Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 11 » Issue 1
Reduced sputum expression of interferon-stimulated genes in severe COPD
Authors Hilzendeger C, da Silva J, Henket M, Schleich F, Corhay JL, Kebadze T, Edwards M, Mallia P , Johnston S , Louis R
Received 6 February 2016
Accepted for publication 7 March 2016
Published 30 June 2016 Volume 2016:11(1) Pages 1485—1494
DOI https://doi.org/10.2147/COPD.S105948
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Clarissa Hilzendeger,1 Jane da Silva,2 Monique Henket,1 Florence Schleich,1 Jean Louis Corhay,1 Tatiana Kebadze,3 Michael R Edwards,3 Patrick Mallia,3 Sebastian L Johnston,3 Renaud Louis1
1Department of Respiratory Medicine, Centre Hospitalier Universitaire (CHU) Liege, Groupe Interdisciplinaire de Génoprotéomique Appliquée (GIGA) I3 University of Liege, Belgium; 2Department of Medicine, Post-graduate Program in Health Sciences, University of Southern Santa Catarina, Palhoça-SC, Brazil; 3Airway Disease Division, Airway Disease Infection Section, National Heart and Lung Institute, Imperial College, London, UK
Background: Exacerbations of COPD are frequent and commonly triggered by respiratory tract infections. The purpose of our study was to investigate innate immunity in stable COPD patients.
Methods: Induced sputum was collected from 51 stable consecutive COPD patients recruited from the COPD Clinic of CHU Liege and 35 healthy subjects. Expression of interferons beta (IFN-β) and lambda1 (IL-29), IFN-stimulated genes (ISGs) MxA, OAS, and viperin were measured in total sputum cells by reverse transcription quantitative polymerase chain reaction (RT-qPCR). The presence of Picornaviruses was assessed by RT-PCR, while potential pathogenic microorganisms (PPM) were identified by sputum bacteriology.
Results: Expression of IL-29 was found in 16 of 51 COPD patients (31%) and in nine of 35 healthy subjects (26%), while IFN-β was detected in six of 51 COPD patients (12%) and in two of 35 healthy subjects (6%). ISGs were easily detectable in both groups. In the whole group of COPD patients, OAS expression was decreased (P<0.05), while that of viperin was increased (P<0.01) compared to healthy subjects. No difference was found with respect to MxA. COPD patients from group D of Global Initiative for Chronic Obstructive Lung Disease (GOLD) had reduced expression of all three ISGs (P<0.01 for MxA, P<0.05 for OAS, and P<0.01 for viperin) as compared to those of group B patients. Picornaviruses were detected in eight of 51 (16%) COPD patients vs four of 33 (12%) healthy subjects, while PPM were detected in seven of 39 (18%) COPD patients and associated with raised sputum neutrophil counts. IFN-β expression was raised when either picornavirus or PPM were detected (P=0.06), but no difference was seen regarding IL-29 or ISGs.
Conclusion: ISGs expression was reduced in severe COPD that may favor exacerbation and contribute to disease progress by altering response to infection.
Keywords: COPD, innate immunity, interferon-β, interferon-λ1, interferon-stimulated genes
Background
COPD is a preventable and treatable disease characterized by persistent airflow limitation that is usually progressive.1 COPD is currently the fourth leading cause of morbidity and mortality worldwide and its prevalence is projected to increase in the coming decades because of continuous exposure to COPD risk factors (eg, tobacco smoke and smoke from biomass fuels) and population aging.2
Episodes of respiratory symptom worsening frequently occur in COPD, and these exacerbations are commonly triggered by respiratory tract viral or bacterial infections. Viruses have been detected in 47%–56% of the patients in exacerbation and approximately half of them are rhinoviruses,3 while bacteria are assumed to be the cause of 50% of exacerbations.4 In addition, coinfection with viruses and bacteria account for ~26% of hospitalized COPD exacerbations.5,6
Airway and lung inflammation is central in COPD,7 but there are contradictory data regarding the innate immunity in COPD. One study using primary bronchial epithelial cells infected in vitro with rhinovirus-1B showed that COPD cells had higher expression of interleukin (IL)-6 and interferons (IFNs)-β and λ1 in comparison to healthy controls.8 In addition, IL-8 was found to be increased in sputum from COPD patients when compared to healthy subjects.9–12 On the other hand, reduced IL-8 production was described in COPD-diseased human bronchial/tracheal epithelial cells treated with cigarette smoke condensate,13 as well as in alveolar macrophages from COPD patients incubated with respiratory pathogens.14
The study of Mallia et al with experimental rhinovirus infection in healthy subjects and COPD patients showed that COPD subjects had increased symptoms, airway obstruction, and inflammation with raised sputum neutrophils and proinflammatory cytokine IL-8 production as compared to healthy subjects.15 They also showed that bronchoalveolar lavage cells from COPD patients produced less IFN-β in response to viral infection in vitro. Interferons and their stimulated genes play a major role in defense against infectious agents.16,17 Much less is known regarding mechanisms of virus-induced exacerbations in COPD compared to asthma,18 and there are limited data on the expression of IFNs and their stimulated genes in stable COPD.
