exactly as described earlier.4 Images of immunofluorescence were visualized using an Observer Z1 optical microscope (Zeiss).4
Image analysis
Image analysis was performed using ImageJ software. The parameters of interest were epidermal thickness for HPS-stained sections, cFos nucleus positive cells, and the surface area of corneodesmosin, LCE1A, and LCE6 immunostaining. Stratum corneum thickness was quantified using Visilog software (FEI, Hillboro, OR, USA). The algorithm followed two steps: segmentation and measurement. The epidermis was removed using a semiautomatic method. A K-mean algorithm was used to segment the image into regions with same characteristics. The region corresponding to the stratum corneum was preserved. The stratum corneum cavities were filled and an average thickness was computed. The quantification of cFos-positive epidermal cells was performed exactly as previously described for other nuclear proteins.4 Data were normalized by basement membrane length and results were expressed as pixel2 per micrometer.
Apoptosis TUNEL assay
To evaluate apoptotic cells, a Click-iT Plus TUNEL Assay was used for in situ apoptosis detection with Alexa Fluor 488 kit (Thermo Fisher Scientific). Paraffin-embedded formalin-fixed samples were cut into 5 μm sections. After deparaffinization and rehydration, TUNEL staining was performed. Nuclear counterstaining using Hoechst 33342 (Thermo Fisher Scientific) was carried out routinely. Negative controls were provided for each section under evaluation by omitting the terminal deoxynucleotidyl transferase enzyme. Positive controls were established by irradiating a skin section with UVB (250 mJ/cm2).
Statistical analysis
For all data, statistical significance was assessed on one-way ANOVA or Student’s t-test. Significant differences are indicated by asterisks, as follows: *p<0.05, **p<0.01, and ***p<0.001.
Results
To analyze the response of human skin to climatic change, a human SE model composed of a porous collagen–glycosaminoglycan-chitosan polymer was used populated by normal fibroblasts, on top of which normal human epidermal keratinocytes were seeded. After 40 days culture, full-thickness reconstructed skin was obtained, which was not only morphologically similar to normal human skin but also expressed all specific dermal and epidermal markers.1 SEs were then exposed to either 40°C and 80% RH (treatment 1: hot and humid) or 10°C and 40 RH (treatment 2: cold and dry) for 30 or 15 min, respectively, at days 40, 42, and 44. They were finally harvested at day 45 and used for RNA extraction and histological examination.
Total RNA was obtained from all SEs and used for transcriptome analysis on Illumina BeadArray, which allows simultaneous profiling of >47,000 transcripts, including coding and noncoding sequences. Transcripts differentially modulated (compared with control conditions) after treatment 1 or 2 were thus identified (fold change >1.5, p<0.05): that is, 349 and 469 transcripts after treatments 1 and 2, respectively (Table S1), with a 55%–45% ratio of downregulated versus upregulated transcripts (Figure 1A). Only 87 transcripts (71 repressed and 16 induced) were modulated after both treatments (Figure 1B). Six transcripts were then selected for validation by qPCR: 3 induced only after treatment 2 (belonging to the late cornified envelope [LCE] gene family of epidermal differentiation markers: LCE1A, LCE1DE, LCE5A); 1 induced by both treatments (FOS, encoding a key regulator of epidermal differentiation); 1 repressed by both treatments (COL4A1, encoding the alpha-1 chain of collagen IV); and 1 repressed only after treatment 2 (LUM, encoding the lumican proteoglycan), these 2 last genes encoding proteins of the dermal extracellular matrix. The qPCR data perfectly matched the microarray data, except for COL4A1, which qPCR did not find significantly repressed by treatment 2 (Figure 1C).
