Back to Journals » OncoTargets and Therapy » Volume 11
Reduced expression of Nrdp1 predicts a poor prognosis in human hepatocellular carcinoma
Authors Shao X, Lu Q, Wang G, Huang W, Yang L, Chen Z
Received 22 December 2017
Accepted for publication 21 May 2018
Published 17 August 2018 Volume 2018:11 Pages 4955—4963
DOI https://doi.org/10.2147/OTT.S160638
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Xian Shao,1,2,* Qian Lu,1,* Gang Wang,1 Wei Huang,1 Linlin Yang,1 Zhong Chen1
1Department of Hepatobiliary Surgery, Affiliated Hospital of Nantong University, Nantong 226001, People’s Republic of China; 2Department of Hepatobiliary Surgery, Affiliated Hospital of Jiangsu University, The First People’s Hospital of KunShan, KunShan 215300, People’s Republic of China
*These authors contributed equally to this work
Background: Hepatocellular carcinoma (HCC) is an aggressive form of liver cancer with particularly poor survival rates for patients. Targeted molecular therapies are lacking, and current treatment is generally limited to surgical resection or liver transplantation. Overexpression and aberrant signaling of the ErbB family of receptors has been implicated in HCC, but the mechanisms underlying ErbB overexpression are unclear. In this study, we investigated the potential role of neuregulin receptor degradation protein-1 (Nrdp1), a regulator of ErbB3 protein stability, in HCC progression.
Methods: We compared the expression of Nrdp1 in various HCC cell lines and in 8 pairs of tumor and peritumor tissue samples using Western blot analysis. Changes in the degree of proliferation were determined before and after small interfering RNA (siRNA)-induced knockdown of Nrdp1 using a cell counting Kit-8 (ccK-8) assay and cell-cycle analysis. The correlation between Nrdp1 expression and prognosis was determined in specimens of 89 HCC patients.
Results: Nrdp1 expression is significantly reduced in HCC tissues compared with adjacent healthy tissues. Higher Nrdp1 expression corresponds to lower maximal tumor size (χ2, P<0.05), lower histological grade (χ2, P<0.05), and higher survival rates by Kaplan–Meier estimate (P<0.05). Higher Nrdp1 expression also corresponds to reduced expression of Ki-67, a marker of cell proliferation (Spearman, r2=0.734; P<0.05). Nrdp1 accumulates in serum-starved HepG2 cancer cells and progressively decreases in expression after re-feeding. Furthermore, depletion of Nrdp1 in healthy L02 cells by siRNA results in enhanced cell proliferation and a greater proportion of cells in S phase.
Conclusions: Our findings suggest an inhibitory role for Nrdp1 in HCC tumorigenesis, and we propose that Nrdp1 may serve as a prognostic biomarker for HCC and as a potential therapeutic target for the treatment of HCC.
Keywords: neuregulin receptor, ErbB3 protein stability, HCC therapeutic target, ErbB, Nrdp1 depletion
Introduction
Hepatocellular carcinoma (HCC) is among the deadliest types of cancer in the world, especially in Eastern Asia. Owing to the high incidence of tumor invasion, frequent intrahepatic spread, and resistance to chemotherapy, the 5-year survival rate of patients with advanced HCC is less than 10%.1 Accumulating data indicate that hepatocarcinogenesis involves the aberrant activation of multiple cancer-related signaling pathways, including the ErbB, Wnt/β-catenin, NF-κB, and Notch pathways.2 However, the precise molecular mechanisms leading to the dysregulation of these pathways in HCC remain poorly understood. Identifying these mechanisms would provide a better understanding of hepatocarcinogenesis and would thereby aid efforts to prevent and treat HCC.
The ErbB receptors – EGFR, ErbB2, ErbB3, and ErbB4 – are a class of receptor tyrosine kinases that regulate many fundamental cellular processes, including proliferation, differentiation, cell survival, and migration.3,4 Aberrant activation of these receptors is thought to play a critical role in the progression of human cancers.5 After ligand binding, ErbB receptors rapidly form homo- or heterodimers, activate their own tyrosine kinase domains, and phosphorylate a variety of ErbB substrates.6 These downstream effectors trigger the activation of important signaling pathways, including PI3k/Akt, JAK/STAT, and MAPKs, that have been implicated in uncontrolled cell proliferation, metastasis, and drug resistance in human cancers.6 Overexpression of ErbB receptors has accordingly been observed in various human cancers, including HCC.7,8 However, the molecular mechanisms that lead to the excessive accumulation of ErbB receptors in HCC are poorly understood.