The purpose of our study was to investigate the innate antiviral immunity in stable COPD patients suffering from variable disease severity. To this end, we have assessed gene expression of IFN-β and IFN-λ1 (IL-29) in sputum cells from 51 stable COPD and 35 healthy subjects. We have also extended our analysis to several IFN-stimulated genes (ISGs) supposed to mediate the cellular response to IFN by assessing three genes involved at different steps of intracellular viral cycle. MxA forms oligomers that trap viral components at early time points of the cellular infection, while OAS activates the latent form of RNaseL leading to viral and host RNA degradation.19 Viperin exerts its effects at the later stages of this life cycle by preventing the release of viral particles apparently by disrupting lipid droplets.20
Methods
Subject characteristics and study design
The study was performed with subjects recruited from the Ambulatory Clinic of CHU Liege. Patient characteristics are given in Table 1. Diagnosis of COPD was performed according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) criteria:1 post-bronchodilator (400 μg salbutamol) forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) ratio <70% in patients who presented dyspnea, chronic cough or sputum production, and a history of exposure to risk factors for the disease (tobacco smoke, occupational dusts or chemicals) with all subjects reporting a smoking history of at last 10 pack-year. On the day of the visit, COPD patients completed the COPD assessment test (CAT questionnaire)21 and were asked about the number of exacerbations in the previous year as defined by an intake of antibiotic and/or methylprednisolone for at least 3 days for worsening of respiratory symptoms. Patients were divided in four groups according to GOLD classification of airway obstruction severity based on post-bronchodilation FEV1: GOLD I (FEV1 ≥80%), GOLD II (FEV1 ≥50% and <80%) were considered as mild to moderate, while GOLD III (FEV1 ≥30% and <50%) and GOLD IV (FEV1 <30%) were considered as severe.1 COPD patients were also classified according to the combined CAT, which takes into account symptoms, spirometric classification, and risk of exacerbations. Patients were divided in four groups: A (low risk and less symptoms), B (low risk and more symptoms), C (high risk and less symptoms), and D (high risk and more symptoms).1 Based on sputum cellularity, patients were classified as neutrophilic phenotype when sputum neutrophil count was ≥76% of sputum total nonsquamous cells.22 All the COPD patients included in this study were in stable state on the day of sputum sampling as none were prescribed antibiotics or methylprednisolone following the visit. In addition, the last exacerbation had occurred more than 4 weeks prior to the visit.
Healthy subjects were recruited by local advertisement in the hospital. None of them exhibited respiratory symptoms or airways responsiveness (provocative concentration of methacholine causing a fall in FEV1 of 20%>16 mg/mL) and all had normal spirometric results (FEV1 >80% predicted value).
The study was approved by the CHU Liege Ethics Committee (ref 2005/181) and all subjects signed an informed consent.
Sputum induction and processing
Sputum was induced and processed as previously reported.23 Cell viability was checked by trypan blue exclusion assay and the differential cell count was performed on cytospins stained with RAPI DIFF II® stain kit (Atom Scientific, Manchester, UK) for 500 total nonsquamous cells counted. Only sputum samples with <30% squamous cell count were considered suitable for this study. Collected cells were centrifuged and the pellet (1–2×106 cells) was mixed with 400 μL of RNAprotect® cell reagent (Qiagen, Hilden, Germany) and kept at −80°C until RNA extraction.