To decipher the cellular functions and pathways potentially impacted by the climatic changes, the Database for Annotation, Vizualisation and Integrated Discovery (DAVID) functional annotation tool was used.5 This analysis revealed (Figure 1D) strong enrichment in genes involved in extracellular matrix maintenance or synthesis for transcripts repressed by treatment 1 (n=22, p=1.1E−12) and, to a lesser extent, for transcripts repressed by treatment 2 (n=15, p=8.4E−5). This result was confirmed by annotation of the 87 transcripts modulated after both treatments, which identified extracellular matrix and collagen genes as being significantly enriched (Figure 1D). The list of genes repressed after treatment 1 and/or 2 and belonging to the extracellular matrix function is given in Table 1. Several genes encoding collagen chains (COL1A2, COL3A1, COL4A1, COL11A1) or various proteins from the dermal extracellular matrix (glypican, fibromodulin, spondin) were repressed after both treatments. Treatment 1 also induced strong repression of genes encoding not only other collagen chains (COL6A3, COL15A1, and COL18A1) but also laminins (LAMA2, LAMB1, and LAMC3) and proteoglycans (tenascin and versican). However, the strongest functional enrichment was found in the list of transcripts induced by treatment 2 for genes involved in keratinocyte differentiation (n=12, p=2.2E−13), especially genes encoding small proline-rich proteins (n=12, p=1.5E−18). Noteworthy, these 12 transcripts belong to the same family of LCE transcripts that were specifically and strongly upregulated after treatment 2 (Table 2). Collectively, these results revealed profound changes in the extracellular matrix and epidermal differentiation after treatment 2 (cold and dry), while treatment 1 also impacted the extracellular matrix, probably more strongly.
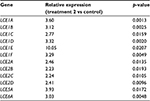  | Table 2 List of LCE transcripts induced after treatment 2 Abbreviation: LCE, late cornified envelope. |
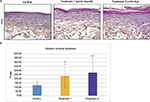
To explore the hypotheses formulated after the transcriptome analysis, the morphology of SEs exposed to the climatic changes described earlier was examined. Both treatments induced global disorganization of equivalent dermis, with apparent reduction in matrix fiber density (Figure 2A). The appearance of the basal epidermal layer was also modified, especially after the cold–dry treatment, with keratinocytes showing heterogeneous morphology, losing their usual sticky columnar organization. The most striking morphological change, however, was the increase in stratum corneum thickness (Figure 2A). Quantification confirmed the visual impression, revealing significant thickening of the stratum corneum after both treatments, especially after the cold–dry one (Figure 2B). Collectively, these observations were in line with the hypothesis based on transcriptome analysis: extracellular matrix remodeling and activation of epidermal differentiation. The latter finding was confirmed by expression study of three epidermal differentiation markers (corneodesmosin, LCE1A, and LCE6A) in control and treated SEs, which showed significant induction of these proteins in cold–dry exposed tissue (Figure 3). The senescence marker cFos was also overexpressed under cold–dry treatment, suggesting that this treatment may induce a complex stress-response phenotype without apoptotic cell death, as shown by negative TUNEL labeling of climatic change-exposed SEs (Figure S1).
Discussion
The present study comprised a gene expression analysis of SE models exposed to repeated short temperature and humidity changes mimicking climatic conditions corresponding to winter and summer outdoor conditions. The aim was to explore how skin, and especially the epidermis, deals with these changes when individuals regularly move from indoors to outdoors. Little is known about the effect of chronic climate conditions, and especially extreme ones, on skin physiology and molecular programs. A few studies examined the effect of RH, temperature, and other parameters, such as altitude or pollution, on skin properties and barrier function. Most of these studies used bioengineering techniques and were performed in climate-controlled chambers for short intervals. They revealed that low RH promotes a hyperproliferative response and pro-inflammatory cytokine release.6,7 No proliferation or cytokine gene response was observed in dry conditions; however, RH was not the only parameter that was modulated in this study, and it is possible that the combination of low temperature and low RH might induce a different skin response. Climate temperature influences skin temperature directly or indirectly. This has been demonstrated by several studies with human subjects exposed to variable ambient temperature: results showed that temperature had stronger effects than RH on skin barrier function8 and especially that low temperature induced transient modification of barrier function.9 More precisely, when skin temperature is decreased, transepidermal water loss is reduced, which may reflect implementation of defense mechanisms against dehydration. The induction of genes encoding proteins from tight junctions such as Claudin (CLDN9 was strongly expressed after treatment 2; see Table S2) could be a sign of this cellular strategy to limit water loss. In SEs exposed to cold–dry conditions, there was a strong increase in the level of genes involved in epidermal differentiation, which correlated with increased stratum corneum thickness. This activation of the differentiation process should strengthen the barrier function and contribute to reducing water loss.
