Back to Journals » International Journal of Nanomedicine » Volume 18
Recombinase Polymerase Amplification-Based Biosensors for Rapid Zoonoses Screening
Authors Feng X , Liu Y, Zhao Y, Sun Z, Xu N , Zhao C, Xia W
Received 2 September 2023
Accepted for publication 21 October 2023
Published 6 November 2023 Volume 2023:18 Pages 6311—6331
DOI https://doi.org/10.2147/IJN.S434197
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor R.D.K. Misra
Xinrui Feng,1,2,* Yan Liu,1,* Yang Zhao,3 Zhe Sun,1,4 Ning Xu,5 Chen Zhao,1 Wei Xia4
1College of Public Health, Jilin Medical University, Jilin, 132013, People’s Republic of China; 2Medical College, Yanbian University, Yanji, 136200, People’s Republic of China; 3Department of Emergency and Intensive Medicine, No. 965 Hospital of PLA Joint Logistic Support Force, Jilin, 132013, People’s Republic of China; 4College of Medical Technology, Beihua University, Jilin, 132013, People’s Republic of China; 5State Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases, Key Laboratory for Zoonosis Research of the Ministry of Education, Institute of Zoonosis, and College of Veterinary Medicine, Jilin University, Changchun, 130062, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chen Zhao; Wei Xia, Email [email protected]; [email protected]
Abstract: Recent, outbreaks of new emergency zoonotic diseases have prompted an urgent need to develop fast, accurate, and portable screening assays for pathogen infections. Recombinase polymerase amplification (RPA) is sensitive and specific and can be conducted at a constant low temperature with a short response time, making it especially suitable for on-site screening and making it a powerful tool for preventing or controlling the spread of zoonoses. This review summarizes the design principles of RPA-based biosensors as well as various signal output or readout technologies involved in fluorescence detection, lateral flow assays, enzymatic catalytic reactions, spectroscopic techniques, electrochemical techniques, chemiluminescence, nanopore sequencing technologies, microfluidic digital RPA, and clustered regularly interspaced short palindromic repeats/CRISPR-associated systems. The current status and prospects of the application of RPA-based biosensors in zoonoses screening are highlighted. RPA-based biosensors demonstrate the advantages of rapid response, easy-to-read result output, and easy implementation for on-site detection, enabling development toward greater portability, automation, and miniaturization. Although there are still problems such as high cost with unstable signal output, RPA-based biosensors are increasingly becoming one of the most important means of on-site pathogen screening in complex samples involving environmental, water, food, animal, and human samples for controlling the spread of zoonotic diseases.
Keywords: recombinase polymerase amplification, biosensor, zoonoses, rapid detection, nanomaterials
Graphical Abstract:
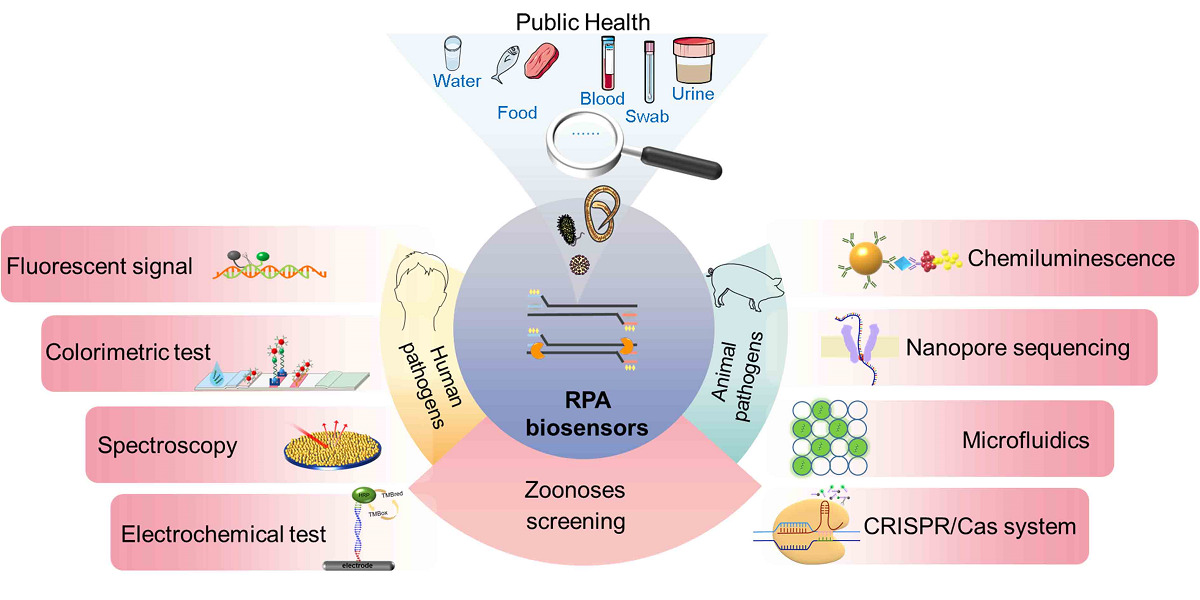
Introduction
Since the emergence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in 2019, this coronavirus has spread to more than 200 countries, cumulatively infecting 672 million people and causing 6.84 million deaths. Moreover, the emergence of numerous variant strains (α, β, γ, δ, and Omicron) presents a serious challenge for controlling the spread of the infectious disease.1 Approximately 60% of new human infectious diseases are zoonoses.2 The WHO proposed the “One Health” Initiative and multidisciplinary integration to address the growing risk of zoonotic diseases.3 The rapid and sensitive detection of zoonotic pathogens from environmental, water, food, animal, and patient samples can effectively control the global spread of zoonotic diseases and reduce their adverse impact on public health. Isothermal amplification technology has attracted much attention from researchers because of its rapidity and simplicity with no need for complex temperature-changing instruments, such as strand displacement amplification (SDA), rolling circle amplification (RCA), nucleic acid sequence-based amplification (NASBA), loop-mediated isothermal amplification (LAMP), helicase-dependent amplification (HDA), crossing priming amplification (CPA), single primer isothermal amplification (SPIA), multiple displacement amplification (MDA), exponential amplification reaction (EXPAR), and whole genome amplification (WGA) and recombinase polymerase amplification (RPA) (Table 1). Isothermal amplification simplifies amplification conditions, and the elimination of thermal cycling reduces the need for instrumentation or a laboratory environment. The warming and cooling process is omitted, effectively reducing the reaction time. Compare with these technologies, RPA technology is highly valued for its advantages, including an undemanding primer design strategy, an easily achieved reaction temperature (25–42 °C), and a short reaction time (20 min). Recently, RPA-based biosensors combined with various nanomaterials have shown more rapid, sensitive, and accurate diagnostic performance than traditional RPA. This review summarizes the principle of RPA technology, the construction principles of various RPA biosensors, and their applications in zoonosis screening.
 |
Table 1 Nucleic Acid Amplification Technology |
The Principle of RPA
The RPA reaction system includes three enzymes: recombinase, single-stranded DNA (ssDNA) binding protein, and strand-displacement DNA polymerase. Recombinant enzymes bind to primers, find the homologous sequence in the double-stranded DNA template, and initiate strand substitution by binding the primer to the homologous sequence. To stabilize the binding of the primer to the homologous sequence, a single-strand binding protein binds the displaced DNA strand. Subsequently, the recombinase dissociates, and the 3’ end of the primer is exposed, recognized, and extended by strand-displacement DNA polymerase. Nucleic acid index amplification is achieved by cycling this process15 (Figure 1). Although some studies recommend slightly longer RPA primers than those used in polymerase chain reaction (PCR),16 RPA primers can initially be designed by the software (eg, Primer Premier 5) used for PCR primer design and experimentally screened according to the guidelines provided by the TwistDx Company.17 The temperature of RPA is usually 25–42 °C,18 but an increased reaction temperature can reduce nonspecific amplification. Because a crowding agent (polyethylene glycol) is added to promote the reaction, continuous shaking is needed for the distribution of components.19 RPA showed good suitability in both DNA20 and RNA21 detection when reverse transcriptase and RNase H were added for one-step reverse transcription amplification.22 With nucleic acid purification, sample types can be common specimens such as saliva, stool, blood, and urine.23,24 The flexible applicability of RPA to a variety of samples shows its great potential for on-site screening.
