Back to Journals » International Journal of Nanomedicine » Volume 19
Recent Advances in Understanding the Molecular Mechanisms of Multidrug Resistance and Novel Approaches of CRISPR/Cas9-Based Genome-Editing to Combat This Health Emergency
Authors Allemailem KS
Received 10 December 2023
Accepted for publication 26 January 2024
Published 7 February 2024 Volume 2024:19 Pages 1125—1143
DOI https://doi.org/10.2147/IJN.S453566
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Eng San Thian
Khaled S Allemailem
Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah 51452, Saudi Arabia
Correspondence: Khaled S Allemailem, Department of Medical Laboratories, College of Applied Medical Sciences, Qassim University, Buraydah 51452, Saudi Arabia, Tel +966163010555, Email [email protected]
Abstract: The rapid spread of multidrug resistance (MDR), due to abusive use of antibiotics has led to global health emergency, causing substantial morbidity and mortality. Bacteria attain MDR by different means such as antibiotic modification/degradation, target protection/modification/bypass, and enhanced efflux mechanisms. The classical approaches of counteracting MDR bacteria are expensive and time-consuming, thus, it is highly significant to understand the molecular mechanisms of this resistance to curb the problem from core level. The revolutionary approach of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated sequence 9 (CRISPR/Cas9), considered as a next-generation genome-editing tool presents an innovative opportunity to precisely target and edit bacterial genome to alter their MDR strategy. Different bacteria possessing antibiotic resistance genes such as mecA, ermB, ramR, tetA, mqrB and blaKPC that have been targeted by CRISPR/Cas9 to re-sensitize these pathogens against antibiotics, such as methicillin, erythromycin, tigecycline, colistin and carbapenem, respectively. The CRISPR/Cas9 from S. pyogenes is the most widely studied genome-editing tool, consisting of a Cas9 DNA endonuclease associated with tracrRNA and crRNA, which can be systematically coupled as sgRNA. The targeting strategies of CRISPR/Cas9 to bacterial cells is mediated through phage, plasmids, vesicles and nanoparticles. However, the targeting approaches of this genome-editing tool to specific bacteria is a challenging task and still remains at a very preliminary stage due to numerous obstacles awaiting to be solved. This review elaborates some recent updates about the molecular mechanisms of antibiotic resistance and the innovative role of CRISPR/Cas9 system in modulating these resistance mechanisms. Furthermore, the delivery approaches of this genome-editing system in bacterial cells are discussed. In addition, some challenges and future prospects are also described.
Keywords: bacteria, multidrug resistance, CRISPR/Cas9, gene editing, nanoparticle, delivery approaches
Introduction
Since the discovery of antibiotics in 1928, commonly used as a weapon of choice against infections, the frequency of antibiotic-resistant infections are increasing rapidly. The widespread use and misuse of antibiotics, inadequate surveillance and regulations has led the bacteria to evolve into antibiotic-resistant strains.1 Some factors like inappropriate and repeated use of similar antibiotic sets, demography, lifestyle and host biology play a significant role in sensitivity of resistance leading to enhancement in antimicrobial resistance (AMR).2,3 Consequently, the therapeutic efficacy of antibiotics is significantly reduced by pathogenic AMR and it has led to the evolution of multidrug resistance (MDR) pathogens.4,5 Therefore, the eradication of MDR complication is utmost important by developing some novel therapies.
MDR is a global health emergency, leading to a huge financial burden, causing substantial morbidity and mortality, threatening to undermine the practice of medicine.6,7 As per the center for disease control and prevention (CDC) records, there is an urgent threat from drug-resistant bacteria, which include Carbapenem-resistant Enterobacteriaceae (CRE), Carbapenem-resistant Acinetobacter baumannii,8 Clostridioides difficile (C. difficile) and Drug-resistant Neisseria gonorrhoeae (N. gonorrhoeae). Some other serious threat bacteria include extended-spectrum beta-lactamase, drug-resistant (Campylobacter, Streptococcus pneumoniae, Shigella, Salmonella serotype typhi, M. tuberculosis, and non-typhoidal Salmonella). In addition, other bacteria in this category include (ESBL)-producing Enterobacteriaceae, multidrug-resistant Pseudomonas aeruginosa, (P. aeruginosa), vancomycin-resistant Enterococci (VRE), and methicillin-resistant Staphylococcus aureus (MRSA). While the concerning threat strains also include Clindamycin-resistant group B Streptococcus and Erythromycin-resistant group A Streptococcus.9 Even though, not all bacterial groups are resistant to antibiotics, six major MDR bacteria, commonly known as ESKAPE bugs are identified which mostly evade the antibiotics action. These include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species and these bacteria are mainly responsible for global nosocomial infections.10,11
Different approaches have been practiced time to time to combat the problems of AMR. These approaches include the use of vaccines, drug repurposing, antimicrobial peptides, phage therapy, anti-virulence compounds, etc. Each of these strategies has some unique features but also suffers from some limitations, thus pushing the researchers to explore some new alternatives with enhanced efficiency. The strategy of RNA interference, restriction endonucleases, and other genome-editing tools have also been practiced recently to combat the problems of MDR.12
The recent breakthrough towards combating the AMR has led to the use of Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated sequence (CRISPR/Cas), primarily a prokaryotic immune system, to overcome the problems of antibiotic resistance challenges.13 CRISPR/Cas system are well-known genome-editing defense systems with different classes, types and subtypes, present in prokaryotes and archaea against mobile genetic elements, plasmids and phages. The discovery of the CRISPR/Cas system has been a major breakthrough in genome-editing technology. This tool allows the bacteria to be specifically modulated genetically for its virulence or resistance.14 Among all CRISPR/Cas types, CRISPR/Cas9 possess some special features, due to its simple structure, high versatility and is thoroughly studied as a genome-editing tool, thus making it highly suitable for MDR applications.15
In this review, the recent updates of some molecular mechanisms of AMR are explored. In this context, the novel approaches of CRISPR/Cas9 as a genome-editing tool to diminish the MDR are discussed. In addition, the delivery approaches of this genome-editing tool to different bacteria are demonstrated. Furthermore, some challenges and future prospects of MDR is also elaborated.
The Biology of Antibiotic Resistance
The ability of bacteria to survive in antibiotics environment, expected to kill them is known as antibacterial resistance or MDR. Even though the MDR is increasing day by day, an increasing expertise about the biological insights and technological advances are also developing in parallel. Bacteria use either an “intrinsic” mechanism or an “acquired” resistance approach for their survival in different antibiotic conditions.16 The resistance mechanisms involve the resistance genes expression against different targets, which can markedly change between varying growth conditions.17
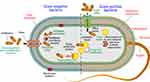
Bacteria often get mutations in their DNA by phages, plasmids and other mobile genetic elements (MGEs).18 The MDR is broadly classified into two types as genetic MDR and phenotypic MDR. The mutations in bacterial DNA or antibiotic resistance genes received from other bacteria give rise to genetic MDR. Clinically, this type of MDR leads to primary treatment failure thus requires alternative antibiotic use or different therapeutic approaches. In contrast to genetic MDR, phenotypic MDR results in changes in bacteria and subside within individual cells, without genetic alteration. This type of MDR against a specific antibiotic does not allow overall growth of the whole bacterial population (Figure 1).
The growth of the overall population of bacteria is not allowed in the case of phenotypic MDR in the presence of a specific antibiotic at or above minimal inhibitory concentration (MIC). This type of AMR can manifest as a minimal killing rate for the bulk bacterial population, sometimes termed as “tolerance”.19 Phenotypic antibiotic resistance, alternatively have a slower killing rate than most of the bacterial population, often termed as “persistence”.
A thorough knowledge about the molecular mechanisms of MDR can help to discover some innovative approaches to treat infectious diseases.20 Recently, considerable progress has been made to understand the antibiotic action and the bacterial inhibitory resistance mechanisms against the killing effects of these antibiotics. The antimicrobial resistance by bacteria is employed by three related approaches, which include tolerance, resistance and persistence.1
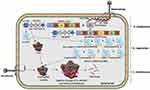
There has been a significant advancement in understanding the biochemical action of different antibiotics and the major mechanism by which bacteria resist the killing potential of antibiotics through different ways. The resistance mechanisms include diverse actions, such as expression of resistance genes, downregulation or modification of porins to decrease the antibiotic influx, antibiotic inactivation, overexpression of active efflux pumps, antibiotic target site modification, target bypass, and target protection (Figure 2).
