Back to Journals » Cancer Management and Research » Volume 12
Regulation of RUNX3 Expression by DNA Methylation in Prostate Cancer
Authors Yang X, Wang S, Reheman A
Received 10 February 2020
Accepted for publication 7 July 2020
Published 27 July 2020 Volume 2020:12 Pages 6411—6420
DOI https://doi.org/10.2147/CMAR.S249066
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Xin Yang, 1,* Shumei Wang, 2,* Alimu Reheman 1
1Department of Urology, The Second Affiliated Hospital of Xinjiang Medical University, Urumqi, Xinjiang Uygur Autonomous Region, People’s Republic of China; 2Urumqi Blood Center, Urumqi, Xinjiang Uygur Autonomous Region, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Alimu Reheman Email [email protected]
Purpose: To investigate the role of DNA methylation in the regulation of Runt-related transcription factor 3 (RUNX3) and the effect of such mechanism on the proliferation of prostate cancer (PCa) cells.
Materials and Methods: The methylation of the RUNX3 in the promoter region in PCa cells was detected by bisulfite-sequencing PCR (BSP). Following treatment of the PCa cells with DNA methylation transferase inhibitor 5-AZA-2ʹ-deoxycytidine (AZA), the effect on methylation level and expression of RUNX3 were analyzed by qRT-PCR, Western blot, and BSP assays. Furthermore, we investigated the effect of the demethylated RUNX3 on proliferation, cell cycle and apoptosis of PCa cells using CCK-8 and flow cytometry assays. Using the DNA methylation transferase (DNMT3b) knockout or overexpression models, the relationship between DNMT3b and RUNX3 methylation was further assessed by qRT-PCR, Western blot and methylation-specific PCR (MSP).
Results: The results indicated that the methylation level of RUNX3 in PCa cell lines was significantly higher than that of normal prostate epithelial (RWPE-1) cells. Furthermore, treatment with AZA not only promoted the demethylation of RUNX3 but also restored the mRNA and protein expression of RUNX3, and the reactivation of expression of the later exhibited its anti-tumor effects through regulation of the cycle progression in PCa cells. Moreover, DNMT3b could regulate the expression level of RUNX3 by altering the DNA methylation of the RUNX3 in PCa cells.
Conclusion: RUNX3 is hypermethylated in a panel of PCa cell lines; inhibition of DNA methylation of RUNX3 could restore its gene expression, which could promote its anticancer effect. Thus, RUNX3 may serve as a novel putative molecular target gene for PCa therapy.
Keywords: prostate cancer, RUNX3, DNA methylation, AZA, DNMT3b
Background
Globally, prostate cancer (PCa) represents the second most frequent malignancy among men. The recent epidemiological survey has recorded an increasing trend in the annual incidence (12.6%) and mortality (5.5%) rates in China during the past decade. With population aging and dietary transitions, the disease burden of PCa will continue to increase in China.1,2 Despite significant research efforts, the pathogenesis of PCa remains poorly understood. Therefore, active investigation on the relevant mechanism of the occurrence and development of PCa still presents significant potential in reducing the PCa-associated morbidity and mortality.
Runt-related transcription factor 3 (RUNX3) gene belongs to the RUNX family of a developmental regulator that orchestrates diverse developmental and cellular processes, including proliferation, differentiation, apoptosis, and cell lineage specification.3 In humans, the gene encoding RUNX3 is located on chromosome 1p36.1. A 4.2 kb cytosine-phosphate-guanine (CpG) island exists near the 5ʹ end promoter of the RUNX3 with a high GC content of 64%. For this reason, genes with this structure are highly susceptible to be regulated by methylation.4,5 DNA methylation is an epigenetic modification that regulates the gene expression and provides a mechanism for conveying and preserving epigenetic information through DNA replication and cell division. Accumulating studies have indicated that aberrant changes in DNA methylation are amongst the most common molecular alterations associated with tumorigenesis and hypermethylation of the promoter region of various cancer suppressor genes is recognized as one of the most frequent mechanisms for loss of gene function.6,7 Moreover, recent advances in epigenetics have provided an improved understanding of molecular mechanisms underlying carcinogenesis. DNA methylation alterations are highly prevalent in prostate cancer, making them a sustained focus of research, with growing evidence supporting their role in progression.8 Furthermore, the inactivation of the RUNX3 is also increasingly implicated in the tumorigenesis of various tumors,9 including gastric cancer,10 liver cancer,11 breast cancer,12 colon cancer13 and melanoma.14 The RUNX3 has two promoters, P1 and P2, and 6 exons. Of these two, the P2 promoter is rich in CpG islands; besides, RUNX3 expression is also predominantly regulated by P2 promoter. Evidently, studies have reported that hypermethylation of the CpG-island adjacent to P2 promoter is associated with leukemia and cancer could lead to DNA-methylated silencing of RUNX3 expression.9,14 In PCa, studies have revealed that RUNX3 was frequently methylated at a frequency of 32.4% in PCa tumor tissues and 14.3% in the prostate intraepithelial neoplasia (PIN) while RUNX3 methylation was not detected in normal or benign prostate tissues, suggesting that RUNX3 methylation was closely associated with prostate tumorigenesis.15,16 However, the role and molecular mechanism underlying aberrant methylation of the RUNX3 in prostate tumorigenesis is rarely reported. Therefore, in this study, we investigated the molecular mechanism by which RUNX3 expression is affected by gene promoter methylation in PCa cells. Furthermore, this study also provided new insights into the diagnostic strategies for PCa and the epigenetic-based therapeutic approaches for PCa based on RUNX3.