 |
Figure 1 The RPA cycle. |
RPA has several advantages: (1) No need for complex temperature-control instruments: A constant temperature of 25–42 °C is easy to achieve. (2) Short reaction time: RPA can be completed within 20 min. (3) High sensitivity and specificity: Its sensitivity and specificity are comparable to those of PCR. (4) Easy to operate: No professional technician is needed, and RPA can be implemented for home testing.25 (5) The reagents are easy to store and transport: RPA reagents are used in lyophilized form with no need for refrigeration during transport.26 (6) Multiplexing: Multiple detection of different pathogens or genes can be performed in one test.27 (7) Robust adaptability: Excellent tolerance to common inhibitors with simple DNA extraction techniques.28 (8) RPA tolerates primer mismatches with high fault tolerance.29 However, this technology has some shortcomings: (1) There is no dedicated software for designing primers or probes. (2) It is easy to produce aerosol pollution. (3) Excessive template concentrations can inhibit RPA reactions.30
Application of the RPA Biosensor in the Detection of Zoonotic Pathogens
Zoonoses are diseases that undergo natural transmission between vertebrates and humans31 via a variety of routes, such as respiratory, fecal-oral, skin contact, sexual, and arthropod routes. Climate and environmental change, agricultural intensification, frequent animal-human interactions, international trade in food, and the use of underfunded medical treatment systems have all exacerbated the prevalence of zoonoses.32 To date, more than 200 zoonoses have been identified in humans.33 Large-scale zoonotic outbreaks pose a serious threat to public health worldwide with negative impacts on the global economy and sustainable development, such as the H1N1 influenza pandemic (2009), the West African Ebola virus disease (EVD) epidemic (2013–2016), the Congo EVD epidemic (2018–2020), and coronavirus disease 2019 (COVID-19).34 With the advantages of high sensitivity, rapid response, and on-site detection of RPA, accurate screening assays can be useful tools for rapidly controlling the spread of zoonotic epidemics. RPA-based biosensors combined with new nanomaterials provided methods for the early and rapid detection of zoonoses from complex samples. These biosensors involve a variety of signal conversions (Figure 2), including fluorescent signals, lateral flow assays (LFAs), enzymatic catalytic reactions, spectroscopic techniques, electrochemical techniques, chemiluminescence, nanopore sequencing techniques, microfluidic digital RPA, and clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) systems.
 |
Figure 2 Principles of RPA biosensors for various signal conversions. |
RPA Biosensors Based on Fluorescent Signal Output Mode
Fluorescent Probes Used in RPA Biosensors
Similar to the TaqMan probe in PCR, the real-time RPA system contains a fluorophore, a tetrahydrofuran (THF) residue, a quencher, and a blocking structure (SpC3, phosphate) located at the 3’ end of the primer,35 in which fluorescence quenching occurs based on fluorescence resonance energy transfer (FRET), which depends on proximity. As nucleic acid exonuclease (Exo) III cleaves the THF site and separates the fluorophore and the quencher, the fluorescent signal accumulates synchronously with the amplification and can be used for real-time35 or end-point detection.36 There is no need to open the lid throughout the process, which reduces the risk of aerosol pollution as well as false positive. In addition, the fluorophore label on the primer effectively prevented false-positive signals caused by the primer dimer and improved the accuracy of detection.37 Unlike fluorescence quantification in PCR, which requires complex and expensive instruments, RPA can be miniaturized for on-site detection due to its low reaction temperature and low energy input required.38 Therefore, numerous portable fluorescence monitors in RPA-based biosensors have been developed for zoonosis screening (Table 2).
 |
Table 2 RPA Biosensors Developed Using Fluorescence Detection |
A porous membrane paper-based RPA correlates the number of fluorescence signal spots with the concentration of the input HIV template (Figure 3a). The quantitative detection of fluorescence nucleation sites using image analysis algorithms is suitable for field testing via mobile phone-based image capture systems. The proposed biosensor also reduces the impact of primer mismatches on amplification efficiency and time to threshold.29 In addition, a 3D printer was reversibly converted into an automated RNA extraction device to extract nucleic acids from urine specimens within 15 min, and the extruder was used as a heat source to perform real-time reverse transcription RPA (RT-RPA).30 Based on the electrophoretic mobility of the microfluidic paper-based isotachophoresis analysis device for nucleic acid extraction from blood samples, RT-RPA can be used to obtain results in 45 min with a limit of detection (LOD) of 5×103 copies/mL in HIV testing (Figure 3b).23 Since RPA reaction reagents do not require cold chain transportation, a variety of RPA-enabled mobile laboratories have been successfully applied in the detection of zoonotic outbreaks.26,49 The RPA mobile suitcase laboratory for parasite detection via real-time fluorescence quantitation (Q-RPA) allows the rapid extraction of nucleic acids by magnetic beads with a sensitivity similar to that of PCR.49
 |
Figure 3 RPA biosensors based on fluorescent signal output mode. (a) Porous membrane paper-based RPA.29 (b) Microfluidic paper-based sotachophoresis RPA.23 (c) A wearable flexible microfluidic RPA device based on SYBR Green I.50 |
The combination of microfluidic technology and Q-RPA promoted the field detection of pathogens. Mycobacterium tuberculosis is detected by a centrifugal microfluidic chip with an LOD of 1×102 CFU/mL.51 A microfluidic device using a single finger press as a driving force can be used for on-site testing without a centrifugation device.52 Since the RPA reaction temperature is close to human body temperature, a watch-type microfluidic device made of polydimethylsiloxane can perform RPA by using hand temperature for heat transfer. The impact of ambient temperature is reduced through an insulated wristband attached to the wrist. A mobile phone detection system was adopted to implement the HIV test, and the LOD of the biosensor was 1×102 copies/mL.53 However, the use of RPA-enabled fluorescent biosensors usually increases the cost of the assay.
Fluorescent Dyes Used in RPA Biosensors
Commonly used fluorescent dyes can be combined with RPA to visualize the amplification results.50 For SYBR Green I can embed into double-stranded DNA (dsDNA), the RPA amplified product binds to it to form a complex, and the single linear oxygen (1O2) produced by photosensitization of the complex is used to oxidize tetramethylbenzidine (TMB) to emit a distinctly visible fluorescence signal under ultraviolet-visible light. This biosensor is used for Salmonella test with an LOD of 10 copies/μL.54 In addition, using SYBR Green I as an indicator combined with a wearable flexible microfluidic device (Figure 3c), an RPA biosensor triggered by body temperature can detect 10 copies/μL zika virus in 10 min.50 However, SYBR green I affects the background signal of visual analysis.50 Moreover, detection with SYBR Green I or other fluorescent dyes is affected by recombinant enzymes, and the accuracy of this biosensor is not high.55
RPA Biosensors Based on Colorimetric Analysis
LFA-Based Visual RPA Biosensors
LFA is widely used in field tests for zoonoses because of its simplicity, convenience, and visualization of results (Table 3), which typically include sample pads, binding pads, nitrocellulose membranes, absorbent pads, plastic backing, and biological reagents.56 RPA products are labeled with biotin and fluorescein (FAM, etc.), which are added to the sample pad and migrate to the binding pad driven by capillary force. The anti-fluorescein antibody labeled on gold nanoparticles (AuNPs) captures the products and continues to flow forward. Then, the complex is intercepted by streptavidin, and the test line appears red.57 In this biosensor, AuNPs coated with antibodies are the key component of signal conversion in the process of converting the increase in nucleic acid amplification products to color changes caused by the aggregation of AuNPs. The color change based on the surface plasmon resonance (SPR) effect on the surface of AuNPs is easily observed by the naked eye. RPA detection by LFA requires amplification products to be diluted 10–50 times to reduce their viscosity and allow them to flow easily.58 Despite the additional dilution step, LFA-based RPA biosensors are currently the most widely used endpoint assay in amplification product tests.5
 |
Table 3 RPA Biosensors Developed Using Colorimetric Analysis |
