Back to Journals » Infection and Drug Resistance » Volume 13
Quorum Quenching: A Potential Target for Antipseudomonal Therapy
Authors Hemmati F, Salehi R, Ghotaslou R , Samadi Kafil H , Hasani A , Gholizadeh P , Nouri R, Ahangarzadeh Rezaee M
Received 28 May 2020
Accepted for publication 7 August 2020
Published 24 August 2020 Volume 2020:13 Pages 2989—3005
DOI https://doi.org/10.2147/IDR.S263196
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Fatemeh Hemmati,1,2 Roya Salehi,3 Reza Ghotaslou,4 Hossein Samadi Kafil,3,4 Alka Hasani,4 Pourya Gholizadeh,1,2 Roghayeh Nouri,1,2 Mohammad Ahangarzadeh Rezaee1,4
1Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran; 2Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran; 3Drug Applied Research Center, Tabriz University of Medical Science, Tabriz, Iran; 4Department of Medical Microbiology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran
Correspondence: Mohammad Ahangarzadeh Rezaee
Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
Tel + 98-4133364661
Email [email protected]
Abstract: There has been excessive rate of use of antibiotics to fight Pseudomonas aeruginosa (P. aeruginosa) infections worldwide, which has consequently caused the increased resistance to multiple antibiotics in this pathogen. Due to the widespread resistance and the current poor effect of antibiotics consumed to treat P. aeruginosa infections, finding some novel alternative therapeutic methods are necessary for the treatment of infections. The P. aeruginosa biofilms can cause severe infections leading to the increased antibiotic resistance and mortality rate among the patients. In this regard, there are no approaches that can efficiently manage these infections; therefore, novel and effective antimicrobial and antibiofilm agents are needed to control and treat these bacterial infections. Quorum sensing inhibitors (QSIs) or quorum quenchings (QQs) are now considered as potential therapeutic alternatives and/or adjuvants to the current failing antibiotics, which can control the virulence traits of the pathogens, so as a result, the host immune system can quickly eliminate bacteria. Thus, the aims of this review article were presenting a brief explanation of the research reports on the natural and synthetic QSIs of P. aeruginosa, and the assessment of the current understanding on the QS mechanisms and various QQ strategies in P. aeruginosa.
Keywords: Pseudomonas aeruginosa, quorum quenchings, quorum sensing, nanoparticle, natural compounds, synthetic compounds
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a Gram-negative pathogen1,2 that causes acute and chronic infections in immunocompromised patients such as cystic fibrosis and burn patients.3 The treatment of P. aeruginosa by conventional antibiotics has become very difficult, due to the rise of multi-drug resistance strains. Therefore, there is an urgent need to find some novel antimicrobial agents and recognize novel approaches to treat or prevent bacterial infections.4–7
The quorum sensing (QS) plays a critical role in multi-drug resistance of P. aeruginosa, which can upregulate both biofilms-associated matrix and efflux pump genes to improve resistance of bacteria against antibacterial agents.8 A new promising approach to treat P. aeruginosa infections is its QS blocking without killing any of the target bacteria.3 Efforts to disturb bacterial biofilms and reduce the expression of efflux pump genes have provided the recognition of molecules produced by prokaryotes and eukaryotes with the capability of inhibiting the QS signals, named as quorum quenchings (QQ) or quorum sensing inhibitor (QSI). Since QQs do not effect the growth of bacterial, they do not inflict potent selective pressures on the increased resistance compared to antibiotics. Therefore, they have been considered as an ideal target for novel anti-virulence drugs.9–11 The QQs could significantly affect the treatment of a broad range of pathogenic bacterial infections.12 Moreover, they can aggressively control the QS signals as well as providing a chance to improve novel agents against QS signals to fight pathogens.13
The aims of this review article were presenting a brief explanation of the research reports on the natural and synthetic QSIs of P. aeruginosa, and the assessment of the current understanding on the QS mechanisms and various QQ strategies in P. aeruginosa.