Materials and Methods
Cell Culture and Transfection
Human normal prostate epithelial cell line RWPE-1 and prostate cancer cell lines PC3, DU145, 22Rv1 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). RWPE-1 cells were cultured in K-SFM special medium (Gbico, Grand Island, NY, USA) supplemented with 2% Bovine Pituitary Extract (BPE) (Invitrogen, CA, USA), 5 mM recombinant epidermal growth factor (rEGF) (Sigma-Aldrich), and 1% penicillin/streptomycin (Invitrogen). While PC3, DU145, and 22Rv1 cells were cultured in RPMI-1640 (Gibco) medium supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Sigma). All the cells were maintained at 37°C in a humidified atmosphere of 5% CO2. For PC3 and DU145 cells in the logarithmic phase were digested with 0.25% trypsin (Gibco), counted and cells were inoculated into a 6-well plate at a cell density of 2 × 105 cells/well, and incubated overnight at 37°C. Using Lipofectamine 3000 transfection reagent (Invitrogen), the cells were transfected with 50 ng of pcDNA3.1-DMNT3b (DMNT3b), Vector, and 100 nM of si-DMNT3b, si-RUNX3, and its negative control (si-NC), respectively. The concentration was determined based on the dose–response experiment. All the plasmids and siRNAs were purchased from Guangzhou Ruibo Biological Co., China. The culture medium was replaced with RPMI-1640 containing 10% FBS following incubation of the transfected cells for 6 h. Then, the cells were cultured at 37°C humidified atmosphere of 5% CO2. The cells were used for subsequent experiments after 48 h of transfection.
Bisulfite Detection and Sequencing (BSP)
PC3 and DU145 cells in the logarithmic growth phase were harvested and genomic DNA was extracted using the Cell DNA extraction kit (Beijing TIANGEN Biological Co., China) following the manufacturer’s instruction. The purity and concentration of the DNA were measured with a UV spectrophotometer. DNA from all cell lines was subjected to bisulfite conversion using the methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturers’ protocols. Then, the bisulfite-treated genomic DNA was amplified with PCR. The amplified PCR was verified using Gel electrophoresis. Subsequently, the PCR product was purified and cloned into a pMD19-T vector, and 10 of the ligated products were transformed into DH5 competent cells. Following transformation, the cells were plated onto X-gal/IPTG- and Amp-coated plates and incubated overnight at 37°C. The three white spot colonies were selected through blue/white colony screening and further inoculated in 2.5 mL of LB liquid medium and incubated overnight at 37°C. Plasmids were extracted using the plasmid extraction kits (TIANGEN) and sequenced in Shanghai Sangon Co., Ltd. The sequences were then analyzed with the UltraEdit Professional Text/Hex Editor.