To detect multiple pathogen targets simultaneously, a variety of different fluorescent groups (eg, FITC, digoxin) can be added to primers. However, this multiplex detection should have similar-sized targets to prevent biased amplification of dsDNA. The reaction balance can be maintained by changing the buffer concentration or the ratio of primers. Notably, multiple test lines must be placed in a specific order to prevent cross-reactivity.65 Jauset-Rubio Miriam used double RPA in combination with LFA to detect Yersinia pestis and Francisella tularensis genomic DNA, obtaining LODs of 243 fg and 4 fg, respectively. The reaction involved a tailing primer, a fishing probe, and proteinase K. The products obtained with the tailing primer can be hybridized with the capture and reporter probes labeled on AuNPs for direct detection. The use of proteinase K and the fishing probe reduces the cost and eliminates the need for centrifugation.66 In addition, LFA-based RPA allows the quantitative detection of nucleic acids after the introduction of a competitive internal amplification control of known copy numbers, which is used as the control in each reaction to ensure that negative results are not due to reaction inhibition.46 By controlling for similar amplification efficiency between the nucleic acids of the control and the sample to be tested, this method can reliably distinguish the threshold of ≤600 or ≥1400 copies of HIV from 1000 copies.67 However, opening the cap in this biosensor increases the risk of aerosol contamination when adding samples to the reaction. The enclosure of the LFA in a microfluidic chip can solve this problem (Figure 4a), reducing the possible exposure to aerosol contaminants in SARS-CoV-2 testing.22 Generally, LFA has slightly lower sensitivity than Q-RPA.15 The use of soluble barriers between the binding pad, the test line, and the control line can effectively improve the sensitivity of LFA-based RPA.68 In addition, two sets of graphene nanosheets of different sizes were used as reporter probes: the smaller one bound to the analyte, and the larger one underwent additional binding to achieve signal amplification of LFA-based RPA.69
 |
Figure 4 RPA biosensors based on colorimetric analysis. (a) RPA and LFA enclosed in a microfluidic chip.21 (b) TiO2 nanoparticle-enhanced RPA biosensor for colorimetric analysis.20 |
Enzymatic Catalytic Reactions Involved in RPA Biosensors
The use of the enzyme-catalyzed color development of specific substrates allows the visualization of results with high specificity and low cost. Methemoglobin chloride induces the formation of a G-quadruplex deoxyribonuclease with similar activity to horseradish peroxidase from a “G”-rich sequence, which catalyzes substrates to produce a visible blue color with characteristic absorbance at 450 nm.17 As a typical DNA nanomaterial with enzymatic catalytic activity, the application of G-quadruplex in RPA biosensors solves the problem of dsDNA-ssDNA conversion and color identification in amplification technology.20 TiO2 nanoparticles used in RPA biosensors achieved an LOD of 4 CFU/mL when detecting Salmonella in colorimetric analysis by adsorbing primers to aggregate into larger nanoclusters, thereby increasing the partial concentration to enhance RPA. The 5’ end of the forward primer contains an endonuclease recognition region and a G-quadruplex sequence (attached to TiO2 nanoparticles). After amplification, the bifunctional structural domain is embedded in dsDNA products. The endonuclease binds to the specific region for cleavage, and the polymerase induces strand shift and extension, generating a single-stranded G-quadruplex DNA. Permethemoglobin chloride induces the formation of G-quadruplex deoxyribonuclease with enzymatic catalytic activity to promote the color development of TMB (Figure 4b). Moreover, a label-free visual RPA biosensor enhanced by a double-strand specific enzyme and terminal deoxynucleotidyl transferase can detect Salmonella at 6 CFU/mL.17 In addition, RPA products recognized by CRISPR-dCas9 and amplified by RCA ultimately produced G-quadruplets, which catalyzed the color development of 2,2’-azino-bis (3-ethyl-benzothiazoline-6-sulfonic acid).70 This biosensor was used for the detection of Leishmania with a sensitivity of 1 zeptomole at 23 °C. However, the variety of substrates currently used for enzymatic reactions is limited, so multiplex assays are challenging.
RPA Biosensors Based on Spectroscopic Analysis
Surface-Enhanced Raman Scattering (SERS) Used in RPA Biosensors
Recently, SERS has become a commonly used technology for inelastic light scattering sensing, and two mechanisms are generally agreed upon: electromagnetic field enhancement (local electric field enhancement on metal surfaces) and chemical enhancement (electron transfer between metals and molecules).71 When molecules are adsorbed on corrugated metal surfaces, such as silver or AuNPs, the signal in SERS can be greatly enhanced by a factor of 108 or more, allowing even single-molecule detection.72 An RPA-integrated microfluidic paper analysis device was designed using SERS and CRISPR/Cas12a, which was applied in Salmonella typhimurium testing with a sensitivity of 1 CFU/mL.73 A conjugated gold nanopillar-4-mercaptobenzoic acid-gold nanoshell probe was designed as a signal conversion element, which linked ssDNA hybridized to the complexes with surface-adsorbed DNA 1 and DNA 2. In the presence of the target gene, the target DNA activates the trans cleavage of CRISPR/Cas12a, resulting in a dispersed colloidal solution of the nanoprobe due to the breakage of ssDNA. In contrast, the nanoprobe is aggregated due to cross-linking via ssDNA in the absence of the target DNA. The change from dispersion to aggregation was determined by SERS, and the quantitative measurement of pathogens was achieved within 45 min.57 However, SERS requires special optical instruments, and the characteristic peaks of labeled amplicons need to be determined experimentally beforehand.74 At present, substrates for SERS are mainly precious metals, which limits the application of SERS in biosensors. A study showed that a conductive glass/Ag/zeolite imidazole skeleton (FTO/Ag/ZIF-8) sandwich structure may serve as a substitute for SERS substrates.75
Surface-Enhanced Infrared Absorption Spectroscopy (SEIRA) Used in RPA Biosensors
SEIRA is an extended infrared spectroscopy technology using molecular vibrations coupled with surface equivalent excitation resonance to enhance the infrared signal of the molecule to be measured by 103–106 times.76 Gold and silver nanomaterials are widely used in SEIRA due to their high stability and proper dielectric properties.77–80 An infrared spectroscopic biosensor based on RPA was proposed to detect SARS-CoV-2 using the SEIRA effect on AuNPs.81 The hybrid was formed by using the complementary DNA probe to capture the target nucleotide, and the SEIRA signal of the chemical environment change of the functional group was detected. When this method is combined with RPA, 2.98 copies/μL SARS-CoV-2 can be detected within 30 min. Moreover, a sufficient distance from the gold surface to the first base of the DNA probe is important to facilitate the hybridization of the probe with the target. However, a disadvantage of this method is the need to purify the RPA product, which adds an additional step. SEIRA suffers from the same problem as SERS in that the substrates are primarily precious metals, although other nanomaterials such as indium tin oxide nanoparticles have been proposed as alternatives.82
Hyperspectral Interferometry Used in RPA Biosensors
Hyperspectral interferometry detects the material with nanometer-scale spectral images based on the fundamental principle of the Fourier transform of spectroscopy, each pixel of which contains a continuous spectral curve that allows identification of the substance corresponding to the object.83 The 3D image data generated by satellite push scans of hyperspectral imaging extend beyond the visible spectrum at very high resolution, allowing finer spectral structures to be measured.84 The solid-phase self-interference RPA chip (1.8 mm × 1.8 mm) was combined with hyperspectral interference. After RPA, the solid-phase primer attached to the chip captured the target fragment, causing a redshift of the hyperspectral interference signal. The increased optical length of the RPA chip was used to demonstrate the existence of the target. The system can detect 6 copies/reaction of Plasmodium falciparum DNA in 20 min, and the cost is only 1/50 of that using Q-RPA. Since the chip is disposable, the cost of a single reaction can be further reduced through a multiplex RPA chip.85 The optical components used in imagers for hyperspectral interferometry are expensive, and the imagers rely on precise scanning to generate 3D data, which is not conducive to field inspection in resource-poor settings, although the spectral reconstruction algorithm to extract spectral information from RGB images may be a new alternative method.83
RPA Biosensors Based on Electrochemical Analysis