Recent advances in different technologies have revealed precise details involved in diverse resistant mechanisms. These include the action of complex efflux systems pointing towards probable routes for inhibitor development.21,22 The mechanism of antibiotic resistance with selected examples of different proteins and enzymes and their gene location is summarized in Table 1.
 |
Table 1 Different Mechanism of Attaining MDR by Bacteria, Selected Examples and Gene Locations |
Different Approaches to Overcome MDR
Different approaches to curb the MDR include the use of combinatorial therapy, vaccines, drug repurposing,44,45 antibodies, antimicrobial peptides,46,47 phage therapy,48,49 anti-virulence compounds,50 and drug loaded nanoparticles (NPs).51–53 Each of these strategies faces some limitations, forcing the exploration for new alternatives with enhanced efficiency.
Combinatorial therapy comprises the practice of using different drug combinations rather than single drug to attain the synergistic effect to kill the bacteria by targeting multiple sites. The rapid occurrence of MDR bacterial strains necessitates the use of combinatorial therapy rather than monotherapy, which is increasingly no longer adequate to threat most of the bacterial infections.54 However, due to some incompatibility complications between different drugs, this procedure affects the pharmacokinetics and pharmacodynamics of the used drugs.55,56
Phage therapy has proven to be efficient to some extent against antibiotic resistant bacteria.57 However, some challenges include their interaction with intracellular bacteria, promote neutralizing antibodies production and the development of phage resistance in bacteria.58 Some bacterial resistance has also been treated with the use of monoclonal antibodies (mAbs). However, some barriers impeding their use include bacterial target selection (eg, lipopolysaccharides possess different serotypes), and degradation by bacterial proteolytic enzymes.59
The approach of RNA interference (RNAi) and steric-blocking oligonucleotides are some RNA based therapeutics used to treat AMR organisms. These approaches exploit the use of oligonucleotides to target bacterial mRNA enzymatically, allowing the removal of genes conferring the resistant phenotypes.60 Antisense RNA-based procedures have also offered the means to keep the track of genes responsible for MDR and growth promotion.61,62 However, RNA-based therapeutics suffer from poor intracellular uptake and some toxicity issues.60
The MDR complications have also been sorted out by the use of gene editing tools like Zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) that can be used to precisely edit DNA of drug resistant bacteria. The employment of TALENs and ZFNs is used against specific DNA sequence for their cleavage.63,64 The TALENs and ZFNs tools have paved novel approach of modern gene editing strategy. Due to some substantial limitations, such as complexity, off-target effects and delivery challenges, these genome-editing approaches have not gained a broad range success. These strategies finally led to the novel use of CRISPR/Cas-based genome-editing for the antibiotic resistance.
Currently, the CRISPR/Cas system is considered as the most innovative approach of genome editing and has been rapidly used for the management of MDR since its discovery. It is extensively used as a genome editing approach since it is fast, less expensive and the most efficient genome-editing tool. It is also used for the removal of bacterial pathogens,65,66 elimination of major infectious viruses,67,68 and improvement of genetic defects.69,70 Different scientific studies support the control of spread of MDR via the use of CRISPR/Cas-based strategy.68,71,72
CRISPR/Cas System and Its Types
The CRISPR/Cas system is an innovative genome-editing tool, which acts as an adaptive immune system in archaea and bacteria protecting them from phages, plasmids and other mobile genetic elements. Almost 75% of archaea and 36% of bacteria possess the CRISPR/Cas system in their genome. Recently, it has been reported that there exists an inverse association between the existence of CRISPR/Cas system with antibiotic resistance, ESBL and carbapenemase production in antibiotic resistant bacteria, such as K. pneumonia.73 In addition, the prevalence of CRISPR/Cas system and its possible association with antibiotic resistance has been reported in E. faecalis and E. faecium.74
The CRISPR/Cas genetic loci integrate the CRISPR array, compromised of repeated sequence and short flanking sequences (spacers). Protospacers constitute the spacers of CRISPR, derived from invading plasmids or phage DNA. Cas proteins, the key components of the CRISPR system, are encoded towards the upstream of the CRISPR array.75,76 The CRISPR array may be clustered on a chromosome in numerous or single loci.77,78 In 2012, there was a great achievement in this genome-editing system when Doudna and Charpentier et al, used CRISPR/Cas9 system having chimeric single guide RNA (sgRNA). The sgRNA is customized by joining CRISPR RNA (crRNA) and transactivating CRISPR RNA (tracrRNA) together.79
The CRISPR/Cas systems are grouped in two classes, six types (type I to type VI) and 33 subtypes. The classification of this genome-editing tool is based on differences in Cas protein content and is briefly summarized in Figure 3.
 |
Figure 3 CRISPR/Cas system classification and functions in bacteria. Based on the generic organization, Cas effectors are classified and functional modules of CRISPR/Cas system are shown. |
In this system of CRISPR/Cas classification, class 1 comprises type I, III and IV together having sixteen subtypes, incorporating effector modules composed of numerous Cas proteins. These effector modules determine crRNA and promote the pre-crRNA processing and its action. However, class 2 is composed of type II, V and VI in addition to seventeen subtypes that further consist of a single crRNA-binding protein, which is large and has multi-domain organization. It is involved in pre-crRNA processing in some variants and interference in all variants.80,81 A unique protein composition is present in each type of CRISPR/Cas system for expression and interference steps.
Although, even with the high diversification of the CRISPR/Cas system, the primary function of this genome-editing tool remains the same. It comprises of three main steps as adaptation, expression and interference (Figures 3 and 4). The adaptation phase includes the consumption of exogenous DNA fragments into bacterial CRISPR array, followed by the expression step, which includes the maturation of CRISPR-RNA (crRNA) from the attained spacers.82,83 The interference step involved the recognition and attachment with complementary nucleotide sequence of attacking genetic elements by crRNA, leading to its cleavage by the help of Cas nuclease84,85 [Figure 4]. Among all different type II CRISPR/Cas systems, S. pyogenes derived CRISPR/Cas9 has been comprehensively studied because of its versatility, simplicity, specificity and efficiency.75,86
Structure of CRISPR/Cas9 System
The CRISPR/Cas9 derived from S. pyogenes (Spy) is the most extensively studied form of the CRISPR/Cas system as the interference step by this genome editing tool requires only single Cas9 protein (Figure 4).87,88 The system is a ribonucleoprotein complex composed of Cas9 protein and single guide RNA (sgRNA). Cas9 protein is the DNA endonuclease enzyme (160 kDa), having 1368 amino acid residues, capable of cleaving precisely each strand of dsDNA.89 This genome-editing tool also requires Csn2, Cas1 and Cas2 for DNA acquisition phase,90 while pre-crRNA processing to mature sgRNA is performed by RNaseIII.91,92
Cas9 endonuclease consists of two lobes, the nuclease (NUC) lobe having amino acid residues from 1 to 55, and 719 to 1368 and the recognition (REC) lobe (residues 56–718).89,93 The REC lobe performs the nucleotide recognition step by three domains named as REC (-I, -II and -III).92,94 Cas9 contains two endonuclease domains, the HNH domain (766–909) and RuvC domain (1–55, 719–765, and 910–1099). Further, the HNH domain is responsible for target the DNA strand cleavage, whereas non-target DNA strand is cleaved by the RuvC domain (Figure 5).93 Further, two key hinge regions as linker L1 and L2 are present in the HNH domain near N and C terminus.95 These linkers create a crosstalk between the HNH and RuvC domains.96 The PAM interacting (PI) domain consists of residues from 1100 to 1368 and confers the PAM specificity, responsibility for the initiation of Cas9 binding to target DNA.89,93 With the initiation of DNA binding, the negatively charged sgRNA:DNA hybrid is stabilized by positively charged residues present at the interface between the NUC and REC lobes97 (Figure 5). However, displaced non-target DNA is stabilized by linker region positively charged amino acid residues (L1 and L2) present between HNH and RuvC domains.92
 |
Figure 5 Three-dimensional structure of SpCas9-sgRNA-DNA ternary complex. (A) Ribbon representation of Cas9-sgRNA-DNA complex at two different angles, obtained from protein data bank (http://www.rcsb.org, PDB ID: 4OO8), edited by UCSF Chimera. (B) Domain organization representing amino acid residue numbers assigned for different protein domains. |
The action mechanism of CRISPR/Cas9 involves different conformational changes in its three-dimensional structure as supported by X-ray crystallography studies obtained at different stages of its activity.89,98,99 The different stages of the S. pyogenes CRISPR/Cas9 gene editing pathway have been resolved as free Cas9 with protein data bank (PDB 4CMQ) (www.rcsb.org),100 sgRNA bound Cas9 (PDB 4ZT0),99 Cas9 in association with target DNA and incomplete non-target DNA with PAM sequence (PDB 4UN3),98 and Cas9 in association with both target DNA and complete non-target DNA (PDB 5F9R).99 The apo Cas9 sgRNA associated Cas9 from S. pyogenes (PDB 4CMQ and PDB 4ZT0)99,100 was obtained at 3.09 Å and 2.9 Å resolution, respectively. Upon sgRNA binding, a significant rearrangement occurs in the REC domain almost 65 Å shift of the REC III domain for the accommodation of sgRNA99 (Figures 4 and 5). A further shift of the REC II domain occurs by the binding of the target DNA and PAM with an incomplete non-target DNA strand to Cas9:sgRNA complex (PDB 4UN3).98 Melting of the foreign DNA occurs by Cas9:sgRNA-based PAM recognition and results in DNA:RNA hybrid formation.98,101