Quorum Sensing
Bacteria at low cell densities behave like single cellular organisms; however, when their population density reaches the concentration threshold, they may change their behavior to the “multicellular” type via sensing. In this step, they communicate via autoinducers (AIs), which enable them to express genes for various phenotypes, particularly those that are responsible for their virulent behavior.14 This system, known as the bacterial QS, can be divided into several steps. In the first step, signaling molecules, also called AIs, are produced by the bacterial cell that are then released either actively or passively into the surrounding environment. After reaching the concentration threshold, signal molecules can be recognized by specific receptors and the signal molecules can lead to some changes in the gene’s expression and regulation.15 Opportunistic bacteria such as P. aeruginosa select to lie “dormant” and postponement their virulent factors until their population density has sufficiently increased to overcome the host’s defense systems.13
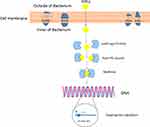
The QS systems operate via a broad range of signals as follows: (a) Oligopeptides,16 (b) N-acyl homoserine lactones (AHLs, AI-1),16 (c) Furanosyl borate (AI-2),13,14 (d) Hydroxyl-palmitic acid methyl ester,13,14 (e) Methyl dodecanoic acid,14 and (f) Diffusible signal factor (DSF); cis-11-methyl-2-dodecenoic acid.17 In addition, the two most broadly studied signaling molecules are as follows: (1) Peptide based QS system or oligopeptides, containing between five and 34 amino acid residue, which are generally involved in intercellular communication in Gram-positive bacteria, and (2) AHLs, which differ in the length and oxidation state of their acyl side chains and are produced by Gram-negative bacteria to screen their population density in QS control of gene expression (Figure 1).4,13,18–20 Also, many other signaling molecules have been known that can act as QS signals. Among these signaling molecules, the cis-2-unsaturated fatty acids are included, often referred to as DSF family signals. The first molecule of the DSF family, cis-11-methyl-2-dodecenoic acid, was discovered in phytopathogen Xanthomonas campestris pv. campestris. Subsequently, other DSF family signals were also described in Burkholderia cenocepacia and P. aeruginosa, which can synthesize cis-2-dodecenoic acid (BDSF) and cis-2-decenoic acid (PDSF), respectively.17,21
There is a significant correlation among QS and the pathogenic factors production, motility, plasmid transfer, antibiotic production, and biofilm formation. Accordingly, the QS system facilitates the population to live and multiply in a better environment with effective intercellular communication, since it can contribute to several behaviors that enable bacteria to resist antibacterial compounds or antibiotics like biofilm development.22 Due to the significance of bacterial communication in the expression of pathogenic factors, QQ can be considered as a potential target to prevent bacterial infection.23
Quorum Sensing Systems in P. aeruginosa
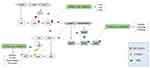
Of Gram-negative bacteria, P. aeruginosa is the common pathogen in the AHL AIs studying and has been extensively used for performing studies on QS.4 The P. aeruginosa has four QS systems as follows: LasI/LasR, RhlI/RhlR systems, which both are two AHL-based signaling systems, as Pseudomonas quinolone signal (PQS), which is a non-AHL-based signaling system, as well as the newly identified QS named integrated QS signal (IQS) (Figure 2).24–26
In P. aeruginosa, Las and Rhl are two critical QS systems represented by LasI/LasR and RhlI/RhlR system. N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) is synthesized by LasI synthase in las system, which forms a LasR-3OC12HSL complex by binding to LasR and can initiate the transcription of many virulence genes such as lasB, apr, and toxA.18,24 This complex indues the expressions of LasI and RhlR; therefore, both AHL-based QS systems can be positively regulated.18 The RhlR is one of the QS transcription factors that is able of autoinduction, as well as replying to N-butanoyl-homoserine lactone (C4-HSL) produced by RhlI. The RhlR-C4HSL complex indues the expression of pathogenesis factors such as pyocyanin, rhamnolipid, and elastase. In addition, RhlR has no direct effect on the LasR system.18,24,27,28