RNA extraction/isolation and cDNA synthesis
After removing RNAprotect cell reagent (Qiagen), the pellet was resuspended in 500 μL of TriPure isolation reagent (Roche Diagnostics GmbH, Mannheim, Germany), to which was added one stainless steel bead (5 mm, Qiagen). The sample was disrupted and homogenized using a TissueLyser system (TissueLyser II; Qiagen), for 2 minutes at 25 Hz. The RNA was separated by phenol-chloroform extraction. The upper aqueous phase (300 μL) was diluted with equal volume of ethanol and transferred to a NucleoSpin RNA binding column (Macherey-Nagel GmbH & Co., Düren, Germany). Washing and drying of silica membrane and RNA elution were executed according to NucleoSpin RNA clean-up protocol (Macherey-Nagel). The residual genomic DNA was eliminated by a DNase treatment with TURBO DNA-free™ Kit of Ambion (Thermo Fisher Scientific, Waltham, MA, USA). The RNA concentration and purity were assessed by NanoDrop ND-1000 spectrophotometer. The cDNA was prepared according to the manufacturer’s instructions from maximum 1 μg of RNA using QuantiTect® Reverse Transcription Kit from Qiagen.
TaqMan real-time qPCR
Quantitative polymerase chain reaction (qPCR) was performed with a 384-well plate (MicroAmp®Optical, Life Technologies Holdings Pte Ltd, Singapore) on a 7900HT system (Thermo Fisher Scientific) using the QuantiTect® Probe PCR Kit (Qiagen). The real-time reaction mixture was prepared in a total volume of 10 μL total PCR reaction and 1 μL cDNA. The cycle parameters were as follows: initial enzyme activation at 95°C for 15 minutes; followed by 40 cycles of sequential incubations at 94°C for 15 seconds and at 60°C for 1 minute; and one cycle at 40°C for 30 seconds.
Primer pairs and probes, all FAM-TAMRA labeled, were selected according to the description of Gielen et al for 18S, IFN-β, MxA, OAS, and viperin.24 For IL-29, probe: 5′-AGTTGCAGCTCTCCTGTCTTCCCCG-3′, forward primer: 5′-CCTTGGAAGAGTCACTCAAGCT-3′, and reverse primer: 5′-AGAAGCCTCAGGTCCCAATT-3′ (accession number: NM_172140.1) were purchased from Eurogentec (Seraing, Belgium). For each sample, a supplementary PCR from RNA for IL-29 and IFN-β was performed to verify the absence of residual genomic DNA. Copy numbers of each gene were determined via standard curves constructed as dsDNA plasmids, and normalized with the housekeeping gene 18S rRNA.
Detection of picornavirus
Qualitative PCR was performed in a volume of 50 μL total PCR reaction and 4 μL cDNA using GoTaq® G2 Flexi DNA Polymerase from Promega Corporation (Fitchburg, WI, USA). The cycle parameters were as follows (2720 Thermal Cycler from Thermo Fisher Scientific): initial enzyme activation at 94°C for 2 minutes; followed by 32 cycles of sequential incubations at 94°C for 30 seconds, at 50°C for 30 seconds, and at 72°C for 2 minutes; and finally one cycle at 72°C for 4 minutes. The primers used were OL26 and OL27 from Thermo Fisher Scientific at 1.5 mM each (product size 380 base pairs).25 PCR products were fractionated on 1.5% agarose (Thermo Fisher Scientific) gels and visualized by ethidium bromide staining and photographed. Any visible band of appropriate size was taken as a positive result. Differentiation of rhinoviruses from enteroviruses is achieved by restriction enzyme digestion of the PCR product. Amplicons generated were digested with Bgl-I from Promega Corporation (10 μL of PCR product plus 10 μL of Bgl-I mix) at 37°C for 1 hour (2720 Thermal Cycler from Thermo Fisher Scientific). PCR products were fractionated on 2.5% agarose (Thermo Fisher Scientific) gels and visualized by ethidium bromide staining and photographed. Any visible single band of appropriate size (189 base pairs) was taken as a positive result for human rhinovirus. Either not digested (product size 380 base pairs) or presenting a double band pattern was taken as a positive result for other picornaviruses.