Mechanistic Action of CRISPR/Cas9
The mechanistic action of CRISPR/Cas9 system involves three stages as adaptation, expression and interference (Figures 3 and 4). The invading foreign DNA (almost 30 bp) is integrated into the CRISPR loci leader side during the adaptation stage. The protospacer adjacent motif (PAM) is decided from the host genome spacer sequence. The RNA transcription from spacers of the CRISPR locus occurs during the expression stage.82,102 The discovery of using sgRNA (tracrRNA and crRNA complex) along with Cas9 has revolutionized the art of CRISPR/Cas9 system for genome engineering to new heights. In addition, as a double check, the PAM sequence is responsible for Cas9 to identify the target DNA.103
It has been reported from the previous study that self-targeting CRISPR/Cas system spacers assist in bacterial cell apoptosis, if the cleaved DNA repair system is delayed.104 Consequently, it is delineated that Cas nuclease reprogramming can help in targeting precise bacterial population. This approach can repurpose the CRISPR/Cas system to kill the specific bacterial population or re-sensitize them for specific antibacterial systems.84
If the gRNA shows enough complementarity with the target DNA, Cas9 continues its function and generates a double stranded blunt cut, three base-pairs upstream of the PAM sequence.105,106 As Cas9 possesses two nuclease domains, the HNH domain cleaves complementary DNA strand, while the RuvC domain cleaves non-complementary DNA strand. The DNA cleavage produces a predominantly blunt-ended double-strand break (DSB).92,107 To repair the DNA breaks, two major repairing mechanism are used as non-homologous end joining (NHEJ) and homology-directed repair (HDR) (Figure 6). The NHEJ approach repairs the DSBs with high efficiency for gene disruptions involving deletions or insertions and occur in absence of homologous DNA sequence. However, the HDR mechanism repairs the DSBs in presence of donor DNA template with low efficiency for gene modifications.108
The sgRNA can be devised systematically to target virulence, MDR or other critical genes specific for pathogenesis. The CRISPR/Cas9 genome-editing system can be further engineered to induce growth inhibition of the target bacterium and its death. In addition, transcriptional repression of target genes and elimination of antibiotic resistance plasmids can be achieved by this genome-editing tool (Figure 4).75,92
The Novel Approaches to Combat the MDR by CRISPR/Cas9-Based Genome Editing
CRISPR/Cas9-based genome-editing is now very well considered as a promising next-generation tool to combat the infectious diseases, especially caused by AMR pathogens.78,80 Based on the location of the target gene, CRISPR/Cas9-based genome editing can be utilized either as a pathogen-targeted approach or as a gene-based approach. The pathogen-based approach is relied on targeting some specific regions of bacterial chromosome. This method leads to bacterial cell destruction and killing of specific pathogenic strains. However, the gene-focused method is carried on by targeting antibiotic resistance genes carried by various plasmid. This approach leads to antibiotics sensitivity towards the bacteria.109 Some target-specific strains of mixed microbial pathogens as well as well-defined infections can be treated by pathogen-focused approaches, whereas the gene-focused tactics are undefined. However, this approach is related to the reduction in antibiotic resistance abundance in microbial community as an approach for the treatment of bacterial infections.110
The CRISPR/Cas9 system is able to distinguish between pathogenic and symbiotic strains by precise sequence targeting, unlike the conventional antimicrobials. Staphylococcus aureus and E. coli have been transformed by this approach by a plasmid encoding Cas9-driven RNA. This method precisely degraded the antibiotic resistance gene expression.110 However, this approach is currently in the preclinical phase, as the overall target is to achieve MDR therapy. However, some clinical studies by this genome-editing tool have also been performed as antimicrobial therapy. Some clinical isolates of S. aureus represented a decline in disease by about 50% after treatment with engineered crRNA and Cas9 for a methicillin resistance gene (mecA).57,79 In parallel, another study demonstrated that this genome-editing tool targets ermB (erythromycin resistance gene) and after treatment, significantly minimize the advancement in intestinal erythromycin-resistant E. faecalis. Furthermore, CRISPR/Cas9 interventions significantly reduced the skin colonization of S. aureus on a mouse skin colonization model.111
Klebsiella pneumonia is one of the ESKAPE pathogens, which can attain multiple MDR through the mutations by horizontal gene transfer.112 In one study, CRISPR/Cas9 genome-editing was introduced in these bacteria to study the functions of ramR, tetA and mgrB genes that mediate tigecycline and colistin resistance in carbapenem-resistant K. pneumonia.113 The increased expression of CRISPR/Cas9 led to the inactivation of these genes, affecting the bacterial susceptibility to colistin and tigecycline, respectively.113
As MDR E. faecalis lacks the complete CRISPR, especially Cas9 system.114 A proficient delivery of whole CRISPR/Cas9 genome-editing tool to these MDR bacteria by the use of pheromone responsive plasmid (PRP) was performed. This approach achieved efficient conjugation with a narrow host range limited to E. faecalis. A constitutively expressed CRISPR/Cas9 system was engineered with pD1, specific for the enterococcal antibiotic resistance genes tetM (encoding tetracycline resistance) and ermB (encoding erythromycin resistance). The erythromycin and tetracycline resistance efficiently depleted from E. faecalis bacteria in vitro. In a parallel context, an in vivo intestinal colonization model showed that donors having PRP targeting ermB potentially reduced the occurrence of erythromycin-resistant intestinal E. faecalis, presenting a support for the utility of engineered PRP in alleviating the multidrug resistance of E. faecalis.115
Enterococcus faecium, the Gram-positive bacteria is becoming a common cause of antibiotic resistance in hospital acquired infection. In one approach of minimizing its antibiotic resistance, high recombination rates of these bacteria by the application of CRISPR/Cas9 DNA-editing led to its targeted mutant generation of MDR clinical E. faecium strain E745. A deletion in the lacL gene (encoding a large subunit of β-galactosidase) was generated. This approach of genome editing can potentially be implemented on other Gram-positive bacteria with better recombination rates, minimizing their antibiotic resistance potential.116
In one study, the transfer efficiency of a conjugative plasmid TP114 containing CRISPR/Cas9 array was optimized. By the use of a single dose, this approach led to the elimination of more than 99.9% of targeted antibiotic resistant E. coli in mouse gut microbiota. This system was also applied to the Citrobacter rodentium infection model and successfully achieved the complete clearance of this infection within few days of treatment.117
Staphylococcus aureus is vulnerable to the powerful staphylolytic enzyme lysostaphin. It is a glycylglycine endopeptidase with significant antimicrobial activity. However, due to its wall teichoic acid, S. aureus shows some resistance to this endopeptidase.118 Some researchers used CRISPR/dCas9, a variant of Cas9, to downregulate the expression of tarG, tarH, and tarO genes, to block the wall teichoic acid formation, and sensitizing S. aureus towards lysostaphin to eliminate these bacteria.119
In one novel study, a CRISPR/Cas9-based plasmid-curing system (pCasCure), was designed to specifically cut carbapenemase genes such as blaKPC, blaNDM and blaOXA-48 in carbapenem-resistant Enterobacteriaceae (CRE). The results demonstrated that pCasCure efficiently cut these genes present in different Enterobacteriaceae species of E. coli, K. pneumonia, E. hormaechei, E. xiangfangensis, and S. marcescens clinical isolates with more than 94% curing efficiency. In addition, parA, repA, and repB genes on pKpQIL plasmid were precisely cleared to prevent the plasmid-based carbapenemase resistance gene to resensitize the effect of carbapenem antibiotic on CRE. This experiment led to the reduction of the MIC value by more than eight times.120
Novel Approaches of Targeting CRISPR/Cas9 System in Bacterial Cells
The targeting methods of CRISPR/Cas9 genome-editing tool to different types of bacteria confronts many challenges, since the active 160 kD ribonucleoprotein complex must pass across the bacterial cell wall to perform its activity. Several research groups have skillfully exploited species-level specificity of plasmids and phage for this genome-editing system delivery. Some novel targeting approaches also involve the nanoparticle encapsulated phage-CRISPR/Cas9 system delivery methods.87,121 However, the search for more innovative targeting approaches is going on to minimize comprehensively the antibiotic resistance complication in the near future. A brief description of some delivery approaches of CRISPR/Cas9 system through phages, plasmids, nanoparticles and extracellular vesicles in different bacterial cells is given below.