CCK-8 Proliferation Experiment
Cell viability was measured using the Cell Counting Kit-8 (CCK-8) assay kit (Sigma). Briefly, PC3 and DU145 cells in the logarithmic growth phase were harvested and transfected with si-RUNX3 and seeded into 96-well plates at a density of 4 × 103 cells/well. Following incubation, 20 μM AZA (Sigma) solution was added to each well for co-culture at 37°C in a humidified atmosphere of 5% CO2 for 24 h. The blank control group was treated with an equal volume of RPMI-1640 (Gibco) culture medium; 10 μL of CCK-8 reagent (Sigma) was added to each well at 0 h, 24 h, 48 h, 72 h, and 96 h and incubated for 2 h in a humidified atmosphere incubator. The absorbance (OD 450) of each well was detected at 450 nm using an automated microplate reader (VersaMax Microplate Reader; Sunnyvale, CA). Average of 5 duplicate wells was used for each time point to draw the growth curve of the cells.
Clone Formation Assay
PC3 and DU145 cells were transfected with si-RUNX3 as described above. After 24 h, transfected cells were seeded into a 6-well plate at a density of 1 × 103 cells/well and incubated for 10 days under the conditions as described in section 1.2.1. Subsequently, the cells were treated with 20 μM of AZA (Sigma) solution on the 4th day of cell culture, and the control group was treated with the equal volume of DMSO (Sigma). The AZA or DMSO was replaced every 24 h and the culture medium was changed every third day. At the indicated times, cells were fixed with 3.7% paraformaldehyde and stained with 0.05% crystal violet for 20 min and dried naturally. The cells were photographed and counted under a microscope, and the number of cell clones was calculated.
Cell Apoptosis Experiment
Apoptotic cell death was measured using a fluorescein isothiocyanate (FITC)-conjugated Annexin V/propidium iodide (PI) assay (BD Bioscience, CA, USA). In brief, AZA-treated PC3 and DU145 were transfected with si-RUNX3, and the cells were washed twice with ice-cooled PBS at 4°C. The cell pellet was collected by centrifugation at 300 r/min for 5 min and re-suspended in 195 μL of Annexin V-FITC binding buffer, and the cell density was adjusted to 7 × 105 cells/mL. Subsequently, the cells were stained with Annexin V-FITC (10 mg/mL) and PI (50 mg/mL) staining solution. Following incubation, the cells were washed with PBS twice and collected at a concentration of 1×106 cells/mL. These cells were incubated for 30 min at room temperature (RT) in the dark and then analyzed with an AccuriTM C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Cells stained with only annexin V were evaluated as being in early apoptosis; cells stained with both annexin V and PI were evaluated as being in late apoptosis or in a necrotic stage. The experiment was repeated 3 times separately.
Cell Cycle Experiment
The cell cycle profile was determined using PI staining. Briefly, AZA-treated PC3 and DU145 cells were transfected with si-RUNX3, washed with ice-cooled PBS, centrifuged at 300 r/min for 5 min and resuspended in PBS. Then, cells were fixed with 70% alcohol in PBS at 4°C. After washing in PBS, cells were treated with PI/RNase staining solution RNase (w/v) at 4°C for 30 min in the dark. The cell cycle distribution was analyzed with flow cytometry (BD Biosciences). The percentages of cells in the G0/G1 phase, S phase, and G2/M phase were determined. The experiment was independently repeated for 3 times.
Methylation-Specific PCR (MSP)
Previous studies have demonstrated that the DNA-methylation of CpG island in the P2 promoter region is the major pathogenic mechanism of RUNX3 silencing.5,17 P2 is located around the exon 2 and P1 is located at the beginning of the exon 6 (Figure 1A). The CpG islands in the P2 promoter region of the RUNX3 were predicted using CpG island prediction software (www.urogene.org). The genomic DNA modified by bisulfite was amplified by the methylation kit (Zymo) according to the manufacturer’s instructions, and the purity and concentration were determined using an ultraviolet spectrophotometer. The DNA was modified with bisulfite using the EpiTect Bisulfite kit according to the manufacturer’s instructions, and the methylated and unmethylated RUNX3 were amplified using the following primers: The sequences of the MSP primers were: RUNX3 methylation primer upstream: upstream 5ʹ-GGTTGTTTGTTTTTTTTGGTTCG-3ʹ, downstream 5ʹ-ATCCT AAAACTACCCAAAATCGTA-3ʹ; unmethylated primers: RUNX3 upstream 5ʹ-GG GATGAGGATTAGGATTTT-3ʹ, downstream 5ʹ-CAAACAAAACACAATAAAAACAAACA-3ʹ. The PCR cycling conditions were: pre-denaturation at 94°C (3 min), followed by 35 cycles of denaturation at 94°C (30s), demethylation at 53°C (30s) and extension at 72°C (90s), and final extension at 94°C for 5 min. Generation of an amplified product with either methylated or unmethylated primers, respectively, indicated presence and absence of methylated sequences in the genome. Generation of amplified products with both primer pairs implied partial methylation. The methylation level of RUNX3 gene was calculated by the ΔΔCt method. The experiment was repeated thrice.