Sputum bacteriology
Sputum samples were collected in sterile sputum cups and sent to the laboratory within 1 hour after expectoration. The bacterial analysis was performed at the Department of Clinical Microbiology, University Hospital of Liège, Belgium. A Gram stain of the sputum in the area of maximal purulence was examined for polymorphonuclear leukocytes, epithelial cells, bacteria, and fungi. Another portion of the sputum was used for microbiological analysis. Sputa were processed according to the standard microbiological methods. Cultures were made on routine media used for the isolation of respiratory pathogens, including blood agar, Mac Conkey agar, colistin nalidixic acid (CNA) agar, Haemophilus influenzae agar, and Sabouraud agar. The plates were incubated at 35°C±2°C in the atmosphere of 5% CO2 except for Mac Conkey and Sabouraud agars that were incubated in an O2 atmosphere. After 24 and 48 hours of incubation, the plates were studied by standard techniques and bacterial colonies were identified by mass spectrometry (Matrix-Assisted Laser Desorption/Ionization Time-of-Flight mass spectrometry [MALDI-TOF]) or phenotypical tests. Respiratory pathogens were reported in a semiquantitative manner and subjected to antibiotic susceptibility testing performed by automated method (VITEK®2; BioMérieux, Marcy l’Etoile, France) or by disc diffusion methods.
Statistical analysis
Data were analyzed using a statistical software package (GraphPad Prism, Version 5). Results were expressed as median (range) for nonparametric data and as mean (± standard deviation) for parametric data. Comparisons between groups for qualitative data were performed by Fisher’s exact test. As for quantitative data, groups were compared by Mann–Whitney for pairwise comparisons or by Kruskal–Wallis with post hoc Dunn’s test for multiple comparisons. Spearman’s rank correlation coefficient was used to identify relationship between variables that showed nonparametric distribution. P-values <0.05 were considered statistically significant.
Results
Patient characteristics
The demographic, functional, and treatment characteristics of subjects are given in Table 1. Healthy subjects and COPD patients were relatively well age–matched even if COPD patients were slightly older. In COPD patients, FEV1/FVC ratio was inversely correlated with age (r=−0.284; P<0.05). The majority of COPD patients (65%) were receiving inhaled corticoids (ICS). Blood cell counts of the patients show raised numbers of circulating neutrophils and monocytes compared to healthy subjects. Likewise, COPD patients had a slight increase in fibrinogen and CRP levels, even if the value remained in the normal range in the large majority of the patients (Table 2). The sputum cell counts are shown in Table 3. COPD patients had higher total nonsquamous sputum cell counts and percentage of neutrophils and eosinophils but lower percentage of macrophages compared to healthy subjects.
  | Table 2 Peripheral blood cell counts and inflammation parameters of healthy subjects and COPD patients |
  | Table 3 Sputum weight and cell counts of healthy subjects and COPD patients |
Detection of IFNs and ISGs in COPD patients
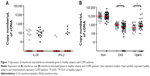
Expression of IL-29 was positive in 16 of 51 COPD patients (31%) and in nine of 35 healthy subjects (26%) (P>0.05). IFN-β was detected in six of 51 COPD patients (12%) and in two of 35 healthy subjects (6%) (P>0.05) (Figure 1A). ISGs were readily detectable in both healthy subjects and COPD patients. OAS expression was decreased in COPD compared to healthy subjects (P<0.05), while that of viperin was increased (P<0.01). There was no difference in MxA expression between the two groups (Figure 1B).
Effect of smoking and inhaled corticosteroids
Among the COPD patients and healthy subjects, no difference was seen in the expression of IL-29 or IFN-β among nonsmokers, ex-smokers, and current smokers. Likewise, current and ex-smoking habit did not influence OAS and MxA expression among the COPD or healthy subjects. Viperin expression in healthy smokers was decreased when compared to healthy nonsmokers (P<0.05), but no such difference was seen between COPD smokers and ex-smokers (P>0.05). There was no significant difference between the COPD patients treated with ICS and those without treatment regarding IFNs and ISGs (P>0.05).
Relationship between IFNs, ISGs, and airway obstruction severity
When classified according to the severity of airway obstruction, there were no differences in IL-29 or IFN-β expressions between the groups (Table 4). However, severe COPD patients showed decreased OAS expression compared to healthy subjects (P<0.01), whereas mild to moderate COPD patients expressed greater level of viperin than healthy subjects (P<0.01). In addition, MxA and OAS expression positively correlated with post-bronchodilator FEV1/FVC ratio in COPD patients (r=0.328; P=0.019 and r=0.341; P=0.014, respectively).