The PQS system can be mediated by 2-alkyl-quinolones.27 In this regard, the pqsABCDE and pqsH systems are two members of quinolone-dependent QS system, which directly synthesize PQS and HHQ (4-hydroxy-2-heptylquinoline) signals, respectively. Although pseudomonas quinolone signal positively regulates the Rhl QS system and indues RhlR expression, it has no direct effect on the LasR system. Also, HHQ and PQS signals link to the transcriptional regulator pqsR, and besides, positively modulate the expression of pathogenic factors such as production of biofilm and swarming and twitching motilities.18,29–31
The 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde is a novel class of QS signal molecules, which belongs to IQS that has been recognized in P. aeruginosa, and is able to integrate the environmental stress cues with the QS system. The ambBCDE cluster encodes the enzymes for L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) biosynthesis occurring through a non-ribosomal peptide synthase (NRPS) pathway that directly synthesize the IQS. When IQS biosynthesis is disturbed, it can disable the Pqs and Rhl systems, which attenuate the pathogenicity of bacteria.25,31,32 In addition, the IQS contributes to the greater pathogenicity of P. aeruginosa in several animal models such as mice,14 zebrafish,14 fruitfly and nematode.32 In addition, IQS is able to sense the phosphate-reduction stress by networking with the QS system. Therefore, in the phosphate reduction stress situation, it can partly take over the purposes of the Las system, which provides some important clues in comprehending the confusing phenomenon.25,32
Overall, the QS signals are hierarchically organized as follows: Las system positively regulates Rhl, Pqs, and Iqs systems.32 Moreover, the RhlI/RhlR and the PqsABCDE/PqsR systems regulate each other, and besides, the ambBCDE/IqsR system regulates the PqsABCDE/PqsR system.26 In addition, each one of these systems is modified by a collection of extra regulators in both transcriptional and post-transcriptional steps.26
Quorum Quenching
Due to the increased antibiotic resistance among the human bacterial pathogens and the current poor effects of antibiotics to treat bacterial infections, finding novel alternative antimicrobial approaches is urgently needed.5–7,33 Correspondingly, one of these approaches is targeting the expression of the QS-regulated pathogenic factors using the analogs of the QS molecules. In this regard, they have been developed as different methods to produce novel antimicrobial agents. This aim can be attained by the control of the synthesis of AIs and their contact with the receptors, as well as the raise of their disintegration.26 In this case, several natural and synthetic compounds have been characterized, which can degrade and also inactivate the QS molecules (Figure 3).
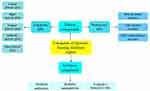 |
Figure 3 Schematic diagram demonstrating quorum sensing inhibitors compounds of P. aeruginosa. |
The system of QS can be disturbed via various methods including: (a) decreasing in the activity of the AHL synthase, (b) inhibition of the production of AHLs, (c) degradation of the AHLs, and (d) the use of various compounds as the antagonists of the signaling molecules.13,34,35
Natural Compounds
Natural-originated compounds are always taken into consideration in medical fields, because they are biodegradable and usually very useful, so they serve as a convenient compound for the inhibition of biological infection. Several studies have shown that the use of natural eukaryotic or prokaryotic derived compounds can reduce the bacterial virulence and modulates QS.4,36-40 In this regard, natural compounds are assumed to be better than other QSIs, so for this reason, they can be used more confidently for a prolonged time and can reach the situation of GRAS (generally recognized as safe).41
Prokaryotic QSIs
In recent years, several QSIs have been reported in bacteria. Singh et al42 in their study showed that Delftia tsuruhatensis SJ01 isolated from the rhizosphere has the AHL degrading activity as well as an anti-biofilm potential. Secondary metabolites of Vibrio alginolyticus inhibit the P. aeruginosa PAO1 virulence factors by downregulating the motility ability, elastase activity, and rhamnolipid production. In addition, the Vibrio alginolyticus extract inhibits the production of biofilm in P. aeruginosa PAO1.43
Moreover, some species of bacteria are capable of producing QQ enzymes that are as follows: (I) Firmicutes: Arthrobacter, Bacillus, and Oceanobacillus; (II) Actinobacteria: Rhodococcus and Streptomyces; (III) Proteobacteria: Acinetobacter, Agrobacterium tumefaciens, Alteromonas, Comomonas, Halomonas, Hyphomonas, Klebsiella pneumoniae, P. aeruginosa, Ralstonia, Stappia, and Variovorax paradoxus; (IV) Bacteroidetes: Tenacibaculum; and (V) Cyanobacteria: Anabaena.14,44-49
Prokaryotes have three types of enzymes that target the AHLs and play an essential role in QQ including AHL lactonases, AHL acylases, and AHL oxidoreductases.40,41
