Back to Journals » Infection and Drug Resistance » Volume 13
Rapid Detection of Ethambutol-Resistant Mycobacterium tuberculosis from Sputum by High-Resolution Melting Analysis in Beijing, China
Authors Wang J, Zhao W, Liu R, Huo F, Dong L, Xue Y, Wang Y, Xue Z, Ma L , Pang Y
Received 6 July 2020
Accepted for publication 29 August 2020
Published 20 October 2020 Volume 2020:13 Pages 3707—3713
DOI https://doi.org/10.2147/IDR.S270542
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Jun Wang,1,* Weijie Zhao,2,* Rongmei Liu,1,* Fengmin Huo,3 Lingling Dong,3 Yi Xue,3 Yufeng Wang,4 Zhongtan Xue,4 Liping Ma,1 Yu Pang3
1Department of Tuberculosis, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis & Thoracic Tumor Research Institute, Beijing 101149, People’s Republic of China; 2Clinical Trial Agency Office, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis & Thoracic Tumor Research Institute, Beijing 101149, People’s Republic of China; 3National Clinical Laboratory on Tuberculosis, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis & Thoracic Tumor Research Institute, Beijing 101149, People’s Republic of China; 4Department of Laboratory Quality Control, Innovation Alliance on Tuberculosis Diagnosis and Treatment (Beijing), Beijing 101149, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yu Pang
National Clinical Laboratory on Tuberculosis, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis & Thoracic Tumor Research Institute, No. 9, Beiguan Street, Tongzhou District, Beijing 101149, People’s Republic of China
Tel/Fax +86-10-8950 9359
Email [email protected]
Liping Ma
Department of Tuberculosis, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis & Thoracic Tumor Research Institute, No. 9, Beiguan Street, Tongzhou District, Beijing 101149, People’s Republic of China
Tel/Fax +86-10-8950 9322
Email [email protected]
Objective: We conducted a retrospective study to evaluate the performance of MeltPro assay for detecting ethambutol (EMB) susceptibility of Mycobacterium tuberculosis (MTB) isolates in sputum specimens in Beijing, China.
Methods: Smear-positive TB patients undergoing MeltPro assay in the Beijing Chest Hospital between January 2019 and December 2019 were included. Phenotypic drug susceptibility testing (DST) was used as the reference standard to calculate the diagnostic accuracy of MeltPro assay for EMB resistance. Sanger sequencing of embB gene was conducted to resolve the discrepancies between MeltPro assay and phenotypic DST.
Results: A total of 222 smear-positive patients were included in our analysis. The overall agreement rate between the two assays was 91.4%, with a kappa value of 0.78. Among 59 EMB-resistant TB cases diagnosed by DST, 49 were identified by MeltPro assay, demonstrating a sensitivity of 83.1%. In addition, 154 out of 163 EMB-susceptible patients diagnosed by DST were correctly detected with MeltPro assay, yielding a specificity of 93.9%. The probe frequency associated with the observed EMB-resistance was as follows: A (45/58), B (7/58), and D (6/58), and no EMB-resistance was associated with probe C. The presence of amino acid substitution was observed among all 9 cases with potentially “false-negative” results, including 7 with Met306Ile, 1 with Met306Val, 1 with Gly406Asp, respectively.
Conclusion: MeltPro assay is a promising diagnostic tool for the detection of EMB resistance in China. The specific amino acid substitution in embB gene is the major reason for discrepancies between MeltPro assay and phenotypic DST.