The principle of electrochemical technologies is to use solid-based electrodes incorporating biosensitive molecules to immobilize one or more reaction components onto the electrode surface, specifically recognizing and trapping target molecules on the electrode surface to initiate redox reactions.86 The electrode acts as a signal transducer to convert the reaction signal into an electrical signal that can be measured. A wash-free and rapid electrochemical method is described to detect RPA-amplified dsDNAs using a zinc finger protein (Figure 5a). Electrochemical detection is achieved using proximity-dependent electron mediation of ferrocenemethanol between a glucose-oxidase label and an electrode, which differentiates the specifically electrode-bound or -unbound labels without a washing or purification step. The LOD of the biosensor was 300 copies (13.2 μL) for the detection of Rickettsia salmonis, with the entire assay completed in 17 min. However, a certain degree of nonspecific amplification is generated at the affinity-modified electrode, with centrifugal filtration required to remove the electroactive dithiothreitol, which adds a reaction step.87 In addition, a reagent-free DNA sensor enhanced by RPA was developed using a combination of ferrocene and reporter probe-modified AuNPs and modified primers (Figure 5b). RPA was performed using primer binding sites and C3 spacer-modified primers to produce amplicons with single-stranded tails at both ends. These tails were designed to be complementary to the capture probe immobilized on the gold electrode and the reporter probe on AuNPs. The demonstrated generic electrochemical genosensor can be exploited for the detection of any DNA sequence.88 In addition, a nanosensor of size 2 mm × 10 mm was immersed in a 150 μL RPA system to analyze E. coli O157:H7 through the change in solution impedance during the reaction (Figure 5c), showing that one copy of gDNA can be detected within 5 min. However, the large reaction system increases the cost of this biosensor.89 RPA is combined with microbead-mediated electrophoresis, and the number of amplicons is detected by dielectrophoretic impedance measurement.90 Biotinylated probes are recognized and captured by streptavidin-labeled magnetic microbeads, with amplicons generated by RPA attached to the beads. Electrical measurements are carried out to obtain amplification results when the beads are added to the microelectrodes. The LOD of this biosensor was 2 copies/reaction with a total detection time of 26 min for antimicrobial-resistant E. coli testing. In addition, using the reaction conditions of RPA close to body temperature, a wearable device based on an electrochemical biosensor with a multimicroelectrode array was established for the detection of SARS-CoV-2, and 0.972 fg/μL (RdRP gene) and 3.925 fg/μL (N gene) genomes can be detected within 40 min.91 The main problems of electrochemical technologies are poor stability and limitations in field applications. The use of micro- and nanoprocessing techniques may be an opportunity to reduce the size of electrochemical biosensors to make them applicable for on-site testing.92
 |
Figure 5 RPA biosensors based on electrochemical analysis. (a) RPA-based washing-free electrochemical biosensor using a zinc finger protein.87 (b) A reagent-free DNA sensor enhanced by RPA based on ferrocene and primer decorated AuNPs.88 (c) RPA-based nanogap impedimetric sensor for real-time DNA monitoring.89 |
Chemiluminescence-Based RPA Biosensors
The chemiluminescence (CL) method uses radioluminescence emitted during the conversion of electrons from the excited state to the ground state during a chemical reaction, with low interference, high specificity, and a higher signal-to-noise ratio than fluorescence.93 A CL microfluidic chip combined with heterogeneous asymmetric RPA achieved highly sensitive detection of Legionella with an LOD of 87 genomic units/μL.94 First, the samples are pretreated with photoactivated propidium monoazide, and this step allows us to distinguish viable and nonviable Legionella. Subsequently, the assay is performed on a microarray, and quantitative detection can be achieved by point amplification. The reverse primer is immobilized on the microarray, and the forward primer is encapsulated with biotin, which is linked to horseradish peroxidase by streptavidin and finally catalyzes the color development of the substrate. However, light emission is completed in a few seconds in most CL systems, so data acquisition and imaging time are limited. In recent years, continuous luminescence emitted for a long time has become a new optical imaging modality. For CL requires high instrumentation, camera equipment with high resolution, high sensitivity, and high interference immunity is necessary for CL imaging applications.95
RPA Biosensors Based on Nanopore Sequencing Technology
Nanopore sequencing technology is a third-generation sequencing method that allows the sequencing of individual DNA or RNA molecules. The principle of this technology is to sequence different nucleotide bases based on changes in electrical signals generated when they pass through a nanopore, which can enable on-site testing by portable nanopore sequencing devices.96 Combining RPA with nanopore sequencing technology for Mycobacterium tuberculosis detection is comparable in accuracy and specificity to PCR technology combined with Illumina sequencing methods. This RPA biosensor is rapid, low-cost and can be automated for resource-poor settings.97 The nanopore sequencing technology R9.4.1 has relatively low processing power for homopolymers, so the sequence accuracy is low. Although the latest Oxford Nanopore R10.4 sequence accuracy is up to 99%, its cost is high.98 These drawbacks limit the wide application of nanopore sequencing technology in RPA-based biosensors.
Microfluidic Digital RPA Biosensors
Digital RPA (dRPA) achieves absolute quantification of nucleic acids by uniformly dividing them into multiple individual reaction units (eg, chambers or droplets), with the advantages of low equipment needs, short elapsed time, and high sensitivity and specificity.6 Combining dRPA with a microfluidic chip can build a low-cost, high-throughput quantifiable thermostatic detection platform. An integrated multidigital RPA microfluidic chip combines DNA extraction, multidigital RPA, and fluorescence detection in a system for quantitative detection with no need to create a standard curve.99 Nucleic acid extraction by magnetic beads can be performed in 15 min without any instrumentation. Reaction components are prepositioned in different areas of the chamber by means of a spiral valve, and the reagents are passively driven to the dRPA zone for detection using a fluorometer. The LOD for E. coli O157:H7, Listeria monocytogenes, and Salmonella enterica is 10 bacterial cells.99 A fully automated digital centrifugation microfluidic platform has been created to automate the entire process from swabbing nucleic acids to reading results in 55 min.100 The difficulty of microfluidic dRPA is accurate initiation, and the additional time of magnesium acetate can seriously affect the amplification reaction. By coupling a microfluidic microinjector with a droplet generator, the addition time of magnesium acetate can be controlled to achieve absolute quantification of nucleic acids.101 The second drawback is the narrow dynamic range. The lower detection limit of microfluidic dRPA is determined mainly by the total number of chambers or droplets and the volume of individual chambers or droplets, and the upper detection limit is determined mainly by the total volume of chambers or droplets. Increasing the number and total volume of chambers in a limited space can help to improve the dynamic range and sensitivity.6
CRISPR/Cas-Enhanced RPA Biosensors
RPA can rapidly amplify DNA exponentially with high sensitivity but is susceptible to nonspecific amplification. Coupling RPA and CRISPR Cas provides a new approach to solve this problem.102 CRISPR is a microbial adaptive immune system in most bacteria and all archaea. CRISPR RNA (crRNA) directs CRISPR-associated Cas proteins to recognize and cleave the target nucleic acid, ultimately separating the fluorescent and bursting moieties and emitting fluorescence.103 The commonly used Cas proteins include Cas9, Cas12a, Cas12b, Cas13a, Cas13b, and Cas14, which exhibit side strand cleavage activity upon binding to their specific targets. After target recognition, the activated Cas nuclease cleaves nearby nucleic acids nonspecifically. By introducing nucleic acid reporters labeled with fluorophores into the reaction, cleavage can be detected with fluorescence intensity or AuNP-based LFA.104 Based on their side chain cleavage activity, universal RNA reporter probes can be designed for the specific detection of target molecules to reduce reaction costs.105 Cas9 specifically binds and cleaves nearby DNA through a guided RNA recognizing the protospacer adjacent motif (PAM) in the target DNA. Cas12a and Cas12b are RNA-directed DNases that recognize T-rich PAM sites and can directly use DNA as a substrate.106 However, Cas12a can only recognize T-rich conventional PAMs, such as TTTV sequences, limiting its application. The Cas12a-based suboptimal PAM approach greatly expands the available options for crRNA.107 Cas13a and Cas13b are RNA-directed RNA enzymes that require transcription by T7 RNA polymerase to convert amplified DNA into RNA for detection and do not require a specific PAM to recognize the target RNA.108 After completing cleavage, they can remain active and cleave other nontarget RNAs. Cas14 is an RNA-primed targeted ssDNA nuclease that has a small molecular weight and excellent specific recognition of ssDNA and is not restricted by the PAM site. When activated, it can be associated with the nonspecific cleavage activity of ssDNA.