The cell-surface expression of ErbB receptors is tightly regulated at multiple levels, with posttranscriptional degradation receiving the greatest attention.9 It is well documented that EGFR is degraded primarily through a ligand-dependent endocytic pathway, but the degradation pathway of other ErbB members is unclear.10 Many studies indicate that the ubiquitin proteasome system may play a role in the proteolysis of ErbB2–4.11 For example, the E3 ubiquitin ligases carboxyl terminus Hsc70-interacting protein (CHIP) and Cullin5 have been proposed to facilitate the ubiquitination and degradation of ErbB2.12,13 In the case of ErbB3, it is thought that the ring finger E3 ubiquitin ligase neuregulin receptor degradation protein-1 (Nrdp1) plays a key role in the ubiquitin-mediated degradation of ErbB3.14,15 Ectopic expression of Nrdp1 leads to significantly reduced ErbB3 expression and markedly impaired proliferation in cancer cells, suggesting that Nrdp1 regulates ErbB3 expression and its downstream signaling. In line with these findings, Nrdp1 has been implicated in the progression of breast cancer.16 In addition to its role in ErbB3 signaling, Nrdp1 may regulate other molecular events involved in cancer, including oxidative stress and the expression of type 1 cytokine receptor.17–19 Nrdp1 may thus exert a broad physiological effect in cancer cells.
Nrdp1 has been implicated in ErbB signaling; however, its expression in HCC and its relevance to HCC progression are unknown. In this study, we investigated the potential role of Nrdp1 in HCC and its prognostic value for HCC patients. We found that Nrdp1 expression is reduced in HCC tissues compared with adjacent healthy tissues. Moreover, higher expression of Nrdp1 correlates with reduced maximal tumor size, lower histological grade, and reduced Ki-67 expression. These findings provide insight into the mechanisms underlying HCC development and identify Nrdp1 as a potential prognostic marker and therapeutic target for patients with HCC.
Materials and methods
Patients and tissue samples
For immunohistochemistry, we included a total of 89 paraffin-embedded HCC and matched-adjacent healthy tissue samples from patients who underwent hepatic resection without preoperative systemic chemotherapy. Samples were collected at the Affiliated Hospital of Nantong University between January 2009 and May 2010. The main clinical and pathologic variables are shown in Table 1. Nineteen fresh HCC tissue specimens were additionally collected from the same sample bank, between January 2015 and July 2015, for Western blotting analysis. Before collecting the samples, written informed consent from patients and approval from the Institutional Ethics Committee of the Affiliated Hospital of Nantong University were obtained.
Immunohistochemistry
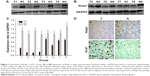
Immunohistochemistry was carried out as described previously.11 Briefly, following endogenous peroxidase neutralization and antigen retrieval by microwave irradiation, slides were preincubated with blocking serum and incubated overnight with rabbit anti-human Nrdp1 polyclonal antibody (diluted 1:200; Santa Cruz Biotechnology) and anti-Ki-67 mouse monoclonal antibody (diluted 1:100; clone 7B11; Zymed Laboratories, San Francisco, CA, USA). Sections were incubated with a secondary antibody and exposed to horseradish peroxidase (HRP)-conjugated streptavidin (Dako, Hamburg, Germany). Photographs of three representative fields were captured using a Leica CCD camera (DFC420) and Leica QWin Plus v3 software. All sections were evaluated by two independent pathologists who had no knowledge of patient characteristics. To assess Nrdp1 expression, five views were chosen per slide, and at least 1,000 cells were counted in each view at high power fields. A score was assigned to each sample based on Nrdp1 staining intensity (0: negative; 1: weak; and 2: strong) and the percentage of Nrdp1-positive cells (1: 0%–25%; 2: 26%–50%; 3: 51%–75%; and 4: ≥75%). A tumor-staining index was calculated for each sample by multiplying the staining intensity score with the percentage score. Samples with an index ≥2 were defined as high-expression samples, whereas those with an index <2 were defined as low-expression samples. For Ki-67 expression, staining was scored semi-quantitatively, using a cutoff of ≥50% positively stained in five field views (≥50%: high-expression sample; <50%: low-expression sample).
Cell culture
One normal hepatocyte cell line (L02) and three HCC cell lines (HepG2, HuH7, and Hep1) were obtained from the Institute of Cell Biology of the Chinese Academy of Sciences. Cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 g/mL streptomycin (Invitrogen, Carlsbad, CA, USA) in 5% CO2 at 37°C.