Phage-Mediated Delivery
As pages are natural bacterial predators and efficiently inject their genomic contents within bacterial cells. Recently modified phages have been incorporated with CRISPR/Cas system to use it as a delivery vehicle.122 The method involves the incorporation of CRISPR-DNA in plasmid constructs within a DNA-phage system, termed as CRISPR-Phage or CR-phage. Since phage makes adhesion on target bacterial cells through different receptors like pili (Gram-negative), flagellin, protein porin OmpC, peptidoglycan, teichoic acids, teichoic acid-peptidoglycan polymer (Gram-positive), and lipopolysaccharide (LPS). The adhesion between phage and pathogenic bacteria leads to the transfer of CRISPR-conjugated materials.123,124 The exogenous Cas enzyme within the pathogenic bacteria reciprocates the genetic changes and reduces its antibiotic resistance.79 In addition, some factors like phage pharmacodynamics, host range, and phage resistance can be modulated to enhance its effectiveness in the system.84 Moreover, the immunodynamics and targeted gene expression in the relationship with host-pathogen can be modulated, thereby making a perfect cooperation for the effective resistance.
E. coli was transformed with a plasmid having CRISPR/Cas9 genome-editing tool against the MDR genes. In the presence of the selection agents, this approach led to an almost one-thousand-fold decrease in transformation efficiency. The novel results led to a new foundation of experiments to use phages to package CRISPR/Cas9 antibiotic resistance gene vectors. It resulted in rapid killing of specific bacteria. In a similar fashion, the virulent strain of S. aureus having antibiotic resistance was targeted by CRISPR/Cas9 encoded in phage.85 This procedure also supported the species-specific targeting and killing of bacteria by this genome-editing tool. For the support of this novel procedure further, an in vivo mouse skin colonization infection model was made. The spread of the colonization of bacteria in this mouse model was significantly reduced by topically applying the CRISPR/Cas9 supported phages. Altogether, these findings reveal that properly selective pathogenic bacteria can be terminated by CRISPR/Cas9 antibacterial tools, while leaving behind the non-targeted bacteria unaffected. This approach is highly significant for the development of antibacterial approach for gut microflora and other environments.125,126 These experiments support the practicability of CRISPR/Cas9 repurposing to attack any antibiotic resistant pathogenic bacteria rather than acting as a defending approach. This approach is expected to work efficiently in the future against newly emerging antibiotic resistant bacterial strains.
The adaptability of CRISPR/Cas9 genome editing by altering just the gRNA sequence enhances its approaches for the management of MDR. Furthermore, the engineering of phage scaffolds can also increase the delivery approaches of CRISPR/Cas9 antibacterial based on species-specific needs. Besides addressing the externally and topically treatable infections, eg, methicillin-resistant Staphylococcus aureus (MRSA), further approaches are required to address intracellular or tissue/organ specific bacterial infections.
Phage-CRISPR/Cas complex can address only external and surface infections, thus limiting its use for tissue specific and intracellular bacterial infections. To overcome all these limitations, CRISPR/Cas-phage complex have been encapsulated within some other nanoformulations to increase their resistance against some unfriendly conditions (eg, pH difference, antibodies etc.) and increase their availability around a specific area. These nanoformulations include liposomes,127–129 fibers, nano-emulsions130 and hydrogels etc.131,132 (Figure 7).
Liposomes have been frequently used as drug delivery vectors and this approach was used to deliver phage to enhance their antibacterial properties. This approach protects the phage from neutralizing antibodies and removes the bacterial biofilm. In a parallel strategy, CRISPR/Cas9-phage complex has been delivered by using alginate hydrogel, to reduce the soft tissue infection with enhanced anti-biofilm effect over time.132 With a suitable nanoformulation and bacteriophage encapsulation approach, the CRISPR/Cas9 system can demonstrate highly efficient against MDR activity; however, there are still many challenges to overcome to achieve the clinical transformation of this novel method for antimicrobial resistance.133
Conjugative Plasmids Mediated Delivery
In addition to phage-mediated delivery approaches, some other alternative delivery means can be used to target CRISPR/Cas9 system in MDR bacterial cells. The approach of using conjugative plasmids as an alternative delivery vehicle can transfer the genetic material between bacterial cells115 (Figure 7). The Enterococcal antibiotic resistance has been reduced significantly by CRISPR/Cas system delivered in bacterial population through plasmids.115 The CRISPR/Cas plasmid delivery approach has been used in carbapenem-resistant Enterobacteriaceae to eliminate its carbapenemase resistance (blaKPC and blaNDM) genes. This approach also re-sensitizes the drug resistant bacteria to carbapenems with better outcomes on carbapenem-resistant Enterobacteriaceae.134 The CRISPR/Cas system plasmid recipient bacteria further transmit this system to other bacteria, thus significantly expanding the application scope of using this genome editing tool to reduce the drug resistant genes.135 The CRISPR/Cas system targeted through plasmids (TAPs) can be efficiently transferred to E. coli and associated Gram-negative Enterobacteriaceae to re-sensitize these bacteria carrying pOXA to prevent the drug resistance spread.136
In a similar fashion, Dong et al designed conjugative CRISPR/Cas9 against mobile E. coli colistin resistance gene (mcr-1). The CRISPR/Cas9 system re-sensitize the pathogens to antibiotics and exhibits the cells to gain immunity against mcr-1. The pMBLcas9-sgRNA recombinant plasmid retained the capacity to transfer into E. coli possessing diverse MCR-1 plasmids, significantly eliminating the MDR plasmids.111
In contrast to phage-based delivery, that requires a specific receptor on the bacterial cell for its attachment, the conjugation does not involve receptors for the plasmid uptake. Thus, due to the emergence of mutations in bacterial receptors, that leads to resistance against phage-mediated delivery, plasmid-based delivery is considered as a preferred procedure.137 However, the plasmid conjugation approach also suffers from some issues, such as low delivery efficiency and narrow host range.138
Nanoparticle Mediated Delivery
Recently, with the rapid innovation in the field of nanotechnology, special types of nanoparticles (NPs) have been used to deliver crRNA and Cas effector molecules in the bacterial cells (Figure 7). Nanoparticles have the capacity to have flexible size and anti-degradation barrier for packaging CRISPR/Cas9 system and can maintain a natural state during gene transfer. Besides this, NPs have the advantage of biocompatibility, smaller immunogenicity, surface functionalization and higher safety as compared to virus vectors.139,140
Inorganic and cationic polymer-based NPs have been used to transfer the CRISPR/Cas genome-editing system components to bacterial cells.141,142 It has been reported that a cationic polymer-based CRISPR genome-editing complex carrying the Cas9 endonuclease and crRNA can be effectively introduced into MRSA and efficiently executes the bacterial killing by targeting the methicillin-resistant gene.66 However, the approach of delivering CRISPR/Cas genome editing system in different antibiotic resistant bacterial cells by using different NPs is still at a very preliminary stage. Many questions also need to be answered to use the nanoformulation strategy of targeting CRISPR/Cas9 system to diverse bacteria. For example, the approach of improving encapsulation rate and the achievement of efficient delivery in some peculiar pathogens such as Mycobacterium tuberculosis, having highly impermeable and usually thick cell wall needs to be resolved walls.143
In one novel study, carbon quantum dots were covalently conjugated to papG-targeted gRNA and Cas9 for the employment of Cri-dot-papG nanoformulation to deliver CRISPR/Cas system in Uropathogenic E. coli (UPEC). This genome editing tool targeted the papG gene (fimbrial adhesion virulence factor), thereby minimizing the pathogenicity of UPEC.144
Extracellular Vesicles-Based Delivery
During the growth phase, Gram-negative microorganisms synthesize outer membrane vesicles (OMVs) that move to extracellular compartment.145,146 These vesicles can be used to deliver antibiotic resistant genes, plasmids or virulence genes as these vesicles are DNase resistant, thus serving as horizontal gene transfer methods of DNA.147,148 Recently, these OMVs were investigated as delivery vesicles for Cas9 RNPs for gene editing purpose.148 It was found that E. coli secreted OMVs can carry CRISPR/Cas9 genome-editing tools to target Streptococcus agalactiae, accomplishing a specific and efficient clearance of S. agalactiae.149 However, the limitation of enriching RNPs within these OMVs limits the delivery efficiency148,150 (Figure 7).