AHL-Lactonase Enzymes
One of the metallo-β-lactamase (MBL) family members is AHL-lactonases, which can hydrolyze the lactone ring.50 Owing to conservation of targeted homoserine lactone ring among all the AHLs and nonspecific interactions within the active-site cavity of the enzymes that are created by variable acyl chains, AHL lactonases have a very broad AHL substrate specificity.44–47 Autoinducer inactivation gene (aiiA), as the first described AHL lactonase, was identified in Bacillus spp. Correspondingly, the expression of aiiA alleles in some pathogenic bacteria such as P. aeruginosa and Burkholderia thailandensis, declined AHL accumulation and also changed the QS-dependent behaviors.51–53
Other AHL-lactonase enzymes could alter the production of virulence factors in P. aeruginosa PAO114 like Mom1 in Muricauda olearia, interfere with swarming motility of P. aeruginosa like HqiA in Pectobacterium carotovorum,18,54 or could decrease the production of pathogenic factors like proteases and pyocyanin, as well as decreasing the biofilm production of P. aeruginosa like Ssopox in Sulfolobus solfactaricus.10
AHL-Acylase Enzymes
The AHL acylases belongs to the novel family of N-terminal nucleophile (NTN) that cleave the acyl side chains in the homoserine lactone, resulting in disabling the AHLs. The acylase enzymes are also recognized as amidase enzymes, which hydrolyze the amide bond between the acyl chain and the homoserine lactone ring.55 Moreover, there are many different types of acylases in terms of the various acyl chain exchanges on AHLs. Also, for the first time, the deacylation activity of AHL was detected in Variovorax paradoxus VAI-C. Accordingly, this strain can use AHLs as a source of nitrogen and energy.56–58
Acyl homoserine lactone acylases have been found in numerous bacteria, such as AhlM in Streptomyces sp. strain M664,59 PvdQ and QuiP in P. aeruginosa PAO1,58,60 AiiC in Anabaena sp. strain PCC712061 and AiiD in Ralstonia sp strain XJ12B.62 Investigating AiiD enzyme of P. aeruginosa PAO1 showed that it is very diverse and shared only 39% similarity at the amino-acid level.13,57 AiiD homologs have also been identified in numerous of Pseudomonas spp., which can have AHL-acylase activity.58,62,63 Notably, the AHL acylases could potentially modulate bacterial behavior by interfering with the production of virulence factors. Also, motility phenotypes in P. aeruginosa PAO1 can be changed by the expression of AHL acylases aiiD.62,64
The P. aeruginosa PAO1 contains four acylases homologues, which belong to NTN hydrolase enzymes including PvdQ (PA2385), QuiP (PA1032), HacB (PA0305), and PA1893. Accordingly, PA1893 is the known member of QS-regulons, while the other homologues have acylase activities.18,57 Also, NTN hydrolase enzymes can decrease the secretion level of pathogenic factors in P. aeruginosa.18,55,65
AHL-Oxidoreductase Enzymes
The AHL oxidoreductase modifies the chemical structure of the AHLs by oxidizing or decreasing the acyl side chain at the third carbon position without damaging the AHLs.57 In general, to the best of our knowledge, there are few studies performed on the disabling of AHLs through the oxidation of the acyl side chain compared to the AHL degradation by lactonases and acylases. Accordingly, these enzymes were firstly observed in Rhodococcus erythropolis that is capable of using a range of AHLs as nitrogen and carbon sources.18,55,66 In recent years, a novel oxidoreductase, known as BpiB09, has been identified that can inactivate 3OC12-HSL. In addition, oxidoreductase BpiB09 in P. aeruginosa PAO1 reduces the accumulation of AHLs, followed by reducing motility phenotypes, pyocyanin secretion, and biofilm production.18,55
Eukaryotic QSIs
The eukaryotic-derived compounds including animals, antibodies, plants, fungus, and algae derived compounds are capable of interfere in bacteria cell-to-cell signalling molecules.4 In this regard, they are usually used in medical field, since they are bio-compatible, usually very efficient, and known as excellent candidates for biological anti-infectious approaches. So, studies have suggested the use of the eukaryotic-derived compounds to decrease bacteria pathogenicity and QS modification.36,67