Nevertheless, activation of epidermal differentiation appears to be a general response of skin to various environmental stresses: it has been observed after ionizing irradiation,10,11 UVB,12,13 and pulsed electric field.14 In these conditions of potentially DNA-damaging stress, it has been suggested that induction of terminal differentiation, finally leading to cell death, is a way to avoid the accumulation of mutations in progenitor cells. The present transcriptome study found no induction of genes linked to the DNA-damaging response (such as DNA repair genes or members of the p53 pathway), which may indicate that the daily climatic changes applied did not act as a genotoxic stress.
The most striking molecular effect was the induction of numerous genes encoding LCE proteins after cold–dry treatment (Table 2). The LCE gene family consists of 18 members subdivided into six groups, LCE1–LCE6, based on amino acid sequence and gene expression pattern.15 These genes are located within the epidermal differentiation complex on chromosome 1q21, neighboring various skin differentiation genes such as small proline-rich region proteins and S100 family proteins.16 Little is known about the function of LCE proteins in the epidermis. Based on expression data, it has been suggested that LCE protein members are likely to have distinct functions in the epidermis that extend beyond putative skin barrier formation.17 The present study found strong induction of the entire LCE1 (1A–1F) and LCE2 (2A–2D) groups of genes after the cold–dry treatment and confirmed LCE1A protein induction in SEs after both cold–dry and warm–humid treatments. The LCE1 and LCE2 genes are the most responsive to calcium treatment,16 indicating a strong link to epidermal differentiation. A strong induction of LCE5A and LCE6A genes and LCE6 proteins was also observed in cold–dry treated skin; both of these LCEs were reported to be induced after epidermal barrier disruption and repair.18 Collectively, these data may indicate that LCE proteins participate in the terminal differentiation response of the treated skin. They also suggest a probable mechanism of cis-gene regulation that may involve common enhancer or promoter sequences able to respond specifically to temperature or humidity changes. Further experiments will be necessary to identify these sequences and clarify the molecular mechanisms allowing correlated induction of 12 members of the LCE gene family.
For the extracellular dermal matrix, results showed a decrease in collagen gene expression. This decrease could contribute to chronological aging effects, as skin collagen levels decrease with aging.19 Precisely, type IV collagen gene repression was observed after climatic stress; it is one of the major components of the dermo–epidermal junction involved in cohesion between the epidermis and dermis, and is diminished in chronological aging and photoaging.20 Climatic stress may thus be implicated in aging effects. Also, various proteoglycans showed decreased gene expression; their specific functions are multiple, but their chemical structures give them an extra function in retention of growth factors and water, whereby they participate in skin hydration. In particular, it was shown that lumican gene expression was decreased by climatic stress. This observation was previously made with other forms of stress such as UV irradiation21 and aging.22 In this model, decreased gene expression of numerous proteoglycans could be linked to skin dehydration.
The present study for the first time provides an exhaustive view of the molecular response elicited by repeated climatic change on a 3D SE model, closer to human skin than monolayered culture. Although SEs lack immune response and important skin structures such as sweat and sebaceous glands, potentially involved in tissue response to climatic change, morphological and biochemical changes were observed, especially in the epidermis, which might be transposable to living human skin. The present study revealed a gene expression pattern allowing specific adaptation of the tissue to these environmental changes. It especially highlighted changes in the epidermal differentiation program and in dermal matrix composition: that is, the two components of the skin that constantly interact to maintain tissue homeostasis and hydration. The results provide insight into optimal skin care for individuals living and working under varying climates.
Acknowledgments
The authors would like to thank Iain McGill for English editing. The support provided by the ProfileXpert genome facility (www.profilexpert.fr) for microarray analysis is acknowledged. This work was supported by Clarins.
Disclosure
An abstract of this paper was presented at the 46th annual ESDR meeting in Munich (September 2016) as a poster presentation with interim findings. The authors report no conflicts of interest in this work.