Currently, the CRISPR/Cas system is widely used in RPA biosensors (Table 4). In the one-step detection system of the biobarcode assay in conjunction with RPA and CRISPR-Cas12a, the signal is triple-amplified to achieve a single-colony limit of detection of Salmonella.109 Although the antigen–antibody reaction in this biosensor is highly specific, the CRISPR/Cas reaction requires repeated lid opening, increasing the risk of aerosol generation. Integrating RPA and the CRISPR/Cas system into a single pot can effectively reduce the problems of nonspecific amplification and cross-contamination. A one-step detection platform (iSCAN-V2) was designed to visualize the easily observed signal of light-emitting diodes through the side chain cleavage of HEX-labeled ssDNA reporter genes.110 CRISPR/Cas12a-assisted one-step RPA in SARS-CoV-2 detection provides approximately 4–5 times higher sensitivity than digital RT-PCR, showing significant advantages in detection time and isothermal reaction conditions.111 In addition, a paper-based multiplex RPA based on a sucrose valve was established to control the opening of the valve between the amplification chamber and the CRISPR detection chamber through the dissolution of sucrose (Figure 6a). By controlling the rate of sucrose dissolution, the valve opening time can be controlled, which has great potential for the rapid, sensitive, and reliable multimolecular diagnosis of infectious diseases in resource-limited environments.102
 |
Figure 6 CRISPR/Cas-enhanced RPA biosensors. (a) CRISPR/Cas12a-enabled autonomous paper-based laboratory platform.102 (b) Single-tube orthogonal Cas12a/Cas13a assay.121 |
 |
Table 4 RPA Biosensors Developed Using the CRISPR/Cas System |
Due to the lack of a specific probe cleavage mechanism for CRISPR/Cas12a and CRISPR/Cas13a, their respective multiplex detection capabilities are limited. Combining Cas12a analysis and T7 transcription-mediated Cas13a analysis allows dual analysis in a single tube (Figure 6b).121 However, when detecting RNA, the hybridization of cDNA-RNA results in slow initiation, the crRNA-Cas12a ribonucleoprotein is gradually inactivated, and nonspecific amplification consumes RPA reagents, resulting in low sensitivity.122 Overcoming this problem requires the rapid production of DNA products from trace RNA templates and the rapid initiation of RPA. The removal of RNA from cDNA-RNA hybrids using the endocytosis function of RNase H solves this problem without compromising the transcriptional activity of Cas12a.123 However, in the one-pot reaction, CRISPR detection competes with RPA, but the final detection signal is dependent on target amplification to generate sufficient substrate for CRISPR detection. Therefore, it is crucial to balance the processes of both.107 In addition, increasing the Mg2+ concentration can improve the sensitivity of the test, as the increase in Mg2+ concentration enhances the hybridization between primers and DNA templates. The statistical design of experiments can also be used to optimize the one-step-based assay.108 Mostly, the detection of CRISPR/Cas systems is based on fluorescence monitoring of the target. However, incomplete quenching of fluorescent moieties and unstable output signals are challenges for the application of this system. The development of CRISPR/Cas-enhanced RPA biosensors combining spectroscopic techniques, chemiluminescence, and electrochemical signals is an effective way to solve these problems.124
Future Perspectives and Conclusions
RPA has gradually become a substitute for PCR and stands out among various nucleic acid technologies because it involves a rapid, compatible, constant temperature reaction (25–42 °C) with a short reaction time (20 min). Due to its natural suitability for field testing, RPA has been extensively studied in the context of zoonoses, although there are still some drawbacks. First, RPA suffers from a certain false-positive rate. An appropriate increase in temperature can reduce this rate. The combination of CRISPR/Cas may also improve the specificity. In addition, there is no universal primer design software, although traditional PCR primers have been shown to work in many studies. Finally, the complex process of nucleic acid extraction from samples limits its application in the field. As RPA has a good tolerance to inhibitors, nucleic acids can be extracted by thermal lysis to simplify the pretreatment step. It is worth noting that the rapid development of new nanomaterials has provided support for the development of integrated detection devices for RPA signal sensitivity and nucleic acid extraction, including functional DNA nanomaterials, precious metal nanomaterials, and composite nanomaterials. By combining fluorescence detection, optical discoloration, spectral analysis, electrochemical technology, chemiluminescence, nanopore sequencing technologies, digital microfluidics or the CRISPR/Cas system, traditional DNA amplification signals in RPA are transformed into fluorescent, optical or electrical signals for rapid, sensitive and accurate detection. The application of novel nanomaterials and multiple signal output modes aims to reduce the nonspecificity, enable multiplexed detection in a one-step reaction, and further enable the portability of detection devices.
Periodic outbreaks of zoonotic diseases pose a serious threat to the global environment, food safety, and animal and human health. The rapid and sensitive screening of zoonoses is essential to control the spread of pathogens from the environment, water, food, and animals to humans. The pretreatment methods of samples from different sources are inconsistent, and the background compositions are complex, including sludge, water, meat, milk, feces, throat swabs, whole blood, and serum. Compared with limited clinical assays for human samples, the detection of zoonotic pathogens faces greater challenges, which puts forward higher requirements for the stability and accuracy of detection methods in zoonosis screening. At present, there are few relevant studies on different sample sources, and the tolerance of RPA biosensors to different types of sample processing needs to be further verified. In the future, research on RPA-based biosensors will mainly focus on the development of wearable sensors, multiplex high-throughput biosensors, one-step biosensors, visualization biosensors, and point-of-care detection to adapt to the increasingly complex zoonosis prevention and control situation. RPA is another leap forward in nucleic acid technology, which is expected to play an important role in the rapid detection of human-animal diseases in the near future, and is widely used in field screening to quickly and accurately identify pathogens and prevent the occurrence of disease pandemics.
Author Contributions
All authors made significant contributions to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Funding
This study was supported by National College Students’ innovation and entrepreneurship training program (S202213706035; S202313706015); the Foundation of Science and Technology Department of Jilin Province (20200404175YY); the Foundation of Ph.D. Research Project of the Jilin Medical University of Jilin Province (2022JYBS011KJ).
Disclosure
The authors have no conflicts of interest to declare for this work.
References
1. Tian D, Sun Y, Xu H, et al. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J Med Virol. 2022;94(6):2376–2383. doi:10.1002/jmv.27643
2. World Health Organization, Food and Agriculture Organization of the United Nations, World Organisation for Animal Health & United Nations Environment Programme. One health joint plan of action (2022‒2026): working together for the health of humans, animals, plants and the environment. Available from: https://www.who.int/publications/i/item/9789240059139.
3. Carpenter A, Waltenburg MA, Hall A, et al. Vaccine Preventable Zoonotic Diseases: challenges and Opportunities for Public Health Progress. Vaccines. 2022;10(7). doi:10.3390/vaccines10070993
4. Zhou D, Wang S, Yang K, et al. Rapid and simultaneous detection of Japanese encephalitis virus by real-time nucleic acid sequence-based amplification. Micro Pathog. 2021;150:104724. doi:10.1016/j.micpath.2020.104724
5. Reid P, Heng N, Hale JD, et al. A TaqMan-based quantitative PCR screening assay for the probiotic Streptococcus salivarius K12 based on the specific detection of its megaplasmid-associated salivaricin B locus. J Microbiol Methods. 2020;170:105837. doi:10.1016/j.mimet.2020.105837
6. Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. Trends Analyt Chem. 2018;98:19–35. doi:10.1016/j.trac.2017.10.015
7. Zhang N, Li C, Dou X, et al. Overview and Future Perspectives of Microfluidic Digital Recombinase Polymerase Amplification (dRPA). Crit Rev Anal Chem. 2022;52(8):1969–1989. doi:10.1080/10408347.2022.2042669
8. Soroka M, Wasowicz B, Rymaszewska A. Loop-Mediated Isothermal Amplification (LAMP): the Better Sibling of PCR? Cells. 2021;10(8). doi:10.3390/cells10081931
9. Xu G, Hu L, Zhong H, et al. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Sci Rep. 2012;2:246. doi:10.1038/srep00246
10. Myrmel M, Oma V, Khatri M, et al. Single primer isothermal amplification (SPIA) combined with next generation sequencing provides complete bovine coronavirus genome coverage and higher sequence depth compared to sequence-independent single primer amplification (SISPA). PLoS One. 2017;12(11). doi:10.1371/journal.pone.0187780
11. He F, Zhou W, Cai R, et al. Systematic assessment of the performance of whole-genome amplification for SNP/CNV detection and β-thalassemia genotyping. J Hum Genet. 2018;63(4):407–416. doi:10.1038/s10038-018-0411-5
12. Carter JG, Orueta Iturbe L, Duprey JHA, et al. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc Natl Acad Sci USA. 2021;118(35). doi:10.1073/pnas.2100347118
13. Wang X, Liu Y, Liu H, et al. Recent advances and application of whole genome amplification in molecular diagnosis and medicine. MedComm. 2022;3(1). doi:10.1002/mco2.116
14. Mota DS, Guimaraes JM, Gandarilla A, et al. Recombinase polymerase amplification in the molecular diagnosis of microbiological targets and its applications. Can J Microbiol. 2022;68(6):383–402. doi:10.1139/cjm-2021-0329
15. Behrmann O, Bachmann I, Spiegel M, et al. Rapid Detection of SARS-CoV-2 by Low Volume Real-Time Single Tube Reverse Transcription Recombinase Polymerase Amplification Using an Exo Probe with an Internally Linked Quencher (Exo-IQ). Clin Chem. 2020;66(8):1047–1054. doi:10.1093/clinchem/hvaa116
16. Zhao L, Wang J, Sun XX, et al. Development and Evaluation of the Rapid and Sensitive RPA Assays for Specific Detection of Salmonella spp. in Food Samples. Front Cell Infect Microbiol. 2021;11:631921. doi:10.3389/fcimb.2021.631921