It has been recognized that animals have evolved multiple defence strategies including anti-microbial peptides, lysozymes, and antibodies to protect themselves against bacterial pathogens. Additionally, interactions between animal hosts and pathogenic bacteria provoke a broad range of reactions, particularly in the presence of QS molecules.13,68 Enzymes of QQ have been discovered in several animals such as mice, rats, and zebrafish. Notably, the enzymes of QQ have been discovered in several animals such as mice, rats, and zebrafish. Acylase I enzyme of porcine kidney could disable QS signals of N-Hexanoyl-L-homoserine lactone (C6HSL) and 3OC12HSL; however, it was shown that it has no effect on C4HSL.14 Acylase I can have effect on diminishing biofilm production by Aeromonas hydrophila and Pseudomonas putida.69 A group of mammalian enzymes known as paraoxonases 1, 2, and 3 (PON) has been found to have hydrolytic activities on esters and lactones, which are relevant to drug metabolism and detoxification of the nerve agents. PON-lactonases vary from prokaryotic lactonases due to the loss of the “HCDH~H~D” motif, and besides, they need calcium ion for their functions.13,70,71 The human epithelial cells and mammalian sera have PON enzymes, which are able to disable and destroy AHLs.18 Several studies have shown that the PON by an active site, can hydrolyze many various substrates such as lactones, esters, and phosphotriesters.18,72,73
Antibody based QSI as one of the methods for anti-infective therapy have been proposed to inhibit QS signals.35 The anti-QS activity of antibodies was firstly reported by Sandra De Lamo Marin’s group.74 They revealed that some of the anti-AHL antibodies could inhibit the 3OC12-AHL-based QS system. In this regard, XYD-11G2 is one of the most efficient antibodies that have capability of inhibiting 3OC12HSL in P. aeruginosa.74 It has been shown that the generation of the monoclonal antibody RS2-IG9 against the 3OC12HSL analog RS2 could be efficient on destroying 3OC12HSL of P. aeruginosa.75 Also, some studies have shown that antibody catalysis could create a novel approach for the inhibition QS in bacteria.35,74
Plant-derived compounds are mostly secondary metabolites that are used for their antibacterial properties, since many years ago. Moreover, they have been used for decreasing bacterial virulence and production of biofilm.4 Notably, many natural compounds have been described as QSIs, and some of the most promising QSI molecules have been found in various plants. These compounds can serve as both autoinducer agonists and antagonists.18,76,77 Also, herbal extracts can act as QSIs, which are structural analogs of AHLs and disrupt the QS system by binding to LuxR/LasR-receptors.78,79 Some of these herbal extracts that act as QSIs as well as their effects on P. aeruginosa are shown in Table 1.
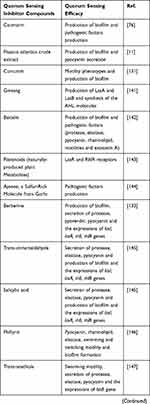 |  | 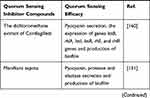 |
Table 1 Herbal Extracts as Quorum Sensing Inhibitors and Their Effects on the P. aeruginosa QS System |
The QSI has been also reported from algae. Some studies have shown that several algal species have natural defense strategies to inhibit microbial accumulation.4,23 Regarding this, halogenated furanones created by the red marine macroalga Delisea pulchra were firstly recognized as anti-QS compounds.80,81 D. pulchra can produce some of the nontoxic and halogenated metabolites, especially brominated furanones that can act as mimics of the bacterial AHL QS signals23 and also inhibit the QS-regulated behaviors by competitively linking to the LuxR-receptors.80,81 Moreover, these compounds are known as structural analogs of AHLs, which control the QS regulation in various bacteria to prevent biofilm formation and subsequent accumulation effectively.23,82 Most of the algal QS modulators investigations have been conducted on the furanones and their derivatives.4 Furanone-mediated inhibition of QS signaling indicated some significant effects on the disruption of expression of P. aeruginosa virulence gene. It is noteworthy that, treatment of bacteria with low concentrations of furanones decrease the secretion of exoprotease enzymes, pyoverdin production, and biofilm formation. The AHL mimic compounds, as halogenated furanones from D. pulchra, serve in the inhibition of the QS-regulated responses.82 The disruption of QS by furanones can result in the improved animal survival after being exposed to the lethal dose of P. aeruginosa inoculations.18
Fungal secondary metabolites can also act as QSI. Penicillic acid and patulin are the secondary metabolites produced by of Penicillium spp., which are able to inhibit the QS system.83 Equisetin is a secondary metabolite of marine-derived fungi that could inhibit the biofilm production, motility phenotypes, and other pathogenic factors in P. aeruginosa. It could also downregulate the expression of lasB, lasI, lasR, pqsA, pqsR, rhlA, rhlI, and rhlR genes.84
Synthetic Compounds