In addition to phages, plasmids, nanoparticles and membrane vesicles, electroporation procedure has also been applied to import plasmid carrying CRISPR/dCas9 into S. aureus and it effectively restored the clearance of Lst on these bacteria.151 However, the complication due to cell damage and other cytotoxicity issues limit this procedure to in vitro tests only. Examples of some delivery strategies of CRISPR/Cas9-based antimicrobial genome editing tools in different types of bacteria with summarized results are listed in Table 2.
 |
Table 2 Examples of Some Novel Delivery Methods of Designed CRISPR/Cas9-Based Antimicrobials in Different Types of Bacteria |
The Challenges and Future Prospects of Mitigating MDR by CRISPR/Cas9
The CRISPR/Cas9 genome editing system holds immense potential in tackling MDR, but its application in this field still comes with a handful of challenges. Many human disease-causing bacteria such as S. enterica, M. tuberculosis, and Burkholderia spp. are well-known intracellular pathogens residing in different host cells. These bacteria offer an additional challenge to be eradicated by CRISPR/Cas9-mediated genome-editing system. Therefore, additional approaches need to be offered to transport CRISPR/Cas9 system delivery vehicles for such microbes. As the delivery of phage to eukaryotic cells by avirulent bacteria and liposomes is well established,160–162 so it is very important to know whether these strategies could also be offered to transport CRISPR/Cas9-phage complex, conjugative plasmids and NPs to eliminate such intracellular pathogens.
Although, CRISPR/Cas9 system has been successfully used to kill bacterial cells, however, the survival of some colonies has been reported by dodging their genome targeting.125,152 Several factors such as spontaneous mutation in spacer sequence, Cas gene or target sequence and the presence of anti-CRISPR (Acr) genes in host genome contribute to emerged resistance against this genome-editing system.163 Thus, to reduce this emerged resistance, future studies need to focus on preventing these spontaneous mutations in the crRNA or Cas gene.164 Target sequence mutations responsible for widespread variants of antibiotic resistant genes, create more challenges for resistance to CRISPR/Cas9 system.
Some more challenges to implement the successful genome-editing by CRISPR/Cas9 also includes the off-target effects for in vivo therapeutic applications. Other challenges regarding the Cas9 from widely studied Streptococcus pyogenes (SpCas9) is its relatively large size, hard to pack in AAV vectors for subsequence bacterial genome-editing.165
The use of CRISPR/Cas9 system along with bioinformatics is emerging as a powerful tool to curb the problems of MDR. Bioinformatics play a significant role in designing and of sgRNA and help in decoding the bacterial genome sequence. The use of innovative computational tools and advanced algorithms in near future will help the researchers to analyze vast amount of antibiotic resistance genome information, track resistance evolution and designing of new antibiotics.
CRISPR/Cas genome-editing system display many potential advantages as compared to conventional antimicrobials. These genome-editing systems are vastly diversified, including almost 33 subtypes.83 It is believed that more efficient CRISPR/Cas types could be exploited in the near future that could eliminate the MDR to maximum. Furthermore, the development of some more innovative CRISPR/Cas9 delivery approaches are expected in near future that will facilitate the effective genome editing in diverse bacteria to curb the MDR problems.
Conclusion
The abusive use of antibiotics in healthcare system has led to a serious impact on human lives by the emergence and wide spread of MDR organisms globally. The modern innovative approach of treating antibiotic resistant organisms by CRISPR/Cas9 genome-editing strategy proves to be a promising method to circumvent the trouble of MDR without targeting beneficial bacteria. Due to some significant features of CRISPR/Cas9, such as high specificity, versatility, efficiency, and programmability, this system can be engineered to target any MDR gene of interest to curb the problems of antibiotic resistance. However, this bacterial genome-targeting approach is still in its infantry stage and faces some challenges in its efficient targeting within bacterial cells. Some delivery approaches such as phages, plasmids and innovative nanoformulations have been engineered to target CRISPR/Cas9 to different bacterial cells, but still the research is proceeding on for better targeting approaches that kill only specific bacteria. In addition, some well-designed bioinformatics tools need to be implemented to advance the decoding of bacterial target sequences and the design of sgRNA and innovative Cas9 variants in near future to minimize further off-target binding sites. This genome-editing tool has a great potential in handling the MDR complications for clinical samples in the near future.
Acknowledgment
Researcher would like to thank the Deanship of Scientific Research, Qassim University for funding publication of this project.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Pacios O, Blasco L, Bleriot I, et al. Strategies to combat multidrug-resistant and persistent infectious diseases. Antibiotics. 2020;9(2):65. doi:10.3390/antibiotics9020065
2. Nath A, Chakrabarti P, Sen S, Barui A. Reactive oxygen species in modulating intestinal stem cell dynamics and function. Stem Cell Rev Rep. 2022;18(7):2328–2350.
3. Brinkac L, Voorhies A, Gomez A, Nelson KE. The threat of antimicrobial resistance on the human microbiome. Microbial Ecology. 2017;74(4):1001–1008.
4. Jacopin E, Lehtinen S, Débarre F, Blanquart F. Factors favouring the evolution of multidrug resistance in bacteria. J Royal Soc Interface. 2020;17(168):20200105.
5. Baym M, Stone LK, Kishony R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science. 2016;351:6268.
6. Roope LS, Smith RD, Pouwels KB, et al. The challenge of antimicrobial resistance: what economics can contribute. Science. 2019;364(6435):eaau4679.
7. Nathan C. Kunkel Lecture: fundamental immunodeficiency and its correction. J Exp Med. 2017;214(8):2175–2191.
8. Al-Ouqaili MT. Identification of an OprD and blaIMP gene-mediated carbapenem resistance in Acinetobacter baumannii and Pseudomonas aeruginosa among patients with wound infections in Iraq. Asian J Pharmaceutics. 2018;12(03):56.
9. Kundar R, Gokarn K. CRISPR-Cas System: a Tool to Eliminate Drug-Resistant Gram-Negative Bacteria. Pharmaceuticals. 2022;15(12):1498.
10. Medina E, Pieper DH. Tackling threats and future problems of multidrug-resistant bacteria. How to overcome the antibiotic crisis: facts, challenges, technologies and future perspectives. Cell Host Microbe. 2016;13:3.
11. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. 2019;10:539.