Real-Time Fluorescence Quantitative PCR (qRT-PCR)
Total RNA was extracted using the TRNzol reagent (Invitrogen Life Technologies, CA, USA) following the manufacturer’s instructions. The purity and the concentration of RNA were measured by a spectrophotometer. Total RNA was reverse transcribed into cDNA using the reverse transcription reagent (Invitrogen Life Technologies) according to the manufacturer’s instructions. qRT-PCR was performed to determine gene expression of target genes using SYBR Green Real-time PCR Kit (Shanghai Solarbio Co., China). The qRT-PCR was performed by initial denaturation at 95 ºC for 10 min, followed by 40 cycles of denaturation at 95 ºC for 7 s, annealing at 60 ºC for 20 s, and annealing at 72°C for 38 s. RT-PCR primers are: RUNX3-F: 5ʹ-TGGCAGGCAATGACGA-3 ‘, RUNX3-R: 5ʹ-TGGTTCGGCAAGGGAC-3ʹ; DMNT3b-F: 5ʹ-TCTGGAAAACCTTCCTGCTG-3 ‘, DMNT3b-R: 5ʹ-CCGGCACATAGGTAAA AGGA-3 ‘; G-AGAAGGCTGGGGCTCATTTG-3ʹ, GAPDH-R:5ʹ-AGGGGCCATCCACAGTCTTC-3 ‘. The 2−ΔΔCT method was used to determine relative gene expression, which was normalized to the amount of GAPDH mRNA. All experiments were performed at least in triplicate for each gene. Data are expressed as the mean ± S.E.M.
Western Blot Experiment
AZA-treated PC3 and DU145 cells were transfected with si-RUNX3. The modified cells were washed with ice-cold PBS and then lysed with RIPA cell lysis buffer supplemented with protease inhibitor cocktail. Protein concentrations were quantified using the BCA protein assay kit (Thermo MA, USA). Proteins were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, MA, USA). Subsequently, the membrane was blocked with 5% skim milk in Tris-buffered saline with Tween-20 (TBST); then PVDF membranes were incubated with primary antibodies including RUNX3 (1:500), Cyclin B1 (1:500), Bax (1:800), Caspase-3 (1:800), Bcl-2 (1:500), p21 (1:500), DNMT3b (1:800), GAPDH (1:1500), respectively, overnight at 4°C (Antibodies to RUNX3, Cyclin B1, Bax, Caspase-3, Bcl-2, p21, DNMT3b and GAPDH were purchased from Abcam, CA, USA). Then, the membrane was washed 3 times with TBST solution for 5 min/time, and membranes were incubated with horseradish enzyme-conjugated secondary antibodies (1:5000) for 1 h at RT. The target bands were visualized using the enhanced chemiluminescence (ECL) reagent (GE Healthcare, NJ, USA) and photographed with Gel Imager. And the gray value analysis was performed using the Image J software. Using GAPDH as the internal reference, the ratio of gray value between the target band and internal reference band was regarded as the relative expression of the target proteins. Experiments for each sample were independently repeated three times.
Statistical Analysis
All data were analyzed using Statistical Package for the Social Sciences (SPSS) 19.0 software and GraphPad Prism 5.0. The data were expressed as mean ± standard deviation (X ± S). The comparison between the two groups was performed by the independent sample t-test. Comparison between multiple groups was performed by one-way analysis of variance (ANOVA) and post hoc Bonferroni correction was applied for multiple comparisons. The significance test level was α=0.05, P <0.05 was considered statistically significant.