Keywords: tuberculosis, ethambutol, molecular diagnostic, embB, China
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB) complex, is the leading cause of death among all infectious diseases.1,2 According to World Health Organization (WHO) estimates, 10.0 million people were ill with TB, and 1.24 million died of TB worldwide in 2018.2 China ranked second among the 30 high-burden countries, behind only India, with approximately 0.87 million reported TB cases in 2018.2 Despite the great achievements in TB control over the past 20 years, the emergence of drug-resistant TB seriously threatens the national TB control effort in China.3,4 Early diagnosis of drug-resistant TB is essential for initiating proper treatment, and also preventing its transmission in the community.5
The routine diagnosis of drug-resistant TB relies on phenotypical in vitro drug susceptibility testing (DST).6 Due to the slow metabolism of the causative MTB, the isolation and DST require several weeks, constituting a major barrier for the timely management of drug-resistant patients with appropriate antibiotics.7 Currently, research on the molecular basis of drug resistance in MTB allows the use of molecular diagnostics to detect drug-resistant MTB by identification of mutations conferring resistance.8 The WHO has approved the use of GenoType® MTBDRplus assay and GeneXpert MTB/RIF for the screening of drug-resistant TB in clinical practice.9,10 The use of molecular diagnostics ensures the availability of results within hours, which would conceivably shorten the time to treatment for individuals affected by TB.8
Ethambutol (EMB) is an important first-line anti-TB drug for treating drug-susceptible TB and preventing the emergence of drug resistance.11 It is also often used to formulate regimens for drug-resistant TB in view of the pronounced synergistic effects of EMB in combination with other drugs.12 The primary target of EMB is the embCAB gene locus, involved in the biosynthesis of the cell wall components arabinogalactan and lipoarabinomannan.13 The emergence of EMB resistance is majorly associated with mutations within embB gene, especially the canonical mutations at codons 306, 406 and 497.11 However, the mutations at these codons are also identified in EMB-susceptible MTB isolates.14 The conflicting results between phenotypic and genotypic resistance testing raise concerns about the clinical significance of EMB mutations for the development of EMB resistance. As a consequence, the molecular diagnostics for EMB susceptibility lag far behind those for other anti-TB drugs, thereby constituting a crucial barrier to optimization of an appropriate treatment regimen. Recently, the MeltPro® MTB/EMB kit (MeltPro, Zeesan Biotech, Xiamen, China), a high-resolution melting curve-based nucleic acid amplification testing (NAAT), was approved by Chinese Food and Drug Administration.15 Given that limited clinical evidence has been reported on the diagnostic accuracy of NAATs on EMB resistance, we conducted a retrospective study to evaluate the performance of this novel molecule method for detecting EMB susceptibility of MTB isolates in sputum specimens from pulmonary TB.
Materials and Methods
Study Design
We conducted a retrospective study in the Beijing Chest Hospital, Capital Medical University between January 2019 and December 2019. Smear-positive TB patients were included in this analysis irrespective of comorbidities. The patients had examination results from sputum specimens by smear microscopy, mycobacterial culture and MeltPro. In vitro drug susceptibility was conducted on the positive cultures. The demographic and clinical information of participants was anonymized and extracted from electronic medical record system.
Laboratory Examination
Briefly, sputum samples were collected and transported to the laboratory for examination. Direct smear from each sputum sample was examined with Auramine O staining for acid fast bacilli (AFB).16 1.0 mL of sputum was digested with N-acetyl-L-cysteine and sodium hydroxide (NALC-NaOH) for 15 minutes, and PBS buffer was added up to a total volume of 45 mL. After centrifugation for 15 min at 3000 × g, the suspension of sediment was inoculated into a BACTEC MGIT tube (BD Microbiology Systems, USA). For smear-positive samples, the remaining 1.0 mL of sputum specimen was detected with MeltPro® MTB/EMB kit according to the manufacturer’s instructions.15 Four dually labeled probe cover regions of embB codons 306, 497, 368, 378, 380 and 406 consisting of the most common mutation types conferring EMB resistance (Figure 1). The probes were perfectly matched with the wild-type sequence of embB gene. The corresponding genotype was interpreted according to the Tm of melting curve of double-stranded structures between probe and targeted embB gene. Fluorescence was recorded at FAM and TET channels. Positive cultures from specimens initially identified as M. tuberculosis complex with the MPT64 antigen kit (Genesis, Hangzhou, China) were subcultured on the Lӧwenstein-Jensen (L-J) medium. The 4-week-old cultures of bacteria were collected for phenotypic DST with MGIT 960 automated system according to the manufacturer’s instructions.15 The drug concentration for EMB was 5.0 μg/mL in MGIT tube.
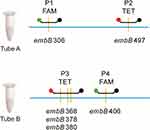 |
Figure 1 The region of embB targeted by the probe of MeltPro TB assay. |
DNA Sequencing
Crude genomic DNA was extracted from the positive culture using boiling method.5 The frozen isolate in 7H9 medium containing 10% oleic acid-albumin-dextrose-catalase (OADC) was firstly subcultured on Löwenstein-Jensen (L-J) medium. After 4-week incubation at 37°C, colonies were harvested from the surface of L-J medium and heated for 30 min at 95°C in 500 μL Tris-EDTA (TE) buffer. The supernatant was used as template for amplification. An 800-bp fragment of embB gene containing EMB resistance determining region was amplified with the primer pair described by our previous report.17 The PCR amplicons were sent to the Tsingke Biotech Company for DNA sequencing service (Tsingke Biotech Company, Beijing, China). We entered the sequencing results into the Basic Local Alignment Search Tool (BLAST) for comparison with the corresponding genes of strain H37Rv (ATCC27294).