Transfection
Four human Nrdp1 siRNA expression vectors and one pSilencer siRNA were constructed with the following sequences: 5′-CAGGCCTTGTCATGATATT-3, 5′-GACGCTACTATGAGAACTA-3′, 5′-TGGAGGAGACAATTGAATA-3′, and 5′-CTATTTGCAGTGGAGTCTT-3′ (labeled siNrdp1-1, siNrdp1-2, siNrdp1-3, and siNrdp1-4, respectively). L02 cells were cultured in 6-cm dishes or 24-well plates to 70%–80% confluency. Cells were then transfected with individual siRNAs using Lipofectamine 2000 transfection reagent (Invitrogen) per the manufacturer’s protocol. Cells were collected for subsequent assays 48 h after transfection.
Western blot
Cells were promptly homogenized in a homogenization buffer and centrifuged at 10,000×g for 30 min to collect the supernatant. Protein concentrations were determined with a BioRad protein assay (BioRad, Hercules, CA, USA). Total protein was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were washed, blocked, and incubated with anti-Nrdp1 antibody (1:1,000; Santa Cruz Biotechnology, CA, USA) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (1:1,000; Sigma). Membranes were washed and incubated with HRP-conjugated secondary antibodies (1:500; Santa Cruz Biotechnology). Proteins were detected using an enhanced chemiluminescence reagent (NEN, Boston, MA). The relative intensity of each band was determined using Image Lab 3.0 (BioRad Laboratories, Inc., Hercules, CA, USA).
Cell Counting Kit-8 (CCK-8) assay
Proliferation and viability of L02 cells were measured using the CCK-8 (Dojindo, Kumamoto, Japan) assay. Briefly, cells transfected with siRNA were seeded in 96-well plates (Corning, NY, USA) at a density of 2×104 cells/well in 100 μL culture medium and incubated overnight. CCK-8 kit reagents were added and cells were incubated for 2 h at 37°C. Absorbance values were measured using an automated plate reader. Each experiment was repeated at least three times.
Cell-cycle analysis
Cells were fixed in 70% ethanol for 24 h at −20°C and then incubated with 1 mg/mL RNase A for 20 min at 37°C. Cells were subsequently stained with propidium iodide (50 Lg/mL; Becton Dickinson, San Jose, CA, USA) in PBS supplemented with 0.5% Tween-20. Stained cells were analyzed by flow cytometry using a Becton Dickinson FACScan (San Jose, CA, USA) and Cell Quest acquisition and analysis software.
Statistical analysis
Statistical analysis was conducted using SPSS 17.0. Associations between Nrdp1/Ki-67 expression and clinicopathological variables were analyzed by χ2 test. The relationship between Nrdp1 and Ki-67 was assessed by Spearman’s correlation. Survivorship was assessed by the Kaplan–Meier method and the log-rank test. Multivariate analysis was conducted using the Cox proportional hazard model. For all statistical analyses, a cutoff value of P<0.05 was used. Results are expressed as the mean ± SD.
Results
Nrdp1 expression in HCC and adjacent healthy tissues
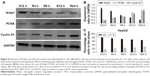
To assess whether Nrdp1 is involved in HCC, we examined levels of Nrdp1 in HCC tissues and corresponding adjacent healthy tissues by Western blotting analysis. Nrdp1 is markedly downregulated in HCC tissues compared with adjacent healthy tissues (Figure 1A–C). In agreement with these observations, immunohistochemical analysis showed that Nrdp1 expression is significantly lower in HCC tissues than in non-tumor tissues. By contrast, Ki-67 expression is elevated in HCC tissues (Figure 1D). Notably, we observed a significant negative correlation between Nrdp1 expression and Ki-67 expression in HCC (r2=0.734; Figure 2B). These findings suggest that Nrdp1 is downregulated in HCC tissues.
Relationship between Nrdp1 expression and clinical pathology of HCC
Based on the level of Nrdp1 observed in immunohistochemical staining, we separated HCC samples into Nrdp1 high-expressing and Nrdp1 low-expressing groups by using a tumor-staining index (see Materials and methods section). We found that the expression level of Nrdp1 in tumor tissue significantly correlates with maximal tumor size, histological grade, and Ki-67 expression, but not with age, gender, serum alpha-fetoprotein level, or tumor metastasis (Table 1). Furthermore, we found that patient survivorship significantly correlates with several clinicopathological parameters, including tumor node number, maximal tumor size, histological grade, Nrdp1 expression, and Ki-67 expression (Table 2). The prognostic value of Nrdp1 in HCC patients was evaluated by survival analysis of patients whose HCC tissues were categorized as Nrdp1 high-expressing versus low-expressing samples. Patients with low Nrdp1 expression had significantly worse survival compared with those expressing high levels of Nrdp1 (Figure 2A).