12. Murugaiyan J, Kumar PA, Rao GS, et al. Progress in alternative strategies to combat antimicrobial resistance: focus on antibiotics. Antibiotics. 2022;11(2):200.
13. Müller V, Rajer F, Frykholm K, et al. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci Rep. 2016;6(1):37938.
14. Schrader SM, Vaubourgeix J, Nathan C. Biology of antimicrobial resistance and approaches to combat it. Sci, trans med. 2020;12:549.
15. Mir A, Edraki A, Lee J, Sontheimer EJ. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol. 2018;13(2):357–365.
16. Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51.
17. Whittle EE, McNeil HE, Trampari E, Webber M, Overton TW, Blair JM. Efflux impacts intracellular accumulation only in actively growing bacterial cells. MBio. 2021;12(5):10–128.
18. Schroeder JW, Yeesin P, Simmons LA, Wang JD. Sources of spontaneous mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2018;53(1):29–48.
19. Balaban NQ, Helaine S, Lewis K, et al. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol. 2019;17(7):441–448.
20. Darby EM, Trampari E, Siasat P, et al. Molecular mechanisms of antibiotic resistance revisited. Nat Rev Microbiol. 2023;21(5):280–295.
21. Alav I, Kobylka J, Kuth MS, et al. Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 2021;121(9):5479–5596.
22. Morgan CE, Glaza P, Leus IV. Cryoelectron microscopy structures of AdeB illuminate mechanisms of simultaneous binding and exporting of substrates. Mbio. 2021;12(1):10–128.
23. Ogawara H. Self-resistance in Streptomyces, with special reference to β-lactam antibiotics. Molecules. 2016;21(5):605.
24. Shinkawa H, Sugiyama M, Nimi O, Nomi R. Molecular cloning and expression in Streptomyces lividans of a streptomycin 6‐phosphotransferase gene from a streptomycin‐producing microorganism. FEBS Lett. 1985;181(2):385–389.
25. Mak S, Xu Y, Nodwell JR. The expression of antibiotic resistance genes in antibiotic‐producing bacteria. Mol Microbiol. 2014;93(3):391–402.
26. Ohnuki TE, Katoh TA, Imanaka TA, Aiba SH. Molecular cloning of tetracycline resistance genes from Streptomyces rimosus in Streptomyces griseus and characterization of the cloned genes. J Bacteriol. 1985;161(3):1010–1016.
27. Reynes JP, Calmels T, Drocourt D, Tiraby G. Cloning, expression in Escherichia coli and nucleotide sequence of a tetracycline-resistance gene from Streptomyces rimosus. Microbiology. 1988;134(3):585–598.
28. Wang TJ, Shan YM, Li H, et al. Multiple transporters are involved in natamycin efflux in S Streptomyces chattanoogensis L10. Mol Microbiol. 2017;103(4):713–728.
29. Yu L, Yan X, Wang L, et al. Molecular cloning and functional characterization of an ATP-binding cassette transporter OtrC from Streptomyces rimosus. BMC Biotech. 2012;12(1):1–2.
30. Li W, Sharma M, Kaur P. The DrrAB efflux system of Streptomyces peucetius is a multidrug transporter of broad substrate specificity. J Biol Chem. 2014;289(18):12633–12646.
31. Ballesta JP, Cundliffe ER. Site-specific methylation of 16S rRNA caused by pct, a pactamycin resistance determinant from the producing organism, Streptomyces pactum. J Bacteriol. 1991;173(22):7213–7218.
32. Kojic M, Topisirovic L, Vasiljevic B. Cloning and characterization of an aminoglycoside resistance determinant from Micromonospora zionensis. J Bacteriol. 1992;174(23):7868–7872.
33. Calcutt MJ, Cundliffe ER. Cloning of a lincosamide resistance determinant from Streptomyces caelestis, the producer of celesticetin, and characterization of the resistance mechanism. J Bacteriol. 1990;172(8):4710–4714.
34. Almutairi MM, Park SR, Rose S, et al. Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides. Proc Natl Acad Sci. 2015;112(42):12956–12961.
35. Ogawara H. Penicillin-binding proteins in Actinobacteria. J Antibiotics. 2015;68(4):223–245.
36. Ogawara H. Distribution of PASTA domains in penicillin-binding proteins and serine/threonine kinases of Actinobacteria. J Antibiotics. 2016;69(9):660–685.
37. Marshall CG, Lessard IA, Park IS, Wright GD. Glycopeptide antibiotic resistance genes in glycopeptide-producing organisms. Antimicrob. Agents Chemother. 1998;42(9):2215–2220.
38. Binda E, Marinelli F, Marcone GL. Old and new glycopeptide antibiotics: action and resistance. Antibiotics. 2014;3(4):572–594.
39. Doyle D, McDowall KJ, Butler MJ, Hunter IS. Characterization of an oxytetracycline‐resistance gene, otrA, of Streptomyces rimosus. Mol Microbiol. 1991;5(12):2923–2933.
40. Prija F, Prasad R. DrrC protein of Streptomyces peucetius removes daunorubicin from intercalated dnrI promoter. Microbiol Res. 2017;202:30–35.
41. Schmutz E, Mühlenweg A, Li SM, Heide L. Resistance genes of aminocoumarin producers: two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob. Agents Chemother. 2003;47(3):869–877.
42. Gatignol A, Durand H, Tiraby G. Bleomycin resistance conferred by a drug‐binding protein. FEBS Lett. 1988;230(1–2):56.
43. Rudolf JD, Bigelow L, Chang C, et al. Crystal structure of the zorbamycin-binding protein ZbmA, the primary self-resistance element in Streptomyces flavoviridis ATCC21892. Biochemistry. 2015;54(45):6842–6851.
44. Liu Y, Tong Z, Shi J, Li R, Upton M, Wang Z. Drug repurposing for next-generation combination therapies against multidrug-resistant bacteria. Theranostics. 2021;11(10):4910.
45. Rangel-Vega A, Bernstein LR, Mandujano-Tinoco EA, García-Contreras SJ, García-Contreras R. Drug repurposing as an alternative for the treatment of recalcitrant bacterial infections. Front Microbiol. 2015;6:282.
46. Hassan M, Kjos M, Nes IF, Diep DB, Lotfipour F. Natural antimicrobial peptides from bacteria: characteristics and potential applications to fight against antibiotic resistance. J Appl Microbiol. 2012;113(4):723–736.
47. Lima PG, Oliveira JT, Amaral JL, Freitas CD, Souza PF. Synthetic antimicrobial peptides: characteristics, design, and potential as alternative molecules to overcome microbial resistance. Life Sci. 2021;278:119647.
48. Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World j Gastrointestinal Pharmacology Therapeutics. 2017;8(3):162.
49. Kortright KE, Chan BK, Koff JL, Turner PE. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232.
50. Ménard R, Schoenhofen IC, Tao L, et al. Small-molecule inhibitors of the pseudaminic acid biosynthetic pathway: targeting motility as a key bacterial virulence factor. Antimicrob. Agents Chemother. 2014;58(12):7430–7440.
51. Rai MK, Deshmukh SD, Ingle AP, Gade AK. Silver nanoparticles: the powerful nanoweapon against multidrug‐resistant bacteria. J Appl Microbiol. 2012;112(5):841–852.
52. Makabenta JM, Nabawy A, Li CH, Schmidt-Malan S, Patel R, Rotello VM. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat Rev Microbiol. 2021;19(1):23–36.
53. Al-Ouqaili MT, Saleh RO, Amin HI, et al. Synthesize of pluronic-based nanovesicular formulation loaded with Pistacia atlantica extract for improved antimicrobial efficiency. Arabian J. Chem. 2023;16(6):104704.
54. Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev. 2012;25(3):450–470.
55. Petrosillo N, Capone A, Di Bella S, Taglietti F. Management of antibiotic resistance in the intensive care unit setting. Expert review of anti-infective therapy. Cell Host Microbe. 2010;8(3):289–302.
56. León-Buitimea A, Garza-Cárdenas CR, Garza-Cervantes JA, Lerma-Escalera JA, Morones-Ramírez JR. The demand for new antibiotics: antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front Microbiol. 2020;1669.
57. Aslam B, Rasool M, Idris A, et al. CRISPR-Cas system: a potential alternative tool to cope antibiotic resistance. Antimicrob Resist Infect Control. 2020;9(1):1–3.