Results
DNA Hypermethylation of the RUNX3 Promoter Region in PCa Cells
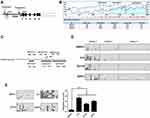
Using CpG island prediction software. The results indicated that this region was rich in CG dinucleotide sequences, which could provide a large number of sites for methylation modification of this gene (Figure 1B). Based on the distribution range of CpG islands in the P2 promoter region of the RUNX3, we precisely designed 3 different pairs of methylation-specific primers, and the three pairs of primers amplify three different regions of P2 as reported in the Figure 1C. Using BSP, we analyzed the DNA methylation status of human normal prostate epithelial cell RWPE-1 and a panel of 3 PCa cell lines PC3, DU145, and 22Rv1. The results indicated that the amplicons of region 1 exhibited high density of CpG sites than those of the two other sets of primers (2 and 3) in four groups of cells, suggesting that this region was the crucial site for methylation in RUNX3 (Figure 1D). Compared with the 7.2% methylation rate of RWPE, the methylation level of PCa cells in the amplicons of primer 1 was significantly increased in the three groups, PC3was 52.1%, DU145 was 33.4%, and 22Rv1 was 40.6% (Figure 1D). The region from −738 to −624 on the P2 promoter of RUNX3 was determined to be the preferred sites for DNA methylation, we next used the Primer 1 to detect the DNA methylation status of RWPE1, PC3, DU145, and 22Rv1 cells. Similarly, the results also indicated that the P2 promoter region of the RUNX3 in the panel of 3 PCa cell lines was significantly hypermethylated compared with normal RWPE1 cells (Figure 1E).
AZA Reverses the Methylation-Mediated Silencing of the RUNX3 and Promoted the RUNX3 Expression in PCa Cells
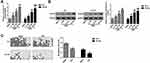
The above experiments indicated that the RUNX3 in the PCa cell lines PC3, DU145, and 22Rv1 presented a high level of methylation, and therefore, we chose PC3 with relatively higher methylation levels and DU145 with lower methylation levels for subsequent related experiments. The PCA and DU145 cells were continuously treated with different concentrations (5 μM, 10 μM, and 20 μM) of DNA methylation transferase inhibitor AZA for 96 h, while the control group was treated with DMSO. The effect of AZA on the gene and protein expression levels of RUNX3 was determined using qRT-PCR and Western blot assays. The results demonstrated that AZA could restore the mRNA and protein expression of RUNX3 in PC3 and DU145 cells in a concentration-dependent manner (Figure 2A, and B). Based on these findings, we further used BSP to detect the DNA methylation level of RUNX3 in PC3 and DU145 cells treated with 20 μM AZA for 96 h. It was found that AZA could significantly reverse the aberrant alterations in the DNA methylation of RUNX3 in PCa cells (Figure 2C). Taken together, the results indicated that the methylation-mediated silencing of the P2 promoter region of RUNX3 significantly down-regulated the expression level of this gene in PCa cells; however, treatment of PCa cells with AZA resulted in a significant increase in RUNX3 mRNA and protein expression levels, suggesting that promoter demethylation by AZA can restore RUNX3 expression.
AZA Inhibited the Proliferation and Induced Apoptosis of PCa Cells by Promoting the Expression of RUNX3
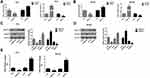
In order to further investigate whether treatment with AZA could affect the biological behavior of PCa cells through promoter demethylation of RUNX3 and elucidate the related mechanism, we first silenced RUNX3 expression in PC3 and DU145 cells using siRNA. The transfection efficiency was verified by qRT-PCR and Western blot assays. The results indicated that compared with the cells transfected with si-NC, the mRNA and protein expression of RUNX3 in PC3 and DU145 cells transfected with si-RUNX3 was significantly down-regulated (Figure 3A, and B). Using CCK-8 cell proliferation, clone formation, cell cycle, and apoptosis assays, we analyzed the effects of treatment with silencing RUNX3 and/or AZA on the proliferation, cell cycle, and apoptosis of PCa cells. The results revealed the DNA methylation transferase inhibitor AZA could inhibit the proliferation of PCa cells, increase the ratio of cells in the G0/G1 phase, reduce the ratio of cells in the S phase, and promote cell apoptosis by restoring the expression of RUNX3 in PC3 and DU145 cells (Figure 3C–F). At the same time, the expression of cell cycle-related protein, cyclin D1 and p21 and apoptosis-related protein, Bax, Bcl-2, and caspase-3 in cells were determined by Western blot assay. The results demonstrated that AZA could promote the expression of proteins p21, Bax, caspase-3, and reduce the protein expression of Bcl-2 and cyclinD1 (Figure 3G). Together, these results indicated that the treatment of PCa cells with AZA may inhibit the proliferation of PCa cells by up-regulating the expression of RUNX3 through inhibition of the cell cycle progression and induction of the intrinsic mechanism of apoptosis.