QSIs production is naturally occurred in a large number of organisms; however, their main limitation is the low levels, in which they are generated as well as the related toxicity in some cases.13 In recent years, control of bacterial virulence using chemical compounds has considerably received many attention. Therefore, these studies aimed to extend targeted synthetic QS regulators.4,13 The process of the inhibition of QS by synthetic compounds can be followed by various mechanisms in P. aeruginosa as follows (Table 2): I) synthetic signal analogs II) modifications in the AHL side-chain, III) modifications in the AHL ring moiety, and IV) antagonists of the receptor-ligand interactions.13,18
 |
Table 2 Synthetic Quorum Sensing Inhibitors, Their Targets, and Effects on the P. aeruginosa QS system |
In recent years, nanoparticles (NPs) have received many consideration, due to their antimicrobial activities. Moreover, the use of NPs is one of the most promising strategies to fight against microbial drug resistance.85 Suitable therapeutic compounds are not only required to display a low toxicity index, but also to have appropriate pharmacokinetic features for clinical usages.86 Nanoparticles are also recognized as synthetic QSIs. However, more studies are required to explore these nanoparticles additionally. Researchers have recently utilized nanotechnology for the extension of innovative nanomaterials targeting QS-regulated pathogenic factors, which create new insights on the expansion of some alternative antibacterial treatments.87,88
Chitosan is a cationic polysaccharide formed by N-acetylglucosamine and glucosamine. Correspondingly, it is linked by β-(1, 4) glycosidic linkages,89 showing many unique properties, such as bio-compatibility,89 biological activity,90 nontoxicity,91 bioadhesion,92 anti-hypercholesterolemia,92 antioxidant93 and antimicrobial activity.94 Moreover, chitosan and its derivatives have an anti-biofilm activity.95,96 Also, the antimicrobial properties of chitosan, due to the presence of its positive charge amino groups, can react with negatively charged lipopolysaccharides of P. aeruginosa, which can consequently inhibit the bacterial proliferation.85,97 Chitosan derivatives such as chitosan NPs have also shown biological activities against microorganisms. Recent studies have shown that the enhanced antimicrobial activity of chitosan NPs is related to the increase of surface area to volume ratio as the particle size decreases. Therefore, the chitosan NPs with vast surface areas modifies the bacterial membrane penetrability via the membrane incorporation and also leads to the death of bacteria.33,97-99 Furthermore, chitosan NPs have shown anti-QS properties by disrupting biofilm production, decreasing the expression of lasR and rhlR genes, and reducing the secretion of pyocyanin and proteases in P. aeruginosa.100 In addition, chitosan NPs have a negative effect on the pathogenic factors produced by P. aeruginosa PAO1.101 In this regard, Muslim et al,100 in their study demonstrated that chitosan significantly decrease the biofilm formation, pyocyanin, protease secretion, as well as the expressions of lasR and rhlR genes in P. aeruginosa. In addition, Ilka et al reported that the loaded kaempferol into chitosan NPs have QSI activity and suggested that a combination of materials with chitosan NPs can inhibit the QS-related encoding genes, which can serve as a new approach for antibacterial treatment acting as the QS-based antibiofilm agents.95
In recent years, zinc oxide (ZnO) NPs have been recognized as efficacious QSIs for P. aeruginosa PAO1, by reducing the generation of different pathogenic factors with no growth-inhibitory efficacy.102 Lee et al102 demonstrated that Zn2+ and ZnO NPs have no bactericidal activities against P. aeruginosa at the concentration levels of <3 mM; however, they have antivirulence activities. In addition, generation of reactive oxygen species (ROS) on the surface of ZnO induce some serious damages to bacteria cell. Also, the attachment of the ZnO NPs on the bacterial surface or cumulating of NPs in the cytoplasm area induces disturbance of cellular action as well as the disorder of the bacterial membrane.103,104 Lara et al reported that ZnO NPs could significantly reduce the secretion of elastase, pyocyanin, and the production of biofilm in P. aeruginosa, which demonstrate that ZnO NPs have a wide spectrum, so it can be considered as an alternative for the treatment of P. aeruginosa infections.105