58. Doss J, Culbertson K, Hahn D, Camacho J, Barekzi N. A review of phage therapy against bacterial pathogens of aquatic and terrestrial organisms. Viruses. 2017;9(3):50.
59. Pelfrene E, Mura M, Sanches AC, Cavaleri M. Monoclonal antibodies as anti-infective products: a promising future? Clin Microbiol Infect. 2019;25(1):60–64.
60. Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11(2):125–140.
61. Kotil S, Jakobsson E. Rationally designing antisense therapy to keep up with evolving bacterial resistance. PLoS One. 2019;14(1):e0209894.
62. Forsyth RA, Haselbeck RJ, Ohlsen KL, et al. A genome‐wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43(6):1387–1400.
63. Zhang HX, Zhang Y, Yin H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol Ther. 2019;27(4):735–746.
64. Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduction Targeted Therapy. 2020;5(1):1.
65. Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci Rep. 2017;7(1):44929.
66. Kang S, Kim J, Hur JK, Lee SS. CRISPR-based genome editing of clinically important Escherichia coli SE15 isolated from indwelling urinary catheters of patients. J Med Microbiol. 2017;66(1):18–25.
67. Redman M, King A, Watson C, King D. What is CRISPR/Cas9? Arch Dis Childhood-Educ Practice. 2016;101(4):213–215.
68. Kim TH, Lee SW. Therapeutic Application of Genome Editing Technologies in Viral Diseases. Int J Mol Sci. 2022;23(10):5399.
69. Mani I. CRISPR-Cas9 for treating hereditary diseases. Progress Mol Biol Translational Sci. 2021;181:165–183.
70. Wu SS, Li QC, Yin CQ, Xue W, Song CQ. Advances in CRISPR/Cas-based gene therapy in human genetic diseases. Theranostics. 2020;10(10):4374.
71. Uribe RV, Rathmer C, Jahn LJ, Ellabaan MM, Li SS, Sommer MO. Bacterial resistance to CRISPR-Cas antimicrobials. Sci Rep. 2021;11(1):17267.
72. Wu Y, Battalapalli D, Hakeem MJ, et al. Engineered CRISPR-Cas systems for the detection and control of antibiotic-resistant infections. J Nanobiotechnol. 2021;19(1):1–26.
73. Jwair NA, Al-Ouqaili MT, Al-Marzooq F. Inverse Association between the Existence of CRISPR/Cas Systems with Antibiotic Resistance, Extended Spectrum β-Lactamase and Carbapenemase Production in Multidrug, Extensive Drug and Pandrug-Resistant Klebsiella pneumoniae. Antibiotics. 2023;12(6):980.
74. Alduhaidhawi AH, AlHuchaimi SN, Al-Mayah TA, et al. Prevalence of CRISPR-cas systems and their possible association with antibiotic resistance in Enterococcus faecalis and Enterococcus faecium collected from hospital wastewater. Infect Drug Resist. 2022;1:1143–1154.
75. Wan F, Draz MS, Gu M, Yu W, Ruan Z, Luo Q. Novel strategy to combat antibiotic resistance: a sight into the combination of CRISPR/Cas9 and nanoparticles. Pharmaceutics. 2021;13(3):352.
76. Zohra T, Numan M, Ikram A, et al. Cracking the challenge of antimicrobial drug resistance with CRISPR/Cas9, nanotechnology and other strategies in ESKAPE pathogens. Microorganisms. 2021;9(5):954.
77. Horodecka K, Düchler M. CRISPR/Cas9: principle, applications, and delivery through extracellular vesicles. Int J Mol Sci. 2021;22(11):6072.
78. Duan C, Cao H, Zhang LH, Xu Z. Harnessing the CRISPR-Cas systems to combat antimicrobial resistance. Front Microbiol. 2021;12:716064.
79. Palacios D, Palmer KL, Duerkop BA. CRISPR-based antimicrobials to obstruct antibiotic-resistant and pathogenic bacteria. PLoS Pathogens. 2021;17(7):e1009672.
80. Bikard D, Barrangou R. Using CRISPR-Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017;37:155–160.
81. Khadempar S, Familghadakchi S, Motlagh RA, et al. CRISPR–Cas9 in genome editing: its function and medical applications. J Cell Physiol. 2019;234(5):5751–5761.
82. Gholizadeh P, Ş K, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;20:1111–1121.
83. Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9(6):467–477.
84. Fage C, Lemire N, Moineau S. Delivery of CRISPR-Cas systems using phage-based vectors. Curr. Opin. Biotechnol. 2021;68:174–180.
85. Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnol. 2014;32(11):1146–1150.
86. Aslam S, Lampley E, Wooten D, et al. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. InOpen Forum Infectious Dis. 2020;7(9):ofaa389.
87. Allemailem KS, Alsahli MA, Almatroudi A, et al. Current updates of CRISPR/Cas9‐mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management. Cancer Commun. 2022;42(12):1257–1287.
88. Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E. CRISPR-Cas in Streptococcus pyogenes. RNA Biology. 2019;16(4):380–389.
89. Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949.
90. Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128.
91. Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340).
92. Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529.
93. Ray A, Di Felice R. Molecular simulations have boosted knowledge of CRISPR/Cas9: a Review. arXiv preprint, arXiv. 2019.
94. Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci. 2017;114(28):7260–7265.
95. Zheng W. Probing the structural dynamics of the CRISPR‐Cas9 RNA‐guided DNA‐cleavage system by coarse‐grained modeling. Proteins: structure. Funct Bioinf. 2017;85(2):342–353.
96. Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. Striking plasticity of CRISPR-Cas9 and key role of non-target DNA, as revealed by molecular simulations. ACS Cent. Sci. 2016;2(10):756–763.
97. Tang L, Zeng Y, Du H, et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomic. 2017;292(3):525–533.
98. Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573.
99. Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9–guide RNA complex preorganized for target DNA recognition. Science. 2015;348(6242):1477–1481.
100. Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997.
101. Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Biophys. J. 2014;106(2):695a.
102. Kamruzzaman M, Iredell JR. CRISPR-Cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front Microbiol. 2020;10:2934.
103. Allemailem KS, Alsahli MA, Almatroudi A, et al. Innovative Strategies of Reprogramming Immune System Cells by Targeting CRISPR/Cas9-Based Genome-Editing Tools: a New Era of Cancer Management. Int j Nanomed. 2023;31:5531–5559.
104. Wimmer F, Beisel CL. CRISPR-Cas systems and the paradox of self-targeting spacers. Front Microbiol. 2020;10:3078.
105. Hussain W, Mahmood T, Hussain J, et al. CRISPR/Cas system: a game changing genome editing technology, to treat human genetic diseases. Gene. 2019;685:70–75.
106. de la Fuente-Núñez C, Lu TK. CRISPR-Cas9 technology: applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. Integr Biol. 2017;9(2):109–122.
107. Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26.
108. Barman NC, Khan NM, Islam M, et al. CRISPR-Cas9: a promising genome editing therapeutic tool for Alzheimer’s disease—A narrative review. Neurol Therapy. 2020;9:419–434.
109. Ekwebelem OC, Aleke J, Ofielu E, Nnorom-Dike O. CRISPR-Cas9 system: a revolutionary tool in the fight against antimicrobial resistance. Infectious Microbes Dis. 2021;3(2):51–56.
110. Shabbir MA, Shabbir MZ, Wu Q, et al. CRISPR-cas system: biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann Clinic Microbiol Antimicrob. 2019;18:1–9.
111. Wang P, He D, Li B, et al. Eliminating mcr-1-harbouring plasmids in clinical isolates using the CRISPR/Cas9 system. J Antimicrob Chemother. 2019;74(9):2559–2565.
112. Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139.
113. Sun Q, Wang Y, Dong N, et al. Application of CRISPR/Cas9-based genome editing in studying the mechanism of pandrug resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2019;63(7):10–128.
114. Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4):10–128.
115. Rodrigues M, McBride SW, Hullahalli K, Palmer KL, Duerkop BA. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob Agents Chemother. 2019;63(11):10–128.