DNMT3b is a Crucial Factor Affecting the Hypermethylation of RUNX3 in PCa Cells
The above results indicated that AZA could promote the expression of RUNX3 through the demethylation of the RUNX3 in PCa cells and inhibit the cell proliferation and promote the apoptosis, thus, AZA played a role of a demethylating agent by regulating the expression of DNA methylation transferase. Previous studies have found a significantly higher expression of DNMT3b in PCa in comparison with normal tissues.20 Therefore, we speculated that DNMT3b functions as a critical factor affecting the hypermethylation of the RUNX3 in PCa cells. To verify this hypothesis, we conducted qRT-PCR and Western blot assays to measure the expression of RUNX3 in PC3 and DU145 cells following transfection with siRNA that targeted the DNMT3b gene and with the overexpression plasmid. The results indicated that the mRNA and protein expression of RUNX3 in the cells were significantly upregulated following DNMT3b knock out, while the mRNA and protein expression of RUNX3 in the cells were significantly down-regulated after the overexpression of DNMT3b (Figure 4A–D). To further verify the molecular mechanism that DNMT3b affected the RUNX3 expression through DNA modification, we performed MSP to detect the DNA methylation level of the P2 promoter region of the RUNX3 after DNMT3b knock out and overexpression. The results demonstrated that the hypermethylation level of RUNX3 in PC3 and DU145 cells was significantly suppressed following DNMT3b knock out, while the overexpression of DNMT3b could significantly promote the methylation level of the gene in the two cells (Figure 4E). Collectively, these findings indicated that DNMT3b could regulate the expression level of the RUNX3 by altering the DNA methylation of the RUNX3 in PCa cells.
Discussion
PCa is the most prevalent malignancy of the male genitourinary system, accounting for high morbidity and mortality, worldwide. Despite significant scientific breakthroughs and technological advancements, the statistics of PCa continue to increase compared to the preceding years as the pathogenesis of PCa remains an elusive concept.18
DNA methylation has long been recognized as one of the hallmarks in cancer and represents an early event in carcinogenesis. Accumulating studies have confirmed hypermethylation of CpG islands in the promoter region of the tumor suppressor gene as in a variety of primary malignant tumors, suggesting that the methylation may function as an essential mechanism involved in the inactivation of tumor suppressor genes, eventually leading to tumorigenesis.19
RUNX3 is a transcription factor that orchestrates various developmental and cellular processes including proliferation, apoptosis, and even in tumorigenesis. RUNX3 is rapidly receiving increasing prominence as it is frequently inactivated by epigenetic mechanisms and the inactivation of the RUNX3 was closely associated with the occurrence and development of diverse cancer types. Because RUNX3 is frequently hypermethylated, its inactivation is considered an early event in tumorigenesis. Furthermore, the DNA methylation rates of the promoter region of RUNX3 in gastric cancer tissues were significantly higher than that in normal control tissues, and the methylation level of RUNX3 was closely associated with the poor prognosis of patients with gastric cancer.10 The methylation level of RUNX3 is reported to be significantly high in a breast cancer patient with positive estrogen receptor expression. In addition, the proliferation, invasion, and migration of cells were significantly decreased following inhibition of the methylation of RUNX3 in tumor cells.12 In prostate cancer, some studies have identified significant hypermethylation of the RUNX3 in tumor tissues, which was associated with the significant under-expression of the RUNX3.15,16 Considering these pieces of evidence, we speculated that the aberrant methylation of the RUNX3 in PCa may be involved in the inactivation of the RUNX3 and the promotion of tumor progression.
To test this hypothesis, our study indicated that the methylation level of PCa cell line was significantly higher than that of normal prostate epithelial cells and AZA could restore RUNX3 expression in PCa cells in a concentration-dependent manner and reduce the methylation level of the promoter region of the RUNX3.