Other attractive compounds are Engineered nanoparticles (ENPs), which are used as QSIs. Several recent researchers have described different mechanisms for the toxicity of ENPs as follows: (a) metal ions toxicity of ENPs,106 (b) enhancement in the generation of reactive oxygen species,107 and (c) the damage to DNA and proteins.108 Also, Li et al109 reported that different ENPs with various physicochemical traits have dissimilar effects on QS systems of P. aeruginosa PAO1. Although upper silver (Ag) ENPs concentrations (mg/L) induce the anti-QS activity in P. aeruginosa PAO1, its lower concentrations (μg/L) have shown to increase its QS activity. In contrast to Ag ENPs, ferrous (Fe) ENPs enhance the concentration level of 3OC12-HSL, but it has no effect on the other QS activities. Mohanty et al110 also showed that Ag ENPs can decrease the release of C4-HSL, C6-HSL, 3OC6-HSL, C8-HSL, and 3OC8-HSL in Pseudomonas syringae. Moreover, Singh et al111 reported that mfAg (mycofabricated Ag NPs) ENPs can decrease C4-HSL and 3OC12-HSL release in P. aeruginosa. In addition, mfAgNPs reduce the biofilm production and QS activities by reducing the expressions of lasIR and rhlIR genes. Furthermore, Wagh et al112 showed that silver nanowires (SNWs) had some hopeful properties of the QS-mediated controlling of biofilm in P. aeruginosa. Notably, when P. aeruginosa was treated in various concentrations of SNWs, it was detected that the most reduction was at 4 mg/mL in the production of biofilm, without inhibiting bacterial growth. Nevertheless, the concentration enhancement (≥5mg/mL) declines the number of viable cells. Therefore, 4 mg/mL of SNWs could dramatically inhibit the production of biofilm without affecting viability, while higher concentrations could inhibit bacterial growth. Gholamrezazadeh et al113 demonstrated that Ag NPs and benzalkonium chloride efficiently induced the rhlR gene expression. Prateeksha et al114 have reported that the selenium NPs harbor a superior QSI property, antibiofilm activity, and antivirulence potential in P. aeruginosa. Moreover, Nafee et al115 reported that ultra-small solid lipid nanoparticles (US-SLNs) inhibited the QS-dependent phenotype like the secretion of pyocyanin in P. aeruginosa.
Antibiotics as QSI
Antibiotics, besides having therapeutic efficacy in killing or inhibiting bacterial proliferation, can also act as signaling molecules, which are capable of reducing the expressions of virulence factors in bacterial populations.116–119
Some studies have demonstrated that several antibiotics are capable of inhibiting virulence factors in P. aeruginosa.13,18,120 In this study, the reply of P. aeruginosa to azithromycin antibiotic was analyzed by the use of the microarray method. Also, its phenotype investigation showed that there is a link between genes regulated by QS and by azithromycin.120 Several studies have revealed that azithromycin significantly has anti-QS activity, and the subinhibitory concentrations (SICs) of azithromycin are capable of blocking many genes regulated by QS.120–122
Sofer et al123 in their study indicated that erythromycin therapy decrease the AHL production in bacteria. Another study has also shown that the treatment of P. aeruginosa with azithromycin reduce the C4HSL and 3OC12HSL production.124 The SICs of macrolides and β-lactam antibiotics can reduce the pathogenic factors’ expression of P. aeruginosa such as the diminished of exotoxin A secretion, pyocyanin, protease, DNase, and phospholipase C, as well as significantly removing the QS activity of P. aeruginosa.125 In addition, SICs of tobramycin could block the expressions of rhlI and rhlR genes by decreasing C4-HSL generation. Previous studies have also confirmed the impact of tobramycin, as a signaling molecule, on virulence genes expression in the transcriptional step.126,127
Skindersoe et al120 investigated several antibiotics for their capabilities in intervening with the bacterial signaling systems. Of the antibiotics used, azithromycin displayed high levels of QSI activity, followed by ciprofloxacin and ceftazidime that had potent QSI activities. Whereas aminoglycoside antibiotics, piperacillin, spectinomycin, and streptomycin had either low levels or no QSI activity. In their study, the protease secretion has been reduced by azithromycin, ciprofloxacin, and ceftazidime, as well as ciprofloxacin and ceftazidime that diminished the elastase activity.120
Furthermore, the nonsteroidal anti-inflammatory drugs (NSAIDs) such as aspirin, piroxicam, and meloxicam are chemical compounds that can be used as the potential inhibitors for controlling the P. aeruginosa QS signaling system as well as biofilm formation. The NSAID drugs can reduce the level of AHL-mediated quorum sensing in P. aeruginosa such as Las, Rhl, and Pqs.77,128,129 Generally, some antibiotics and drugs have QSI potentials that can reduce the levels of AHL synthesis in P. aeruginosa. Therefore, diminishing in the bacterial population by antibiotics can result in the decreased levels of pathogenic factors.