17. TwistAmp® manuals. Available from: https://www.twistdx.co.uk/en/support/manuals/twistamp-manuals.
18. Zhang Y, Tian J, Li K, et al. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal Chim Acta. 2019;1075:144–151. doi:10.1016/j.aca.2019.05.020
19. Tran DH, Tran HT, Pham T, et al. Direct multiplex recombinase polymerase amplification for rapid detection of S taphylococcus aureus and P seudomonas aeruginosa in food. Mol Biol Res Commum. 2022;11(1):1–10. doi:10.22099/mbrc.2021.41503.1664
20. Tian J, Chu H, Zhang Y, et al. TiO2 Nanoparticle-Enhanced Linker Recombinant Strand Displacement Amplification (LRSDA) for Universal Label-Free Visual Bioassays. ACS Appl Mater Interfaces. 2019;11(50):46504–46514. doi:10.1021/acsami.9b16314
21. Liu D, Shen H, Zhang Y, et al. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab Chip. 2021;21(10):2019–2026. doi:10.1039/d0lc01222j
22. Qian J, Boswell SA, Chidley C, et al. An enhanced isothermal amplification assay for viral detection. Nat Commum. 2020;11(1):5920. doi:10.1038/s41467-020-19258-y
23. Bender AT, Sullivan BP, Zhang JY, et al. HIV detection from human serum with paper-based isotachophoretic RNA extraction and reverse transcription recombinase polymerase amplification. Analyst. 2021;146(9):2851–2861. doi:10.1039/d0an02483j
24. Chen J, Xu Y, Yan H, et al. Sensitive and rapid detection of pathogenic bacteria from urine samples using multiplex recombinase polymerase amplification. Lab Chip. 2018;18(16):2441–2452. doi:10.1039/c8lc00399h
25. Tan KK, Azizan NS, Yaacob VN, et al. Operational utility of the reverse-transcription recombinase polymerase amplification for detection of dengue virus. BMC Infect Dis. 2018;18(1):169. doi:10.1186/s12879-018-3065-1
26. Weidmann M, Faye O, Faye O, et al. Development of Mobile Laboratory for Viral Hemorrhagic Fever Detection in Africa. J Infect Dis. 2018;218(10):1622–1630. doi:10.1093/infdis/jiy362
27. Xiong E, Jiang L, Tian T, et al. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew Chem Int Ed Engl. 2021;60(10):5307–5315. doi:10.1002/anie.202014506
28. Moore MD, Jaykus LA. Development of a Recombinase Polymerase Amplification Assay for Detection of Epidemic Human Noroviruses. Sci Rep. 2017;7:40244. doi:10.1038/srep40244
29. Sullivan BP, Chou YS, Bender AT, et al. Quantitative isothermal amplification on paper membranes using amplification nucleation site analysis. Lab Chip. 2022;22(12):2352–2363. doi:10.1039/d2lc00007e
30. Chan K, Weaver SC, Wong PY, et al. Rapid, Affordable and Portable Medium-Throughput Molecular Device for Zika Virus. Sci Rep. 2016;6:38223. doi:10.1038/srep38223
31. Leal FW, Ternova L, Parasnis SA, et al. Climate Change and Zoonoses: a Review of Concepts, Definitions, and Bibliometrics. Int J Environ Res Public Health. 2022;19(2). doi:10.3390/ijerph19020893
32. Fouque F, Reeder JC. Impact of past and on-going changes on climate and weather on vector-borne diseases transmission: a look at the evidence. Infect Dis Poverty. 2019;8(1):51. doi:10.1186/s40249-019-0565-1
33. Conrady B. Epidemiological, Mitigation and Economic Impact of Zoonoses. Int J Environ Res Public Health. 2021;18(21). doi:10.3390/ijerph182111704
34. Judson SD, Rabinowitz PM. Zoonoses and global epidemics. Curr Opin Infect Dis. 2021;34(5):385–392. doi:10.1097/QCO.0000000000000749
35. Yang M, Ke Y, Wang X, et al. Development and Evaluation of a Rapid and Sensitive EBOV-RPA Test for Rapid Diagnosis of Ebola Virus Disease. Sci Rep. 2016;6:26943. doi:10.1038/srep26943
36. Ahn H, Batule BS, Seok Y, et al. Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal Chem. 2018;90(17):10211–10216. doi:10.1021/acs.analchem.8b01309
37. Wang L, Zhao P, Si X, et al. Rapid and Specific Detection of Listeria monocytogenes With an Isothermal Amplification and Lateral Flow Strip Combined Method That Eliminates False-Positive Signals From Primer-Dimers. Front Microbiol. 2019;10:2959. doi:10.3389/fmicb.2019.02959
38. Davi SD, Kissenkotter J, Faye M, et al. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn Microbiol Infect Dis. 2019;95(1):41–45. doi:10.1016/j.diagmicrobio.2019.03.015
39. Dieng I, Hedible BG, Diagne MM, et al. Mobile Laboratory Reveals the Circulation of Dengue Virus Serotype I of Asian Origin in Medina Gounass (Guediawaye), Senegal. Diagnostics. 2020;10(6). doi:10.3390/diagnostics10060408
40. Faye M, Abd EWA, Faye O, et al. A recombinase polymerase amplification assay for rapid detection of rabies virus. Sci Rep. 2021;11(1):3131. doi:10.1038/s41598-021-82479-8
41. Yehia N, Eldemery F, Arafa AS, et al. Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid Detection of Avian Influenza Virus H9N2 HA Gene. Vet Sci. 2021;8(7). doi:10.3390/vetsci8070134
42. Tomar PS, Kumar S, Patel S, et al. Development and Evaluation of Real-Time Reverse Transcription Recombinase Polymerase Amplification Assay for Rapid and Sensitive Detection of West Nile Virus in Human Clinical Samples. Front Cell Infect Microbiol. 2020;10:619071. doi:10.3389/fcimb.2020.619071
43. Yoo H, Lee JY, Park KS, et al. Lead-start isothermal polymerase amplification controlled by DNAzymatic switches. Nanoscale. 2022;14(21):7828–7836. doi:10.1039/d1nr07894a
44. Archer J, Barksby R, Pennance T, et al. Analytical and Clinical Assessment of a Portable, Isothermal Recombinase Polymerase Amplification (RPA) Assay for the Molecular Diagnosis of Urogenital Schistosomiasis. Molecules. 2020;25(18). doi:10.3390/molecules25184175
45. Yang X, Zhang X, Wang Y, et al. A Real-Time Recombinase Polymerase Amplification Method for Rapid Detection of Vibrio vulnificus in Seafood. Front Microbiol. 2020;11:586981. doi:10.3389/fmicb.2020.586981
46. Garrido-Maestu A, Azinheiro S, Fucinos P, et al. Comparative study of multiplex real-time recombinase polymerase amplification and ISO 11290-1 methods for the detection of Listeria monocytogenes in dairy products. Food Microbiol. 2020;92:103570. doi:10.1016/j.fm.2020.103570
47. Guo M, Feng P, Zhang L, et al. Rapid Detection of Clostridium tetani by Recombinase Polymerase Amplification Using an Exo Probe. J Microbiol Biotechnol. 2022;32(1):91–98. doi:10.4014/jmb.2109.09022
48. Zheng Y, Hu P, Ren H, et al. RPA-SYBR Green I based instrument-free visual detection for pathogenic Yersinia enterocolitica in meat. Anal Biochem. 2021;621:114157. doi:10.1016/j.ab.2021.114157
49. Gunaratna G, Manamperi A, Böhlken-Fascher S, et al. Evaluation of rapid extraction and isothermal amplification techniques for the detection of Leishmania donovani DNA from skin lesions of suspected cases at the point of need in Sri Lanka. Parasit Vectors. 2018;11(1):665. doi:10.1186/s13071-018-3238-1
50. Yang B, Kong J, Fang X. Bandage-like wearable flexible microfluidic recombinase polymerase amplification sensor for the rapid visual detection of nucleic acids. Talanta. 2019;204:685–692. doi:10.1016/j.talanta.2019.06.031
51. Law I, Loo J, Kwok HC, et al. Automated real-time detection of drug-resistant Mycobacterium tuberculosis on a lab-on-a-disc by Recombinase Polymerase Amplification. Anal Biochem. 2018;544:98–107. doi:10.1016/j.ab.2017.12.031
52. Wang Z, Wang Y, Lin L, et al. A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis. Biosens Bioelectron. 2022;195:113663. doi:10.1016/j.bios.2021.113663
53. Kong M, Li Z, Wu J, et al. A wearable microfluidic device for rapid detection of HIV-1 DNA using recombinase polymerase amplification. Talanta. 2019;205:120155. doi:10.1016/j.talanta.2019.120155