The down-regulation and the loss of function of the RUNX3 caused by the promoter methylation were associated with the occurrence of various tumors. The decreased expression of RUNX3 in the intestinal tumors weakened the inhibition of the Wnt signaling pathway, which leads to the up-regulation of cells cycle-related proteins Cyclin D1, c-Myc, CDK4, and in turn promoted the cell proliferation.20 Indeed, the increased expression of RUNX3 in gastric cancer cell lines could down-regulate Cyclin D1 expression and up-regulate the expression of Bax and Caspase-3/7/8 to induce apoptosis and eventually inhibit tumor growth and distant metastasis.21 Previous studies have indicated that promoter hypermethylation of the RUNX3 was noticeably related to the poor prognosis of patients with PCa; however, the underlying molecular mechanism is rarely reported. Thus, to explore whether the suppression of the methylation of the RUNX3 could inhibit the progression of PCa, we identified that AZA could promote the expression of Bax, Caspase-3, p21 protein and suppress the protein expression of Bcl-2 and CyclinD1 by restoring the expression of RUNX3, which markedly inhibited the proliferation of PCa cells, increased the ratio of cells in the G0/G1 phase, reduced the ratio of cells in the S phase and induced apoptosis. Overall, these findings suggested that following treatment with AZA treatment, the aberrant methylation of the RUNX3 was gradually reversed to increase the expression level of RUNX3 so as to exert the antitumor effect of the gene, which is consistent with its role in other tumors.
AZA, an inhibitor of DNMTs, can reduce the methyltransferase activity of the DNMTs through covalent binding, resulting in DNA hypomethylation, the activation of tumor suppressor genes in vivo or in vitro and restore the normal anticancer function.22–24 DNMT3b is a member of the DNA methylation transferase family, which catalyzes the methylation of specific CpG sites on the DNA chain and maintains the original pattern of DNA methylation in a cell lineage to ensure the replication fidelity of epigenetic patterns, thus playing a critical role in the methylation process.25 Some pieces of the literature suggested that DNMT3b is an essential molecule that regulates the methylation of the RUNX3 in colon cancer,25,26 and that DNMT3b expression was significantly increased in PCa.16,27 Previous studies have found the significantly higher expression of DNMT3b in PCa in comparison with normal tissues and were correlated with Gleason score, postoperative survival of patients with PCa, and tumor metastasis.28,29 Thus, to investigate the mechanism by which AZA up-regulates the RUNX3 expression in PCa, we constructed siRNA and cDNA clones targeting DNMT3b and transfected PCa cells, and the results demonstrated that the DNMT3b knockout in PCa cells could induce RUNX3 re-expression through a reduction in the methylation level; however, the increased expression of DNMT3b inhibited the expression of DNMT3b by increasing the methylation level of RUNX3. These results indicated that DNMT3b may function as a critical factor that alters the DNA methylation of the RUNX3 in PCa cells to regulate the expression level of RUNX3.
Conclusion
The results of this investigation suggested that the reduced expression of RUNX3 in PCa was significantly associated with hypermethylation of CpG islands at the P2 promoter region of RUNX3. Furthermore, the treatment of PCa cells with AZA could restore the expression of RUNX3, and the reactivation of expression of the later exhibited anti-tumor effects through regulation of the cycle progression of PCa cells. Besides, DNMT3b may function as a critical epigenetic regulator that may alter the DNA methylation of RUNX3 in PCa cells. Taken together, the results of this study further clarified the contribution of epigenetic regulatory mechanism in the occurrence and development of prostate tumorigenesis and provided a theoretical basis and experimental findings for the development of RUNX3 as a novel putative molecular target gene for PCa therapy.
Disclosure
The authors declare no conflicts of interest.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
3. Mangan JK, Speck NA. RUNX1 mutations in clonal myeloid disorders: from conventional cytogenetics to next generation sequencing, a story 40 years in the making. Crit Rev Oncog. 2011;16(1–2):77–91. doi:10.1615/CritRevOncog.v16.i1-2.80
4. Kataoka J, Shiraha H, Horiguchi S, et al. Loss of Runt-related transcription factor 3 induces resistance to 5-fluorouracil and cisplatin in hepatocellular carcinoma. Oncol Rep. 2016;35(5):2576–2582. doi:10.3892/or.2016.4681
5. Bangsow C, Rubins N, Glusman G, et al. The RUNX3 gene – sequence, structure and regulated expression. Gene. 2001;279(2):221–232. doi:10.1016/S0378-1119(01)00760-0
6. Pechalrieu D, Etievant C, Arimondo PB. DNA methyltransferase inhibitors in cancer: from pharmacology to translational studies. Biochem Pharmacol. 2017;129:1–13. doi:10.1016/j.bcp.2016.12.004
7. Li Q, Wei W, Jiang Y, et al. Promoter methylation and expression changes of BRCA1 in cancerous tissues of patients with sporadic breast cancer. Oncol Lett. 2015;9(4):1807–1813. doi:10.3892/ol.2015.2908
8. Fraser M, Sabelnykova VY, Yamaguchi TN, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017;541(7637):359–364. doi:10.1038/nature20788
9. Chuang LSH, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29(18):2605–2615. doi:10.1038/onc.2010.88
10. Wang N, Sui F, Ma J, et al. Site-specific hypermethylation of RUNX3 predicts poor prognosis in gastric cancer. Arch Med Res. 2016;47(4):285–292. doi:10.1016/j.arcmed.2016.07.011
11. El-Bendary M, Nour D, Arafa M, et al. Methylation of tumour suppressor genes RUNX3, RASSF1A and E-Cadherin in HCV-related liver cirrhosis and hepatocellular carcinoma. Br J Biomed Sci. 2019;77:35–40.