Statistical Analysis
The phenotypic DST was used as the reference standard to calculate the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The Kappa value was used to assess the agreement between MeltPro and phenotypic DST. A Kappa value more than 0.75 was taken to indicate excellent agreement. In addition, Chi-squared test was conducted to compare the performance of MeltPro stratified to different initial drug-resistant profiles. All calculations were completed in SPSS 20.0 (IBM Corp., Armonk, NY, USA). A P value less than 0.05 was considered statistically significant.
Results
Patient Enrollment
A total of 249 patients underwent MeltPro assay for EMB resistance between January 2019 and December 2019. Of these patients, 27 (10.8%) were excluded due to being culture negative (6/249, 2.4%), culture contamination (10/249, 4.0%) and detection failure by MeltPro (9/249, 3.6%), respectively (Figure 2). Finally, 222 patients were included in our final analysis. The demographic and clinical characteristics are summarized in Table 1. The mean age was 46.7 years (range, 15 to 88 years), 165 patients (74.3%) were male, and 123 (55.4%) were new cases. The most frequently observed underlying disease was diabetes (60/222, 27.0%), followed by cardiopathy (12/222, 5.4%) and liver diseases (6/222, 2.7%). Resistance was noted in 30.6% of patients (68/222) for rifampicin, 37.4% (83/222) for isoniazid, 17.6% (39/222) for levofloxacin, and 7.2% (16/222) for amikacin. In addition, 53 (53/222, 23.9%) patients were afflicted with multidrug-resistant tuberculosis and 11 (11/222, 5.0%) had extensively drug-resistant tuberculosis.
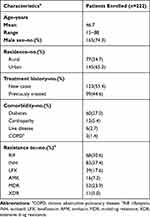 |
Table 1 Demographic and Clinical Characteristics of Patients Enrolled in This Study |
 |
Figure 2 Enrollment of participants. |
MeltPro Assay
We firstly evaluated the performance of MeltPro assay versus phenotypic DST for detecting EMB resistance. As shown in Table 2, the overall agreement rate between the two assays was 91.4% [203/222, 95% confidence interval (95% CI): 87.8–95.1], with a kappa value of 0.78. Among 59 EMB-resistant TB cases diagnosed by DST, 49 were identified by MeltPro assay, demonstrating a sensitivity of 83.1% (95% CI: 73.5–92.6). In addition, 154 out of 163 EMB-susceptible patients diagnosed by DST were correctly detected with MeltPro assay, yielding a specificity of 93.9% (95% CI: 90.2–97.6). The most common probe for EMB resistance detection was probe A (n=45, 77.6%). The frequency of probe B and D conferring EMB resistance was 7 (12.1%) and 6 (10.3%), respectively. In contrast, no isolate harboring mutation was detected within probe C (Table 3).
 |
Table 2 Performance of MeltPro for Detecting Ethambutol Resistance |
 |
Table 3 Distribution of embB Mutations Stratified to MeltPro Assay |
Sequencing results
We further conducted Sanger DNA sequencing analysis to resolve the discrepancies between MeltPro assay and phenotypic DST. DNA sequencing results of 19 discordant isolates are listed in Table 4. The presence of amino acid substitution was observed among all 9 cases with potentially “false-negative” results, including 7 (7/9) with Met306Ile, 1 (1/9) with Met306Val, 1 (1/9) with Gly406Asp, respectively. In addition, out of 10 cases with MeltPro S/MGIT R results, no genotypic mutations were detected in 9 cases by a screen of the embB gene, whereas one case harbored mutation in codon 328, which fell outside of probe-covering regions.