Relationship between IFNs, ISGs, and COPD exacerbations
COPD patients were classified according to the number of exacerbations during the last 12 months recorded by history taking. When compared to healthy subjects, COPD patients with less than two exacerbations displayed higher viperin expression while the group that had ≥2 exacerbations showed decreased OAS expression (Table 5).
IFNs and ISGs according to ABCD GOLD groups
Among the 45 patients with CAT score, the distribution of COPD patients in ABCD groups was 1, 27, 0, and 17, respectively. Therefore, we only compared healthy subjects, and B and D groups. There was no difference with respect to IFN-β and IL-29 expression between the groups (Figure 2A). By contrast, we found a decrease in all three ISGs in patients from group D as compared to group B (P<0.01 for MxA, P<0.05 for OAS, and P<0.01 for viperin) (Figure 2B). OAS expression in group D was also decreased compared to healthy subjects, and viperin expression in group B was significantly higher when compared to healthy controls.
Relationship between IFNs, ISGs, and COPD sputum cell counts
IL-29 and IFN-β expressions positively correlated with the percentage of sputum neutrophils in COPD patients (r=0.30 and 0.29, respectively, P<0.05 for both). By contrast, IL-29 and IFN-β negatively correlated with the percentage of sputum macrophages in COPD patients (r=−0.31, P<0.05 and r=−0.40, P<0.01, respectively). MxA expression was inversely related to the percentage of bronchial epithelial cells (r=0.29, P<0.05).
Relationship between IFNs, ISGs, and sputum bacteriology
Bacterial culture was performed for the sputum samples of 39 COPD patients to identify colonization with potential pathogenic microorganisms (PPM). Seven out of 39 subjects (18%) had positive bacteriology despite being in stable state. Among them, three (8%) were positive for Pseudomonas aeruginosa, two (5%) for H. influenza, and two (5%) for Streptococcus pneumoniae. Sputum neutrophil counts were significantly higher when sputum was positive for PPM compared to when without PPM (median of 97%, ranging from 62% to 99% vs median of 76%, ranging from 11% to 95%, P<0.05). Positive bacteriology was not associated with raised IFNs or ISGs expression (data not shown).
Relationship between IFNs and ISGs and detection of picornavirus in sputum samples
The detection of picornaviruses was performed by reverse transcription q-PCR (RT-qPCR) in all COPD patients (n=51) and healthy subjects, apart from two healthy subjects (n=33). Only 12 sputum samples out of 84 were positive for picornaviruses (14%), including eight COPD patients (16%) and four healthy subjects (12%). Human rhinovirus was detected in seven COPD and one healthy subject, while other picornaviruses were detected in one COPD and three healthy subjects. Only one COPD patient had concomitant detection of both rhinovirus and PPM. Sputum neutrophil count was significantly higher in samples positive for picornavirus compared to those without virus detection (median of 87%, ranging from 54% to 97% vs median of 74%, ranging from 11% to 99%, P<0.05). Similarly, positive samples for PPM and/or picornavirus had significantly higher sputum neutrophils when compared to negative samples (median of 87%, ranging from 54% to 99% vs median of 64%, ranging from 11% to 95%, P<0.001). There was no difference in IFNs and ISGs between those positive vs negative for viruses (data not shown). However, there was a trend to have more often detectable IFN-β in those patients positive to either picornaviruses or PPM (4/18 vs 4/66; P=0.06).
Relationship between IFNs, ISGs, and systemic inflammation
CRP and fibrinogen levels were in the normal range for the majority of COPD patients. Only 15 and 13 COPD patients had CRP and fibrinogen above the upper limit of the normal range (>6 mg/L and >4.3 g/L, respectively). There was no correlation between fibrinogen or CRP levels and IFNs or ISGs expression (P>0.05, data not shown). In addition, there was no difference in the level of fibrinogen or CRP in the blood between positive and negative samples for virus and/or bacteria in sputum samples (P>0.05, data not shown).