54. Li X, Zheng T, Xie YN, et al. Recombinase Polymerase Amplification Coupled with a Photosensitization Colorimetric Assay for Fast Salmonella spp. Testing. Anal Chem. 2021;93(16):6559–6566. doi:10.1021/acs.analchem.1c00791
55. Cao Y, Zheng K, Jiang J, et al. A novel method to detect meat adulteration by recombinase polymerase amplification and SYBR green I. Food Chem. 2018;266:73–78. doi:10.1016/j.foodchem.2018.05.115
56. Zhang Y, Chai Y, Hu Z, et al. Recent Progress on Rapid Lateral Flow Assay-Based Early Diagnosis of COVID-19. Front Bioeng Biotechnol. 2022;10:866368. doi:10.3389/fbioe.2022.866368
57. Liu HB, Du XJ, Zang YX, et al. SERS-Based Lateral Flow Strip Biosensor for Simultaneous Detection of Listeria monocytogenes and Salmonella enterica Serotype Enteritidis. J Agric Food Chem. 2017;65(47):10290–10299. doi:10.1021/acs.jafc.7b03957
58. Li J, Zhong Q, Shang MY, et al. Preliminary Evaluation of Rapid Visual Identification of Burkholderia pseudomallei Using a Newly Developed Lateral Flow Strip-Based Recombinase Polymerase Amplification (LF-RPA) System. Front Cell Infect Microbiol. 2021;11:804737. doi:10.3389/fcimb.2021.804737
59. Xu Y, Wei Y, Cheng N, et al. Nucleic Acid Biosensor Synthesis of an All-in-One Universal Blocking Linker Recombinase Polymerase Amplification with a Peptide Nucleic Acid-Based Lateral Flow Device for Ultrasensitive Detection of Food Pathogens. Anal Chem. 2018;90(1):708–715. doi:10.1021/acs.analchem.7b01912
60. Xi Y, Xu CZ, Xie ZZ, et al. Rapid and visual detection of dengue virus using recombinase polymerase amplification method combined with lateral flow dipstick. Mol Cell Probes. 2019;46:101413. doi:10.1016/j.mcp.2019.06.003
61. Huang P, Jin H, Zhao Y, et al. Nucleic acid visualization assay for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) by targeting the UpE and N gene. PloS Negl Trop Dis. 2021;15(3):e9227. doi:10.1371/journal.pntd.0009227
62. James AS, Todd S, Pollak NM, et al. Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: preparing for the future. Virol J. 2018;15(1):75. doi:10.1186/s12985-018-0985-8
63. Nie Z, Zhao Y, Shu X, et al. Recombinase polymerase amplification with lateral flow strip for detecting Babesia microti infections. Parasitol Int. 2021;83:102351. doi:10.1016/j.parint.2021.102351
64. Lai MY, Ooi CH, Lau YL. Recombinase Polymerase Amplification Combined with a Lateral Flow Strip for the Detection of Plasmodium knowlesi. Am J Trop Med Hyg. 2018;98(3):700–703. doi:10.4269/ajtmh.17-0738
65. Wang P, Liao L, Ma C, et al. Duplex On-Site Detection of Vibrio cholerae and Vibrio vulnificus by Recombinase Polymerase Amplification and Three-Segment Lateral Flow Strips. Biosensors. 2021;11(5). doi:10.3390/bios11050151
66. Jauset-Rubio M, Tomaso H, El-Shahawi MS, et al. Duplex Lateral Flow Assay for the Simultaneous Detection of Yersinia pestis and Francisella tularensis. Anal Chem. 2018;90(21):12745–12751. doi:10.1021/acs.analchem.8b03105
67. Hull IT, Kline EC, Gulati GK, et al. Isothermal Amplification with a Target-Mimicking Internal Control and Quantitative Lateral Flow Readout for Rapid HIV Viral Load Testing in Low-Resource Settings. Anal Chem. 2022;94(2):1011–1021. doi:10.1021/acs.analchem.1c03960
68. Alam N, Tong L, He Z, et al. Improving the sensitivity of cellulose fiber-based lateral flow assay by incorporating a water-dissolvable polyvinyl alcohol dam. Cellulose. 2021;28(13):8641–8651. doi:10.1007/s10570-021-04083-3
69. Napione L. Integrated Nanomaterials and Nanotechnologies in Lateral Flow Tests for Personalized Medicine Applications. Nanomaterials. 2021;11(9). doi:10.3390/nano11092362
70. Bengtson M, Bharadwaj M, Franch O, et al. CRISPR-dCas9 based DNA detection scheme for diagnostics in resource-limited settings. Nanoscale. 2022;14(5):1885–1895. doi:10.1039/d1nr06557b
71. Serebrennikova KV, Berlina AN, Sotnikov DV, et al. Raman Scattering-Based Biosensing: new Prospects and Opportunities. Biosensors. 2021;11(12). doi:10.3390/bios11120512
72. Langer J, Jimenez DAD, Aizpurua J, et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano. 2020;14(1):28–117. doi:10.1021/acsnano.9b04224
73. Zhuang J, Zhao Z, Lian K, et al. SERS-based CRISPR/Cas assay on microfluidic paper analytical devices for supersensitive detection of pathogenic bacteria in foods. Biosens Bioelectron. 2022;207:114167. doi:10.1016/j.bios.2022.114167
74. Wang J, Koo KM, Wee EJ, et al. A nanoplasmonic label-free surface-enhanced Raman scattering strategy for non-invasive cancer genetic subtyping in patient samples. Nanoscale. 2017;9(10):3496–3503. doi:10.1039/c6nr09928a
75. Xue X, Chen L, Zhao C, et al. Tailored FTO/Ag/ZIF-8 structure as SERS substrate for ultrasensitive detection. Spectrochim Acta A Mol Biomol Spectrosc. 2022;282:121693. doi:10.1016/j.saa.2022.121693
76. Qi W, Tian Y, Lu D, et al. Detection of glutathione in dairy products based on surface-enhanced infrared absorption spectroscopy of silver nanoparticles. Front Nutr. 2022;9:982228. doi:10.3389/fnut.2022.982228
77. Okoro G, Husain S, Saukani M, et al. Emerging Trends in Nanomaterials for Photosynthetic Biohybrid Systems. Acs Mater Lett. 2023;5(1):95–115. doi:10.1021/acsmaterialslett.2c00752
78. Kuo JC, Tan SH, Hsiao YC, et al. Unveiling the Antibacterial Mechanism of Gold Nanoclusters via In Situ Transmission Electron Microscopy. Acs Sustain Chem Eng. 2022;10(1):464–471. doi:10.1021/acssuschemeng.1c06714
79. Chang TK, Cheng TM, Chug HL, et al. Metabolic Mechanism Investigation of Antibacterial Active Cysteine-Conjugated Gold Nanoclusters in. Acs Sustain Chem Eng. 2019;7(18):15479–15486. doi:10.1021/acssuschemeng.9b03048
80. Dizaji AN, Ozek NS, Yilmaz A, et al. Gold nanorod arrays enable highly sensitive bacterial detection via surface-enhanced infrared absorption (SEIRA) spectroscopy. Colloids Surf B Biointerfaces. 2021;206:111939. doi:10.1016/j.colsurfb.2021.111939
81. Yao Z, Zhang Q, Zhu W, et al. Rapid detection of SARS-CoV-2 viral nucleic acids based on surface enhanced infrared absorption spectroscopy. Nanoscale. 2021;13(22):10133–10142. doi:10.1039/d1nr01652k
82. Ma Y, Li Q, Wang S, et al. Observation of tunable surface plasmon resonances and surface enhanced infrared absorption (SEIRA) based on indium tin oxide (ITO) nanoparticle substrates. Spectrochim Acta A Mol Biomol Spectrosc. 2022;271:120914. doi:10.1016/j.saa.2022.120914
83. Zhang J, Su R, Fu Q, et al. A survey on computational spectral reconstruction methods from RGB to hyperspectral imaging. Sci Rep. 2022;12(1):11905. doi:10.1038/s41598-022-16223-1
84. Hedde PN, Cinco R, Malacrida L, et al. Phasor-based hyperspectral snapshot microscopy allows fast imaging of live, three-dimensional tissues for biomedical applications. Commun Biol. 2021;4(1):721. doi:10.1038/s42003-021-02266-z
85. Jin X, Fu R, Du W, et al. Rapid, Highly Sensitive, and Label-Free Pathogen Assay System Using a Solid-Phase Self-Interference Recombinase Polymerase Amplification Chip and Hyperspectral Interferometry. Anal Chem. 2022;94(6):2926–2933. doi:10.1021/acs.analchem.1c04858
86. Reid MS, Le XC, Zhang H. Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: an EXPAR Example. Angew Chem Int Ed Engl. 2018;57(37):11856–11866. doi:10.1002/anie.201712217
87. Fang CS, Kim KS, Ha DT, et al. Washing-Free Electrochemical Detection of Amplified Double-Stranded DNAs Using a Zinc Finger Protein. Anal Chem. 2018;90(7):4776–4782. doi:10.1021/acs.analchem.8b00143