 |
Table 4 Analysis on Discordant Results Between MeltPro and MGIT for EMB Resistance |
Discussion
Early detection of drug resistance is essential to initiate appropriate anti-TB therapy regimens.8 Molecular diagnosis techniques for EMB resistance have lagged behind other drugs. In the present study, we assessed the diagnostic accuracy of MeltPro assay for detection of EMB resistance among TB patients. Our data have demonstrated that MeltPro assay provides an accurate option for the diagnosis of EMB-resistance among smear-positive patients. In a recent meta-analysis on the diagnostic value of GenoType® MTBDRsl, its pooled sensitivity was 67.9% for EMB resistance,18 which is significantly lower than our observation (83.1%). Several plausible explanations may be responsible for the difference. On the one hand, the GenoType® MTBDRsl only uses the embB306 mutations as molecular markers for predicting EMB resistance.18 The inclusion of more mutations in the detection panel of MeltPro assay, including embB codons 497, 368, 378, 380 and 406 conferring EMB resistance, therefore increased its sensitivity to identify EMB resistance. On the other hand, the main drawback of GenoType® MTBDRsl is its poor capability to detect less than 50% heteroresistance. In contrast, MeltPro outperforms GenoType® MTBDRsl in diagnosis of EMB-resistant patients due to the cooccurrence of susceptible and resistant bacteria population, which serves as another explanation for its increased sensitivity.19
Mutation in emb306 is a major mechanism of EMB resistance in MTB.14 Previous studies showed that the percentage of emb306 mutations presented geographic diversity, varying from 30% in Kuwait to 68% in Germany.14,20 In a recent study from Li et al, the amino acid substitution at codon 306 was noted in 69% of EMB-resistant isolates from China.21 Similarly, emb306 occurred only in 63% of EMB-resistant MTB isolates in our cohort. Additionally, exchange wild-type embB497 and embB406 with mutant codons serve as another predominant mechanism, accounting for ~20% EMB resistance in the present study, while no mutation was found within the codons 368, 378 and 380 covered by probe C. Based on our experience, multiple-site detection panel of embB 306, 406, and 497 (probe A, B and D) provides an effective solution for detection of EMB-resistant mutants in China. In view of the difference in molecular characteristics of EMB-resistant MTB isolates across regions, the diagnostic accuracy of MeltPro for EMB resistance should be systematically evaluated in high-burden settings.
Our results support the findings of previous studies that phenotypically EMB-susceptible isolates carried mutations at codon 306 of the embB gene.14,22 There is strong evidence that the embB mutants had increased minimum inhibitory concentrations (MICs) compared to the wild-type tubercle bacilli;23 but the wide range of increased MICs makes it difficult to identify EMB-resistant isolates with MICs close to the critical concentration of phenotypic DST method.6 These findings raise concerns regarding the standardization of phenotypic DST for EMB. As a bacteriostatic agent, the culture-based in vitro phenotypic DST methods for EMB can be notoriously problematic because of small difference in MICs between EMB-susceptible and EMB-resistant strains.6,24 Another barrier for accurate EMB susceptibility testing is the poor stability of this drug during incubation at 37°C.25 Indeed, the results on the proficiency testing on DST in supranational TB reference laboratories have demonstrated that the performance of EMB testing was the worst among all the first line drugs tested.26 We speculate that the presumably inaccurate methodology for phenotypic DST is the major reason for the “EMB-susceptible” isolates with embB mutations; thus the molecular tests are more likely to warrant accurate EMB susceptibility results for MTB.
We also acknowledge several limitations to our study. First, the present study was retrospective in design with all of the inherent limitations of such studies. The potential bias in patient inclusion may limit the significance of our conclusion. Second, only the sequences of embB gene were analyzed in our experiments. Therefore, it is difficult to identify EMB-resistant isolates that do not carry mutations in this locus. Further whole-genome sequencing will be conducted to elucidate the molecular mechanism in EMB-resistant isolates without embB mutations. Third, in view of the consistency between phenotypic and genotypic assays, the cases with MeltPro S/MGIT S and MeltPro R/MGIT R were not analyzed using Sanger sequencing. It may weaken the significance of our conclusion. Fourth, embB306 mutation is considered as the predictive marker for MDR-TB,27 which was not assessed in the present study. Despite these limitations, our data provide important evidence for further integrating MeltPro into the diagnosis of drug-resistant TB by national TB programs in China.
In conclusion, our results demonstrate that MeltPro assay is a promising diagnostic tool for the detection of EMB resistance in China. The specific amino acid substitution in embB gene is the major reason for discrepancies between MeltPro assay and phenotypic DST. In view of presumably inaccurate methodology for phenotypic DST, molecular tests are more likely to warrant accurate EMB susceptibility results for MTB.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of Beijing Chest Hospital affiliated to Capital Medical University, and each participant signed a written informed consent to agree with the anonymous use of clinical data.
Acknowledgments
This work was supported by the National Major Project (2018ZX10103001-004) and the Beijing Hospitals Authority’ Ascent Plan (DFL20191601).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Raviglione M, Sulis G. Tuberculosis 2015: burden, Challenges and Strategy for Control and Elimination. Infectious Disease Reports. 2016;8(2):6570. doi:10.4081/idr.2016.6570
2. World Health Organization. (2019) Global Tuberculosis Report 2019. 2019.
3. Wang L, Zhang H, Ruan Y, et al. Tuberculosis prevalence in China, 1990–2010; a longitudinal analysis of national survey data. Lancet. 2014;383(9934):2057–2064. doi:10.1016/S0140-6736(13)62639-2
4. Zhao Y, Xu S, Wang L, et al. National survey of drug-resistant tuberculosis in China. N Engl J Med. 2012;366(23):2161–2170. doi:10.1056/NEJMoa1108789
5. Zhang Z, Lu J, Liu M, et al. Genotyping and molecular characteristics of multidrug-resistant. Mycobacterium Tuberculosis Isolates from China J Infect. 2015;70(4):335–345.