Reduced expression of Nrdp1 in proliferating HCC cells
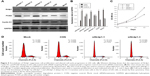
To better understand the relationship between Nrdp1 and HCC development, we assessed the expression of Nrdp1 in the context of HCC cell proliferation. We first measured the expression of Nrdp1 in three HCC cell lines (HepG2, HuH7, and Hep1) and in one normal hepatocyte cell line (L02) by Western blot. We found that L02 hepatocytes have the highest expression of Nrdp1 among all cell lines examined (Figure 3A and B). We next analyzed whether Nrdp1 expression is altered as a function of the proliferation status of HCC cells by using a serum starvation and re-feeding assay. After serum starvation for 72 h, HepG2 cells arrested at the G1 phase. The cells then progressively entered S phase upon serum re-feeding, as indicated by progressive accumulation of proliferating cell nuclear antigen (PCNA) and Cyclin D1 (Figure 4C). Intriguingly, Nrdp1 showed marked accumulation in serum-starved HepG2 cells, and declined after serum re-feeding in a time-dependent manner (Figure 4A and B). These data demonstrate that Nrdp1 expression is suppressed in proliferating HCC cells, suggesting that Nrdp1 may be involved in HCC proliferation.
Depletion of Nrdp1 enhances cell proliferation and accelerates cell-cycle progression in hepatocytes
The observation that Nrdp1 accumulates in cells arrested in G1 phase and declines during S phase suggested a possible functional role for Nrdp1 in cell proliferation. We, therefore, used siRNA to deplete Nrdp1 in normal L02 hepatocytes to assess the impact on proliferation. We first transfected four different siRNAs (siNrdp1-1, siNrdp1-2, siNrdp1-3, and siNrdp1-4) into L02 cells and measured interference efficiencies by Western blotting. Transfection of siNrdp1-1 and siNrdp1-3 resulted in efficient Nrdp1 depletion, whereas siNrdp1-2 and siNrdp1-4 had only minor effects on expression (Figure 5A and B). We then assessed the impact of Nrdp1 depletion on L02 cell proliferation using the CCK-8 assay. We found that, when Nrdp1 is depleted, L02 cells exhibit enhanced cell growth compared with mock-transfected or non-transfected cells (Figure 5C). When we analyzed the cell-cycle distribution of L02 cells, we found that depleting Nrdp1 results in a significantly elevated proportion of cells in S phase (Figure 5D). These findings suggest that Nrdp1 depletion may promote the proliferation of hepatocytes.
Discussion
Currently, the most effective treatment approaches for HCC are surgical resection and transplantation; however, the 5-year survival rate of patients is poor.20 Researchers have recently focused on developing targeted molecular therapies for HCC.21 Understanding the molecular mechanisms of HCC is, therefore, key to developing novel and effective therapeutic strategies. In this study, we found that Nrdp1 expression is significantly reduced in HCC tissues compared with paired adjacent healthy tissues. Moreover, HCC patients with low Nrdp1 expression show shorter postoperative survival than patients with high Nrdp1 expression. We thus propose that downregulation of Nrdp1 may contribute to the progression of HCC.
As a putative tumor suppressor, Nrdp1 has been found to participate in the progression of several types of cancers, including breast, colorectal, and prostate cancer.16,22,23 Nrdp1 was assumed to negatively regulate the cell-surface expression of ErbB3, thereby influencing cell proliferation, differentiation, migration, and invasion. Nrdp1 suppresses cellular ErbB3 by marking the receptor for proteolytic degradation, thereby negatively regulating the transduction of ErbB3 signaling. A recent study showed that the Hepatitis B virus X protein can reduce the stability of Nrdp1 to upregulate ErbB3 in HCC cells.24 However, the prognostic significance of Nrdp1 expression in tumor specimens has not been documented. We report that low expression of Nrdp1 predicts poor prognosis and a markedly shorter survival rate in HCC patients. To our knowledge, this is the first report indicating that Nrdp1 may serve as prognostic biomarker for patients with HCC.