References
Black AF, Bouez C, Perrier E, Schlotmann K, Chapuis F, Damour O. Optimization and characterization of an engineered human skin equivalent. Tissue Eng. 2005;11(5–6):723–733. | ||
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. | ||
Gras B, Jacqueroud L, Wierinckx A, et al. Snail family members unequally trigger EMT and thereby differ in their ability to promote the neoplastic transformation of mammary epithelial cells. PLoS One. 2014;9(3):e92254. | ||
Dos Santos M, Metral E, Boher A, Rousselle P, Thepot A, Damour O. In vitro 3-D model based on extending time of culture for studying chronological epidermis aging. Matrix Biol. 2015;47:85–97. | ||
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. | ||
Denda M, Sato J, Tsuchiya T, Elias PM, Feingold KR. Low humidity stimulates epidermal DNA synthesis and amplifies the hyperproliferative response to barrier disruption: implication for seasonal exacerbations of inflammatory dermatoses. J Invest Dermatol. 1998;111(5):873–878. | ||
Denda M. Influence of dry environment on epidermal function. J Dermatol Sci. 2000;24(Suppl 1):S22–S28. | ||
Cravello B, Ferri A. Relationships between skin properties and environmental parameters. Skin Res Technol. 2008;14(2):180–186. | ||
Thorleifsson A, Wulf HC. Emollients and the response of facial skin to a cold environment. Br J Dermatol. 2003;148(6):1149–1152. | ||
Lamartine J, Franco N, Le Minter P, et al. Activation of an energy providing response in human keratinocytes after gamma irradiation. J Cell Biochem. 2005;95(3):620–631. | ||
Acheva A, Ghita M, Patel G, Prise KM, Schettino G. Mechanisms of DNA damage response to targeted irradiation in organotypic 3D skin cultures. PLoS One. 2014;9(2):e86092. | ||
Hong SP, Kim MJ, Jung MY, et al. Biopositive effects of low-dose UVB on epidermis: coordinate upregulation of antimicrobial peptides and permeability barrier reinforcement. J Invest Dermatol. 2008;128(12):2880–2887. | ||
Lee JH, An HT, Chung JH, Kim KH, Eun HC, Cho KH. Acute effects of UVB radiation on the proliferation and differentiation of keratinocytes. Photodermatol Photoimmunol Photomed. 2002;18(5):253–261. | ||
Arai KY, Nakamura Y, Hachiya Y, et al. Pulsed electric current induces the differentiation of human keratinocytes. Mol Cell Biochem. 2013;379(1–2):235–241. | ||
Bergboer JG, Tjabringa GS, Kamsteeg M, et al. Psoriasis risk genes of the late cornified envelope-3 group are distinctly expressed compared with genes of other LCE groups. Am J Pathol. 2011;178(4):1470–1477. | ||
Jackson B, Tilli CM, Hardman MJ, et al. Late cornified envelope family in differentiating epithelia – response to calcium and ultraviolet irradiation. J Invest Dermatol. 2005;124(5):1062–1070. | ||
Niehues H, van Vlijmen-Willems IM, Bergboer JG, et al. Late cornified envelope (LCE) proteins: distinct expression patterns of LCE2 and LCE3 members suggest nonredundant roles in human epidermis and other epithelia. Br J Dermatol. 2016;174(4):795-802. | ||
de Koning HD, van den Bogaard EH, Bergboer JG, et al. Expression profile of cornified envelope structural proteins and keratinocyte differentiation-regulating proteins during skin barrier repair. Br J Dermatol. 2012;166(6):1245–1254. | ||
Callaghan TM, Wilhelm K. A review of ageing and an examination of clinical methods in the assessment of ageing skin. Parvt I: Cellular and molecular perspectives of skin ageing. Int J Cosmet Sci. 2008;30(5):313–322. | ||
Feru J, Delobbe E, Ramont L, et al. Aging decreases collagen IV expression in vivo in the dermo-epidermal junction and in vitro in dermal fibroblasts: possible involvement of TGF-beta1. Eur J Dermatol. 2016;26(4):350–360. | ||
Shin JE, Oh JH, Kim YK, Jung JY, Chung JH. Transcriptional regulation of proteoglycans and glycosaminoglycan chain-synthesizing glycosyltransferases by UV irradiation in cultured human dermal fibroblasts. J Korean Med Sci. 2011;26(3):417–424. | ||
Vuillermoz B, Wegrowski Y, Contet-Audonneau JL, Danoux L, Pauly G, Maquart FX. Influence of aging on glycosaminoglycans and small leucine-rich proteoglycans production by skin fibroblasts. Mol Cell Biochem. 2005;277(1–2):63–72. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.