12. Liu H, Yan Z, Yin Q, et al. RUNX3 epigenetic inactivation is associated with estrogen receptor positive breast cancer. J Histochem Cytochemi. 2018;66(10):709–721. doi:10.1369/0022155418797315
13. Kim BR, Kang MH, Kim JL, et al. RUNX3 inhibits the metastasis and angiogenesis of colorectal cancer. Oncol Rep. 2016;36(5):2601–2608. doi:10.3892/or.2016.5086
14. Kang S, Wang Z, Li B, et al. Anti-tumor effects of resveratrol on malignant melanoma is associated with promoter demethylation of RUNX3 gene. Die Pharmazie. 2019;74(3):163–167.
15. Kang GH, Lee S, Lee HJ, et al. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202(2):233–240. doi:10.1002/path.1503
16. Kobayashi Y, Absher DM, Gulzar ZG, et al. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21(7):1017–1027. doi:10.1101/gr.119487.110
17. Woolf E, Xiao C, Fainaru O, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Nat Acad Sci. 2003;100(13):7731–7736. doi:10.1073/pnas.1232420100
18. Chen R, Sjoberg DD, Huang Y, et al. Prostate specific antigen and prostate cancer in Chinese men undergoing initial prostate biopsies compared with Western cohorts. J Urol. 2017;197(1):90–96. doi:10.1016/j.juro.2016.08.103
19. Singh RR, Reindl KM, Jansen RJ. Epigenomics of pancreatic cancer: a critical role for epigenome-wide studies. Epigenomes. 2019;3(1):5. doi:10.3390/epigenomes3010005
20. Ito K, Lim AC-B, Salto-Tellez M, et al. RUNX3 attenuates β-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14(3):226–237. doi:10.1016/j.ccr.2008.08.004
21. Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317(24):2532–2542. doi:10.1001/jama.2017.7248
22. Wang S, Liu H, Wang Z, et al. Effects of 5-azacytidine on RUNX3 gene expression and the biological behavior of esophageal carcinoma cells. Mol Med Rep. 2014;9(4):1259–1265. doi:10.3892/mmr.2014.1945
23. Kang H-F, Dai Z-J, Bai H-P, et al. RUNX3 gene promoter demethylation by 5-Aza-CdR induces apoptosis in breast cancer MCF-7 cell line. Onco Targets Ther. 2013;6:411.
24. Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer. 2008;123(1):8–13. doi:10.1002/ijc.23607
25. Nosho K, Shima K, Irahara N, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15(11):3663–3671. doi:10.1158/1078-0432.CCR-08-2383
26. Peng Z, Wei D, Wang L, et al. RUNX3 inhibits the expression of vascular endothelial growth factor and reduces the angiogenesis, growth, and metastasis of human gastric cancer. Clin Cancer Res. 2006;12(21):6386–6394. doi:10.1158/1078-0432.CCR-05-2359
27. Zelic R, Fiano V, Ebot EM, et al. Single-nucleotide polymorphisms in DNMT3B gene and DNMT3B mRNA expression in association with prostate cancer mortality. Prostate Cancer Prostatic Dis. 2019;22(2):284–291. doi:10.1038/s41391-018-0102-5
28. Xue G, Ren Z, Chen Y, et al. A feedback regulation between miR-145 and DNA methyltransferase 3b in prostate cancer cell and their responses to irradiation. Cancer Lett. 2015;361(1):121–127. doi:10.1016/j.canlet.2015.02.046
29. Gillio-Tos A, Fiano V, Zugna D, et al. DNA methyltransferase 3b (DNMT3b), tumor tissue DNA methylation, Gleason score, and prostate cancer mortality: investigating causal relationships. Cancer Causes Control. 2012;23(9):1549–1555. doi:10.1007/s10552-012-0032-9
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.