6. Kim SJ. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J. 2005;25(3):564–569.
7. Pang Y, Xia H, Zhang Z, et al. Multicenter evaluation of genechip for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2013;51(6):1707–1713.
8. Miotto P, Zhang Y, Cirillo DM, Yam WC. Drug resistance mechanisms and drug susceptibility testing for tuberculosis. Respirology. 2018;23(12):1098–1113.
9. World Health Organization. Policy Update: Xpert MTB/RIF Assay for the Diagnosis of Pulmonary and Extrapulmonary TB in Adults and Children. Geneva, Switherland: World Health Organization; 2013.
10. World Health Organization. The Use of Molecular Line Probe Assay for the Detection of Resistance to Second-Line Anti-Tuberculosis Drugs. Geneva, Switherland: World Health Organization; 2013.
11. Zhao LL, Sun Q, Liu HC, et al. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother. 2015;59(4):2045–2050.
12. Zhu C, Liu Y, Hu L, Yang M, He ZG. Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis. J Biol Chem. 2018;293(43):16741–16750.
13. Telenti A, Philipp WJ, Sreevatsan S, et al. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat Med. 1997;3(5):567–570.
14. Plinke C, Rusch-Gerdes S, Niemann S. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2006;50(5):1900–1902.
15. Pang Y, Dong H, Tan Y, et al. Rapid diagnosis of MDR and XDR tuberculosis with the MeltPro TB assay in China. Sci Rep. 2016;6:25330.
16. Xia H, Song YY, Zhao B, et al. Multicentre evaluation of Ziehl-Neelsen and light-emitting diode fluorescence microscopy in China. Int J Tuberc Lung Dis. 2013;17(1):107–112.
17. Chen Q, Pang Y, Liang Q, et al. Molecular characteristics of MDR Mycobacterium tuberculosis strains isolated in Fujian. China Tuberculosis. 2014;94(2):159–161.
18. Feng Y, Liu S, Wang Q, et al. Rapid diagnosis of drug resistance to fluoroquinolones, amikacin, capreomycin, kanamycin and ethambutol using genotype MTBDRsl assay: a meta-analysis. PLoS One. 2013;8(2):e55292.
19. Hu S, Li G, Li H, et al. Rapid detection of isoniazid resistance in Mycobacterium tuberculosis isolates by use of real-time-PCR-based melting curve analysis. J Clin Microbiol. 2014;52(5):1644–1652.
20. Ahmad S, Jaber AA, Mokaddas E. Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis. 2007;87(2):123–129.
21. Li MC, Chen R, Lin SQ, et al. Detecting Ethambutol Resistance in Mycobacterium tuberculosis Isolates in China: A Comparison Between Phenotypic Drug Susceptibility Testing Methods and DNA Sequencing of embAB. Front Microbiol. 2020;11:781.
22. Zhang D, Liu B, Wang Y, Pang Y. Rapid molecular screening for multidrug-resistant tuberculosis in a resource-limited region of China. Trop Med Int Health. 2014;19(10):1259–1266.
23. Plinke C, Cox HS, Kalon S, Doshetov D, Rusch-Gerdes S, Niemann S. Tuberculosis ethambutol resistance: concordance between phenotypic and genotypic test results. Tuberculosis. 2009;89(6):448–452.
24. Zhang Z, Wang Y, Pang Y, Kam KM. Ethambutol resistance as determined by broth dilution method correlates better than sequencing results with embB mutations in multidrug-resistant Mycobacterium tuberculosis isolates. J Clin Microbiol. 2014;52(2):638–641.
25. Piersimoni C, Olivieri A, Benacchio L, Scarparo C. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. J Clin Microbiol. 2006;44(1):20–28.
26. Van Deun A, Wright A, Zignol M, Weyer K, Rieder HL. Drug susceptibility testing proficiency in the network of supranational tuberculosis reference laboratories. Int J Tuberc Lung Dis. 2011;15(1):116–124.
27. Shi D, Li L, Zhao Y, et al. Characteristics of embB mutations in multidrug-resistant Mycobacterium tuberculosis isolates in Henan. China J Antimicrob Chemother. 2011;66(10):2240–2247.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