The last two decades have witnessed astonishing progress in the development of targeted molecular therapies for a variety of human diseases, especially cancers. Owing to the well-documented role of ErbB receptors in cancer development, intensive efforts have been made to produce new therapies that target these receptors. Although therapies based on EGFR and ErbB2 have achieved great success in the treatment of malignancies, there have not been significant improvements in ErbB3-based therapies. ErbB3 has, nevertheless, attracted significant attention for its potential therapeutic value in cancer therapy. ErbB3 preferentially forms heterodimers with ErbB2 and EGFR to activate downstream events. Overexpression of ErbB3 has been confirmed in various cancer types, including HCC. Intriguingly, we found that Nrdp1 expression is reduced in cancer specimens. Other studies have shown that ectopic expression of Nrdp1 can abolish ErbB3 expression in cancer cells. These data imply that the upregulation of ErbB3 in cancer cells may be caused by a loss of Nrdp1 expression or function. Restoring Nrdp1 expression or activity might, therefore, serve as an alternative approach to inactivating ErbB3, offering a potential therapeutic strategy for patients with HCC. Further studies might help to clarify the therapeutic value of Nrdp1 in HCC.
In summary, we provide compelling evidence that Nrdp1 plays a tumor inhibitory role in HCC. These findings identify Nrdp1 as a potential new biomarker for HCC progression and prognosis, and may provide a novel target for clinical treatment. However, the complex molecular mechanisms of Nrdp1 and the contribution of Nrdp1 to HCC require further investigation.
Acknowledgments
This work was supported by the clinical medical science and technology division of Jiangsu Technical Department (BL2014060); the project “medical leading talent and innovation team” (LJ201134) of Jiangsu Province. The authors also thank Travis J Bernardo, PhD, from Liwen Bianji, Edanz Editing China, for editing the English language and grammar of a draft of this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27(9):1485–1491. | ||
Moeini A, Cornellà H, Villanueva A. Emerging signaling pathways in hepatocellular carcinoma. Liver Cancer. 2012;1(2):83–93. | ||
Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(Suppl 2):21–26. | ||
Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284(1):99–110. | ||
Lee Y, Ma J, Lyu H, Huang J, Kim A, Liu B. Role of erbB3 receptors in cancer therapeutic resistance. Acta Biochim Biophys Sin (Shanghai). 2014;46(3):190–198. | ||
Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells. 2014;3(2):304–330. | ||
Berasain C, Avila MA. The EGFR signalling system in the liver: from hepatoprotection to hepatocarcinogenesis. J Gastroenterol. 2014;49(1):9–23. | ||
Liu J, Ahiekpor A, Li L, et al. Increased expression of ErbB-2 in liver is associated with hepatitis B x antigen and shorter survival in patients with liver cancer. Int J Cancer. 2009;125(8):1894–1901. | ||
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–516. | ||
Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274(5295):2086–2089. | ||
Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. | ||
Zhou P, Fernandes N, Dodge IL, et al. ErbB2 degradation mediated by the co-chaperone protein CHIP. J Biol Chem. 2003;278(16):13829–13837. | ||
Ehrlich ES, Wang T, Luo K, et al. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106(48):20330–20335. | ||
Diamonti AJ, Guy PM, Ivanof C, Wong K, Sweeney C, Carraway KL 3rd. An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc Natl Acad Sci U S A. 2002;99(5):2866–2871. | ||
Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A. 2002;99(23):14843–14848. | ||
Yen L, Cao Z, Wu X, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 2006;66(23):11279–11286. | ||
Zhou A, Pan D, Yang X, Zhou J. Overexpression of Nrdp1/FLRF sensitizes cells to oxidative stress. Biochem Biophys Res Commun. 2011;410(4):771–774. | ||
Wauman J, De Ceuninck L, Vanderroost N, Lievens S, Tavernier J. RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J Cell Sci. 2011;124(Pt 6):921–932. | ||
Wang C, Chen T, Zhang J, et al. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10(7):744–752. | ||
Kudo M. Hepatocellular carcinoma 2009 and beyond: from the surveillance to molecular targeted therapy. Oncology. 2008;75(Suppl 1):1–12. | ||
Patel A, Sun W. Molecular targeted therapy in hepatocellular carcinoma: from biology to clinical practice and future. Curr Treat Options Oncol. 2014;15(3):380–394. | ||
Chen L, Siddiqui S, Bose S, et al. Nrdp1-mediated regulation of ErbB3 expression by the androgen receptor in androgen-dependent but not castrate-resistant prostate cancer cells. Cancer Res. 2010;70(14):5994–6003. | ||
Lu H, Li H, Mao D, Zhu Z, Sun H. Nrdp1 inhibits growth of colorectal cancer cells by nuclear retention of p27. Tumour Biol. 2014;35(9):8639–8643. | ||
Cao K, Gong H, Qiu Z, et al. Hepatitis B virus X protein reduces the stability of Nrdp1 to up-regulate ErbB3 in hepatocellular carcinoma cells. Tumour Biol. 2016;37(8):10375–10382. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.