Discussion
The purpose of our study was to investigate some components of the innate immune response in airways from stable COPD patients. It appeared that ISGs were expressed to a greater extent than IFN-β and IL-29 in both COPD and healthy subjects. While ISGs were expressed in almost all subjects, IFNs were detected in less than one-third of the patients and healthy subjects alike. Our study shows that COPD patients had altered IFN-induced gene expression in their sputum compared to healthy subjects. The altered expression was inconsistent for the whole group with decreased OAS but increased viperin and MxA. However, the most severe patients, classified as group D according to the GOLD 2015 guidelines, had clearly reduced ISGs expression compared to those from group B. On the other hand, COPD patients with the most intense airway neutrophilic inflammation had raised expression of IL-29 and IFN-β, the latter being more often expressed when microbes were detected in sputum samples.
Our data showed that several ISGs may be differently activated. Interestingly, when considering the most severe COPD patients from group D of GOLD, it turned out that all three ISGs were clearly reduced as compared to mild to moderate COPD and also compared to healthy subjects for OAS. In addition, the reduction of OAS expression was consistent when considering the most severe COPD, with regard to airflow limitation, number of exacerbations in the last 12 months, and the combined assessment in ABCD groups when compared to healthy subjects. The reduced OAS expression is in keeping with recently published data from the ECLIPSE study where a decreased expression of OASL, another member of OAS family, was found by microarray analysis in sputum samples from COPD patients prone to exacerbations.26 Zheng et al showed that there was increased viral replication in primary airway epithelial cells from patients with cystic fibrosis (CF-AECs) caused by the lack of NOS2 and OAS induction in response to virus or IFN-γ.27 They also suggest that the cause of this event is possibly an impairment of activation of STAT 1 in CF-AECs compared to healthy airway epithelial cells. In our study, it is worth noting that viperin seems to be particularly overexpressed in mild to moderate COPD while decreased when disease becomes more severe and prone to exacerbate. This finding is intriguing and may indicate vigorous immune defense mechanisms in mild to moderate COPD that diminishes when disease progresses. On the other hand, it is worth emphasizing that MxA, OAS, and viperin act at different successive steps of the intracellular viral cycle. MxA represents the first line of defense against viral infection inside the cell. It forms oligomers that can bind viral components and degrade them at early time points of the viral cycle. The low constitutive expression levels of OAS are amplified by IFNs and can detect viral double-stranded RNAs (dsRNA).19 After activation by dsRNA, OAS oligomerizes and synthesizes 2′,5′-oligoadenylates that activate RNaseL. Dimers of RNaseL can then cleave viral and cellular RNA in the cytoplasm.19 Viperin might exert its antiviral activity by blocking viral budding from lipid rafts as well as by inhibiting the formation of viral replication complex in lipid droplets, acting overall in later stages of the viral cycle inside the cell.20 Combined reduced expression of all these three ISGs might strongly weaken the antiviral mechanisms in the most severe COPD. Indeed, Hurst et al demonstrated that COPD patients with more exacerbations had more frequent episodes of naturally occurring colds as compared to patients with infrequent exacerbations.28
Further highlighting the link between COPD severity and ISGs, our results also show that MxA and OAS expressions are directly correlated with post-bronchodilator FEV1/FVC ratio, thereby demonstrating that expression of ISGs diminishes as airway obstruction progresses.
The reasons why severe COPD patients have reduced ISGs gene expression remain unclear. Although ICS are powerful drugs to reduce immune system activation, our cross-sectional study does not provide evidence for an impact of ICS on IFNs and ISGs expression. A definitive answer to this question would, however, require a longitudinal intervention study. Likewise, we did not find an effect of active smoking on the gene expression. By contrast, it seems that the presence of either picornaviruses or PPM, besides being associated with intense sputum neutrophilia, favors IFN-β expression (P=0.06).