88. Al-Madhagi S, O’Sullivan CK, Prodromidis MI, et al. Combination of ferrocene decorated gold nanoparticles and engineered primers for the direct reagentless determination of isothermally amplified DNA. Mikrochim Acta. 2021;188(4):117. doi:10.1007/s00604-021-04771-8
89. Lee H, Yi SY, Kwon JS, et al. Rapid and highly sensitive pathogen detection by real-time DNA monitoring using a nanogap impedimetric sensor with recombinase polymerase amplification. Biosens Bioelectron. 2021;179:113042. doi:10.1016/j.bios.2021.113042
90. Nakano M, Kalsi S, Morgan H. Fast and sensitive isothermal DNA assay using microbead? dielectrophoresis for detection of anti-microbial resistance genes. Biosens Bioelectron. 2018;117:583–589. doi:10.1016/j.bios.2018.06.063
91. Kim HE, Schuck A, Lee SH, et al. Sensitive electrochemical biosensor combined with isothermal amplification for point-of-care COVID-19 tests. Biosens Bioelectron. 2021;182:113168. doi:10.1016/j.bios.2021.113168
92. Singh A, Sharma A, Ahmed A, et al. Recent Advances in Electrochemical Biosensors: applications, Challenges, and Future Scope. Biosensors. 2021;11(9). doi:10.3390/bios11090336
93. Hai Z, Li J, Wu J, et al. Alkaline Phosphatase-Triggered Simultaneous Hydrogelation and Chemiluminescence. J Am Chem Soc. 2017;139(3):1041–1044. doi:10.1021/jacs.6b11041
94. Kober C, Niessner R, Seidel MQ. Quantification of viable and non-viable Legionella spp. by heterogeneous asymmetric recombinase polymerase amplification (haRPA) on a flow-based chemiluminescence microarray. Biosens Bioelectron. 2018;100:49–55. doi:10.1016/j.bios.2017.08.053
95. Yan Y, Shi P, Song W, et al. Chemiluminescence and Bioluminescence Imaging for Biosensing and Therapy: in Vitro and In Vivo Perspectives. Theranostics. 2019;9(14):4047–4065. doi:10.7150/thno.33228
96. Latorre-Perez A, Gimeno-Valero H, Tanner K, et al. A Round Trip to the Desert: in situ Nanopore Sequencing Informs Targeted Bioprospecting. Front Microbiol. 2021;12:768240. doi:10.3389/fmicb.2021.768240
97. Gliddon HD, Frampton D, Munsamy V, et al. A Rapid Drug Resistance Genotyping Workflow for Mycobacterium tuberculosis, Using Targeted Isothermal Amplification and Nanopore Sequencing. Microbiol Spectr. 2021;9(3):e61021. doi:10.1128/Spectrum.00610-21
98. Wang Y, Zhao Y, Bollas A, et al. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348–1365. doi:10.1038/s41587-021-01108-x
99. Yin J, Zou Z, Hu Z, et al. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab Chip. 2020;20(5):979–986. doi:10.1039/c9lc01143a
100. Schulz M, Calabrese S, Hausladen F, et al. Point-of-care testing system for digital single cell detection of MRSA directly from nasal swabs. Lab Chip. 2020;20(14):2549–2561. doi:10.1039/d0lc00294a
101. Cui JQ, Liu FX, Park H, et al. Droplet digital recombinase polymerase amplification (ddRPA) reaction unlocking via picoinjection. Biosens Bioelectron. 2022;202:114019. doi:10.1016/j.bios.2022.114019
102. Yin K, Ding X, Li Z, et al. Autonomous lab-on-paper for multiplexed, CRISPR-based diagnostics of SARS-CoV-2. Lab Chip. 2021;21(14):2730–2737. doi:10.1039/d1lc00293g
103. Huang M, Liu S, Xu Y, et al. CRISPR/Cas12a Technology Combined With RPA for Rapid and Portable SFTSV Detection. Front Microbiol. 2022;13:754995. doi:10.3389/fmicb.2022.754995
104. Talwar CS, Park KH, Ahn WC, et al. Detection of Infectious Viruses Using CRISPR-Cas12-Based Assay. Biosensors. 2021;11(9). doi:10.3390/bios11090301
105. An B, Zhang H, Su X, et al. Rapid and Sensitive Detection of Salmonella spp. Using CRISPR-Cas13a Combined With Recombinase Polymerase Amplification. Front Microbiol. 2021;12:732426. doi:10.3389/fmicb.2021.732426
106. Xiong D, Dai W, Gong J, et al. Rapid detection of SARS-CoV-2 with CRISPR-Cas12a. PloS Biol. 2020;18(12):e3000978. doi:10.1371/journal.pbio.3000978
107. Lu S, Tong X, Han Y, et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat Biomed Eng. 2022;6(3):286–297. doi:10.1038/s41551-022-00861-x
108. Wang B, Wang R, Wang D, et al. Cas12aVDet: a CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal Chem. 2019;91(19):12156–12161. doi:10.1021/acs.analchem.9b01526
109. Cai Q, Wang R, Qiao Z, et al. Single-digit Salmonella detection with the naked eye using bio-barcode immunoassay coupled with recombinase polymerase amplification and a CRISPR-Cas12a system. Analyst. 2021;146(17):5271–5279. doi:10.1039/d1an00717c
110. Aman R, Marsic T, Sivakrishna RG, et al. iSCAN-V2: a One-Pot RT-RPA-CRISPR/Cas12b Assay for Point-of-Care SARS-CoV-2 Detection. Front Bioeng Biotechnol. 2021;9:800104. doi:10.3389/fbioe.2021.800104
111. Park JS, Hsieh K, Chen L, et al. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv Sci. 2021;8(5):2003564. doi:10.1002/advs.202003564
112. Ali Z, Sanchez E, Tehseen M, et al. Bio-SCAN: a CRISPR/dCas9-Based Lateral Flow Assay for Rapid, Specific, and Sensitive Detection of SARS-CoV-2. ACS Synth Biol. 2022;11(1):406–419. doi:10.1021/acssynbio.1c00499
113. Zong N, Gao Y, Chen Y, et al. Automated Centrifugal Microfluidic Chip Integrating Pretreatment and Molecular Diagnosis for Hepatitis B Virus Genotyping from Whole Blood. Anal Chem. 2022;94(12):5196–5203. doi:10.1021/acs.analchem.2c00337
114. Yin D, Yin L, Wang J, et al. Visual Detection of Duck Tembusu Virus With CRISPR/Cas13: a Sensitive and Specific Point-of-Care Detection. Front Cell Infect Microbiol. 2022;12:848365. doi:10.3389/fcimb.2022.848365
115. Park BJ, Park MS, Lee JM, et al. Specific Detection of Influenza A and B Viruses by CRISPR-Cas12a-Based Assay. Biosensors. 2021;11(3). doi:10.3390/bios11030088
116. Jiao Z, Yang J, Long X, et al. CRISPR/Cas12a-Assisted Visual Logic-Gate Detection of Pathogenic Microorganisms Based on Water-Soluble DNA-Binding AIEgens. Front Chem. 2021;9:801972. doi:10.3389/fchem.2021.801972
117. You Y, Zhang P, Wu G, et al. Highly Specific and Sensitive Detection of Yersinia pestis by Portable Cas12a-UPTLFA Platform. Front Microbiol. 2021;12:700016. doi:10.3389/fmicb.2021.700016
118. Liu L, Duan JJ, Wei XY, et al. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci Total Environ. 2022;838(Pt 2):156048. doi:10.1016/j.scitotenv.2022.156048
119. Xu JH, Kang L, Yuan B, et al. Development and evaluation of a rapid RPA/CRISPR-based detection of Francisella tularensis. Front Microbiol. 2022;13:901520. doi:10.3389/fmicb.2022.901520
120. Jirawannaporn S, Limothai U, Tachaboon S, et al. Rapid and sensitive point-of-care detection of Leptospira by RPA-CRISPR/Cas12a targeting lipL32. PloS Negl Trop Dis. 2022;16(1):e10112. doi:10.1371/journal.pntd.0010112
121. Tian T, Qiu Z, Jiang Y, et al. Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens Bioelectron. 2022;196:113701. doi:10.1016/j.bios.2021.113701
122. Feng W, Peng H, Xu J, et al. Integrating Reverse Transcription Recombinase Polymerase Amplification with CRISPR Technology for the One-Tube Assay of RNA. Anal Chem. 2021;93(37):12808–12816. doi:10.1021/acs.analchem.1c03456
123. Malci K, Walls LE, Rios-Solis L. Rational Design of CRISPR/Cas12a-RPA Based One-Pot COVID-19 Detection with Design of Experiments. ACS Synth Biol. 2022;11(4):1555–1567. doi:10.1021/acssynbio.1c00617
124. Zavvar TS, Khoshbin Z, Ramezani M, et al. CRISPR/Cas-engineered technology: innovative approach for biosensor development. Biosens Bioelectron. 2022;214:114501. doi:10.1016/j.bios.2022.114501
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