116. de Maat V, Stege PB, Dedden M, et al. CRISPR-Cas9-mediated genome editing in vancomycin-resistant Enterococcus faecium. FEMS Microbiol Lett. 2019;366(22):fnz256.
117. Neil K, Allard N, Roy P, et al. High‐efficiency delivery of CRISPR‐Cas9 by engineered probiotics enables precise microbiome editing. Mol Syst Biol. 2021;17(10):e10335.
118. Wanner S, Schade J, Keinhörster D, et al. Wall teichoic acids mediate increased virulence in Staphylococcus aureus. Nature Microbiol. 2017;2(4):1–2.
119. Wu X, Zha J, Koffas MA, Dordick JS. Reducing Staphylococcus aureus resistance to lysostaphin using CRISPR‐dCas9. Biotechnol. Bioeng. 2019;116(12):3149–3159.
120. Hao M, He Y, Zhang H, et al. CRISPR-Cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2020;64(9):10–128.
121. Allemailem KS, Almatroodi SA, Almatroudi A, et al. Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: a Future Perspective of Cancer Management. Int J Mol Sci. 2023;24(8):7052.
122. Hatoum-Aslan A. Phage genetic engineering using CRISPR–Cas systems. Viruses. 2018;10(6):335.
123. Fagen JR, Collias D, Singh AK, Beisel CL. Advancing the design and delivery of CRISPR antimicrobials. Curr Opin Biomed Eng. 2017;4:57–64.
124. Dowah AS, Clokie MR. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018;10:535–542.
125. Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5(1):10–128.
126. Ando H, Lemire S, Pires DP, Lu TK. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Systems. 2015;1(3):187–196.
127. Chadha P, Katare OP, Chhibber S. Liposome loaded phage cocktail: enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns. 2017;43(7):1532–1543.
128. Colom J, Cano-Sarabia M, Otero J, Cortés P, Maspoch D, Llagostera M. Liposome-encapsulated bacteriophages for enhanced oral phage therapy against Salmonella spp. Appl Environ Microbiol. 2015;81(14):4841–4849.
129. Singla S, Harjai K, Katare OP, Chhibber S. Encapsulation of bacteriophage in liposome accentuates its entry in to macrophage and shields it from neutralizing antibodies. PLoS One. 2016;11(4):e0153777.
130. Esteban PP, Jenkins AT, Arnot TC. Elucidation of the mechanisms of action of Bacteriophage K/nano-emulsion formulations against S. aureus via measurement of particle size and zeta potential. Colloids Surf., B. 2016;139:87–94.
131. Abdelsattar AS, Abdelrahman F, Dawoud A, Connerton IF, El-Shibiny A. Encapsulation of E. coli phage ZCEC5 in chitosan–alginate beads as a delivery system in phage therapy. Amb Express. 2019;9(1):1–9.
132. Cobb LH, Park J, Swanson EA, et al. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS One. 2019;14(11):e0220421.
133. Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16(6):387–399.
134. Kang YK, Kwon K, Ryu JS, Lee HN, Park C, Chung HJ. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjugate Chem. 2017;28(4):957–967.
135. Gama JA, Zilhão R, Dionisio F. Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid. 2017;93:6–16.
136. Cabezón E, Ripoll-Rozada J, Peña A, De La Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015;39(1):81–95.
137. Pereira HS, Tagliaferri TL, Mendes TA. Enlarging the toolbox against antimicrobial resistance: aptamers and CRISPR-Cas. Front Microbiol. 2021;12:606360.
138. Pursey E, Sünderhauf D, Gaze WH, Westra ER, van Houte S. CRISPR-Cas antimicrobials: challenges and future prospects. PLoS Pathogens. 2018;14(6):e1006990.
139. Riley MK, Vermerris W. Recent advances in nanomaterials for gene delivery—a review. Nanomaterials. 2017;7(5):94.
140. Sahu R, Verma R, Dixit S, et al. Future of human Chlamydia vaccine: potential of self-adjuvanting biodegradable nanoparticles as safe vaccine delivery vehicles. Exp Rev Vaccines. 2018;17(3):217–227.
141. Lee K, Conboy M, Park HM, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1(11):889–901.
142. Rahimi H, Salehiabar M, Charmi J, et al. Harnessing nanoparticles for the efficient delivery of the CRISPR/Cas9 system. Nano Today. 2020;34:100895.
143. Chiaradia L, Lefebvre C, Parra J, et al. Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci Rep. 2017;7(1):12807.
144. Gupta S, Kumar P, Rathi B, et al. Targeting of Uropathogenic Escherichia coli papG gene using CRISPR-dot nanocomplex reduced virulence of UPEC. Sci Rep. 2021;11(1):17801.
145. Collins SM, Brown AC. Bacterial outer membrane vesicles as antibiotic delivery vehicles. Front Immunol. 2021;12:733064.
146. Martora F, Pinto F, Folliero V, et al. Isolation, characterization and analysis of pro-inflammatory potential of Klebsiella pneumoniae outer membrane vesicles. Microb Pathogenesis. 2019;136:103719.
147. Dell’Annunziata F, Folliero V, Giugliano R, et al. Gene transfer potential of outer membrane vesicles of gram-negative bacteria. Int J Mol Sci. 2021;22(11):5985.
148. Yao X, Lyu P, Yoo K, et al. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J Extracell Vesicles. 2021;10(5):e12076.
149. Fantappiè L, de Santis M, Chiarot E, et al. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J Extracell Vesicles. 2014;3(1):24015.
150. Lu M, Xing H, Yang Z, et al. Recent advances on extracellular vesicles in therapeutic delivery: challenges, solutions, and opportunities. Eur. J. Pharm. Biopharm. 2017;119:381–395.
151. Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio. 2012;3(2):e00277–11.
152. Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature Biotechnol. 2014;32(11):1141–1145.
153. Wan P, Cui S, Ma Z, et al. Reversal of mcr-1-mediated colistin resistance in Escherichia coli by CRISPR-Cas9 system. Infect Drug Resist. 2020;22:1171–1178.
154. Reuter A, Hilpert C, Dedieu-Berne A, et al. Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Res. 2021;49(6):3584–3598.
155. Dong H, Xiang H, Mu D, Wang D, Wang T. Exploiting a conjugative CRISPR/Cas9 system to eliminate plasmid harbouring the mcr-1 gene from Escherichia coli. Int J Antimicrob Agents. 2019;53(1):1–8.
156. Wu ZY, Huang YT, Chao WC, Ho SP, Cheng JF, Liu PY. Reversal of carbapenem-resistance in Shewanella algae by CRISPR/Cas9 genome editing. J Adv Res. 2019;18:61–69.
157. Hamilton TA, Pellegrino GM, Therrien JA, et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat Commun. 2019;10(1):4544.
158. Ram G, Ross HF, Novick RP, Rodriguez-Pagan I, Jiang D. Conversion of staphylococcal pathogenicity islands to CRISPR-carrying antibacterial agents that cure infections in mice. Nature Biotechnol. 2018;36(10):971–976.
159. Kim HY, Chang RY, Morales S, Chan HK. Bacteriophage-delivering hydrogels: current progress in combating antibiotic resistant bacterial infection. Antibiotics. 2021;10(2):130.
160. Broxmeyer L, Sosnowska D, Miltner E, et al. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium: a model for phage therapy of intracellular bacterial pathogens. J Infect Dis. 2002;186(8):1155–1160.
161. Nieth A, Verseux C, Barnert S, Süss R, Römer W. A first step toward liposome-mediated intracellular bacteriophage therapy. Expert Opin Drug Delivery. 2015;12(9):1411–1424.
162. Yan W, Banerjee P, Xu M, et al. Formulation strategies for bacteriophages to target intracellular bacterial pathogens. Adv. Drug Delivery Rev. 2021;176:113864.
163. Wu X, Zhu J, Tao P, Rao VB. Bacteriophage T4 escapes CRISPR attack by minihomology recombination and repair. MBio. 2021;12(3):10–128.
164. Csörgő B, León LM, Chau-Ly IJ, et al. A compact Cascade–Cas3 system for targeted genome engineering. Nature Methods. 2020;17(12):1183–1190.
165. Uddin F, Rudin CM, Sen T. CRISPR gene therapy: applications, limitations, and implications for the future. Front Oncol. 2020;10:1387.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.