When combining PPM and picornaviruses, we found that approximately one-third of our patients had pathogens detected and those patients had raised sputum neutrophil counts. It is worth noting that all patients were in stable state and had overall CRP and fibrinogen levels in the normal range as opposed to what happens during an exacerbation.29 The minority of patients with raised CRP and fibrinogen levels showed no differences in the expression of IFNs and ISGs in sputum cells. This finding points to the possibility of other causes of systemic inflammation than microbial airways contamination. Although the presence of microbes in the airways was associated with high sputum neutrophil counts, some subjects had very high sputum neutrophil counts without evidence of bacterial colonization with PPM or infection with picornaviruses. Even if conventional bacterial culture lacks sensitivity to assess airway bacterial load,30 this might also be seen as reflecting innate immune activation following exposure to a wide range of irritants, including smokes,10,31 chemical dusts, or air pollutants.32–34
One limitation of our study is its cross-sectional design precluding any conclusion as to whether the deficiency in ISGs found in severe COPD is a phenomenon that is stable over time and if this may persist when patients exacerbate making infection clearance more difficult.
Conclusion
We conclude that ISGs may be readily detected in stable COPD and healthy subjects, while expression of IFNs was less clear. ISGs expression was reduced in COPD patients prone to exacerbation and with severe airway obstruction. The reduced innate immunity in COPD may contribute to disease progress by altering response to infection.
Acknowledgments
The authors thank the clinical microbiology laboratory at the Centre Hospitalier Universitaire (University of Liege, Belgium), Unilab, for the bacteriology tests in sputum. The work was supported by federal research grant IAP P7/30.
Author contributions
CH participated in the design of the study, carried out RT-qPCR and FACS studies, performed the statistical analysis, and drafted the manuscript. JDS participated in the design of the study and helped to draft the manuscript. MH participated in subjects’ recruitment, RT-qPCR, and FACS studies. FS and JLC participated in subjects’ recruitment, analysis, and interpretation of data. TK carried out picornaviruses study. MRE, PM, SLJ, and RL conceived the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
SLJ is an author of patents relating to the use of inhaled IFNs as treatments for exacerbations of airway disease. SLJ has research grants and/or has consulted for GlaxoSmithKline, Astrazeneca, Merck, Boehinger Ingelheim, Centocor, Amgen, Roche/Genentech, Biota, Kyorin, Novartis, Sanofi, and Synairgen. RL has research grants and/or has consulted for GlaxoSmithKline, Astrazeneca, Novartis, and Chiesi. The other authors report no conflicts of interest in this work.
References
From the Global Strategy for the Diagnosis, Management and Prevention of COPD, Global Initiative for Chronic Obstructive Lung Disease (GOLD); 2015. Available from: http://www.goldcopd.org/. Accessed April 7, 2015. | ||
Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. | ||
Beasley V, Joshi PV, Singanayagam A, Molyneaux PL, Johnston SL, Mallia P. Lung microbiology and exacerbations in COPD. Int J Chron Obstruct Pulmon Dis. 2012;7:555–569. | ||
Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359(22):2355–2365. | ||
Papi A, Bellettato CM, Braccioni F, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173(10):1114–1121. | ||
Perotin JM, Dury S, Renois F, et al. Detection of multiple viral and bacterial infections in acute exacerbation of chronic obstructive pulmonary disease: a pilot prospective study. J Med Virol. 2013;85(5):866–873. | ||
Caramori G, Kirkham P, Barczyk A, Di Stefano A, Adcock I. Molecular pathogenesis of cigarette smoking-induced stable COPD. Ann N Y Acad Sci. 2015;1340:55–64. | ||
Baines KJ, Hsu AC, Tooze M, Gunawardhana LP, Gibson PG, Wark PA. Novel immune genes associated with excessive inflammatory and antiviral responses to rhinovirus in COPD. Respir Res. 2013;14:15. | ||
Baines KJ, Simpson JL, Gibson PG. Innate immune responses are increased in chronic obstructive pulmonary disease. PLoS One. 2011;6(3):e18426. | ||
Keatings VM, Collins PD, Scott DM, Barnes PJ. Differences in interleukin-8 and tumor necrosis factor-alpha in induced sputum from patients with chronic obstructive pulmonary disease or asthma. Am J Respir Crit Care Med. 1996;153(2):530–534. | ||
Moermans C, Bonnet C, Willems E, et al. Sputum cytokine levels in patients undergoing hematopoietic SCT and comparison with healthy subjects and COPD: a pilot study. Bone Marrow Transplant. 2014;49(11):1382–1388. | ||
Zhang X, Zheng H, Zhang H, et al. Increased interleukin (IL)-8 and decreased IL-17 production in chronic obstructive pulmonary disease (COPD) provoked by cigarette smoke. Cytokine. 2011;56(3):717–725. | ||
Nadigel J, Audusseau S, Baglole CJ, Eidelman DH, Hamid Q. IL-8 production in response to cigarette smoke is decreased in epithelial cells from COPD patients. Pulm Pharmacol Ther. 2013;26(5):596–602. | ||
Berenson CS, Kruzel RL, Eberhardt E, et al. Impaired innate immune alveolar macrophage response and the predilection for COPD exacerbations. Thorax. 2014;69(9):811–818. | ||
Mallia P, Message SD, Gielen V, et al. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am J Respir Crit Care Med. 2011;183(6):734–742. | ||
Contoli M, Message SD, Laza-Stanca V, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med. 2006;12(9):1023–1026. | ||
Edwards MR, Slater L, Johnston SL. Signalling pathways mediating type I interferon gene expression. Microbes Infect. 2007;9(11):1245–1251. | ||
Singanayagam A, Joshi PV, Mallia P, Johnston SL. Viruses exacerbating chronic pulmonary disease: the role of immune modulation. BMC Med. 2012;10:27. | ||
Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. | ||
Mattijssen S, Pruijn GJ. Viperin, a key player in the antiviral response. Microbes Infect. 2012;14(5):419–426. | ||
Jones PW, Harding G, Berry P, Wiklund I, Chen WH, Kline Leidy N. Development and first validation of the COPD Assessment Test. Eur Respir J. 2009;34(3):648–654. | ||
Schleich FN, Manise M, Sele J, Henket M, Seidel L, Louis R. Distribution of sputum cellular phenotype in a large asthma cohort: predicting factors for eosinophilic vs neutrophilic inflammation. BMC Pulm Med. 2013;13:11. | ||
Quaedvlieg V, Sele J, Henket M, Louis R. Association between asthma control and bronchial hyperresponsiveness and airways inflammation: a cross-sectional study in daily practice. Clin Exp Allergy. 2009;39(12):1822–1829. | ||
Gielen V, Johnston SL, Edwards MR. Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur Respir J. 2010;36(3):646–654. | ||
Papadopoulos NG, Hunter J, Sanderson G, Meyer J, Johnston SL. Rhinovirus identification by BglI digestion of picornavirus RT-PCR amplicons. J Virol Methods. 1999;80(2):179–185. | ||
Singh D, Fox SM, Tal-Singer R, Bates S, Riley JH, Celli B. Altered gene expression in blood and sputum in COPD frequent exacerbators in the ECLIPSE cohort. PLoS One. 2014;9(9):e107381. | ||
Zheng S, De BP, Choudhary S, et al. Impaired innate host defense causes susceptibility to respiratory virus infections in cystic fibrosis. Immunity. 2003;18(5):619–630. | ||
Hurst JR, Donaldson GC, Wilkinson TM, Perera WR, Wedzicha JA. Epidemiological relationships between the common cold and exacerbation frequency in COPD. Eur Respir J. 2005;26(5):846–852. | ||
Corhay JL, Moermans C, Henket M, Nguyen Dang D, Duysinx B, Louis R. Increased of exhaled breath condensate neutrophil chemotaxis in acute exacerbation of COPD. Respir Res. 2014;15:115. | ||
Garcha DS, Thurston SJ, Patel AR, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67(12):1075–1080. | ||
Doz E, Noulin N, Boichot E, et al. Cigarette smoke-induced pulmonary inflammation is TLR4/MyD88 and IL-1R1/MyD88 signaling dependent. J Immunol. 2008;180(2):1169–1178. | ||
Nightingale JA, Rogers DF, Barnes PJ. Effect of inhaled ozone on exhaled nitric oxide, pulmonary function, and induced sputum in normal and asthmatic subjects. Thorax. 1999;54(12):1061–1069. | ||
Nordenhall C, Pourazar J, Blomberg A, Levin JO, Sandstrom T, Adelroth E. Airway inflammation following exposure to diesel exhaust: a study of time kinetics using induced sputum. Eur Respir J. 2000;15(6):1046–1051. | ||
Wallace J, D’Silva L, Brannan J, Hargreave FE, Kanaroglou P, Nair P. Association between proximity to major roads and sputum cell counts. Can Respir J. 2011;18(1):13–18. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.