Synergism Between QSIs and Antibiotics
A single QSI can not be very effective on bacteria, and could also lead to resistance against QSI. Therefore, a combination therapy of QSIs and antibiotics are suggested. Accordingly, this combination therapy prevents the resistance to a single QSI and can also increase the effectiveness of therapy without enhancing the toxicity of the antibiotics.41,130
Vadekeetil et al41 also reported the synergistic interaction between proanthocyanidin active fraction and ciprofloxacin against P. aeruginosa QS. Moreover, there was a considerable decrease in the amount of the expressions of several pathogenic factors such as motility phenotypes and biofilm formation; however, it did not affect the secretion of elastase and protease. The significant suppressing effect of ciprofloxacin has been also observed on the twitching motility of P. aeruginosa. In addition, their study demonstrated that ciprofloxacin combined with proanthocyanidin active fraction have a higher anti-motility property on all three forms of motilities (ie, swimming, swarming, and twitching).
The inhibitory effects of ciprofloxacin on biofilm formation were also observed at SICs. However, it was found that, in the presence of proanthocyanidin active fraction, ciprofloxacin decreases biofilm production up to fivefold compared to the control sample.41
The synergistic efficacy of curcumin with ceftazidime and ciprofloxacin on signaling system in P. aeruginosa PAO1, was investigated by Roudashti et al.131 Their findings indicated that SICs of curcumin, ceftazidime, and ciprofloxacin both alone and in combination can dramatically decrease motility phenotypes and the production of biofilm. Furthermore, these compounds, alone and in combination, can also reduce the expression of the genes regulated by QS.131 Bahari et al132 evaluated the synergistic efficacy of curcumin combined with azithromycin and gentamicin on signaling system in P. aeruginosa PAO1, and reported that the curcumin in combination with antibiotics drastically decline 3OC12-HSL and C4-HSL signals. Moreover, the above-mentioned compounds, alone and in combination, can considerably decrease motility phenotypes and biofilm production of P. aeruginosa PAO1.132
Li et al133 also found that azithromycin and berberine could significantly reduce the production of several pathogenic factors such as biofilm production, as well as secretion of pyocyanin and elastase, and remarkably inhibition of the QS system and the expressions of the genes regulated by QS. In their study, it was also demonstrated that LasA activity was drastically decreased after the administration of azithromycin and berberine, separately and in combination.
Chanda et al134 showed that linolenic acid and tobramycin (LNA+TOB) had significant impacts on downregulating the QS-mediated genes. Also, they have revealed that LNA+TOB therapy could inhibit motility phenotypes and reduce the development of infection. Therefore, it can be deduced that LNA+TOB is more effective on the inhibition of the pathogenic secretion and biofilm production compared to alone LNA or TOB in targeting the QS system of P. aeruginosa. Similar to these QSIs, it was observed that, Aminoglycosides in combination with resveratrol dramatically decrease the production of biofilm in comparison to each one of the agents alone, besides, it could significantly inhibit the expression of the QS regulatory genes.135
Bacterial Resistance to QSIs
The proposal reporting that bacteria may develop resistance to the QSIs compounds was offered for the first time in 2010.136 The base for this assumption came from some studies demonstrating that the expression of the central QS genes was extremely diverse among various strains of the bacteria such as Vibrio spp. and P. aeruginosa.137 Lately, resistance mechanisms to the best-QSIs have been observed in the in vitro, and also in clinical isolates indicating that the increased resistance to these types of compounds is the facility. Brominated furanone C-30 is one of the best QSIs that is effluxed by the MexAB-OpmR pump. Bacterial species that have mutations in their efflux pump-encoding genes mexR and nalC are also resistant to C-30, which was observed in P. aeruginosa for the first time.138 5-fluorouracil also is another QSI that some of the clinical isolates of P. aeruginosa are resistant to it.137,139,140 It has been proposed that the probabilities of QSIs resistance are lower compared to those for conventional antibiotics. In this regard, the combination of QSIs and antibiotics to hamper biofilms formation and reduce the pathogenic factors in bacteria, can be considered as an alternative approach.
Conclusion
The QS inhibition is a broadly accepted anti-virulence and non-bactericidal mechanism. The development of diverse QS suppressing agents and the inhibition of QS mediators might be known as an evolutionary alteration that can decrease the resistance of the fouling bacteria. In addition, QS inhibition alone cannot affect the antibiotic susceptibility of bacteria. Therefore, more studies are required to demonstrate their mechanisms of action and the optimal amounts of the QS inhibitory compounds that are safe and applicable.
Acknowledgment
This article was financially supported by the Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hancock RE, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Update. 2000;3(4):247–255. doi:10.1054/drup.2000.0152
2. Engel LS, Hill JM, Caballeero AR, Green LC, O’Callaghan RJ, Protease IV, a unique extracellular protease and virulence factor from Pseudomonas aeruginosa. J Biol Chem. 1998;273(27):16792. doi:10.1074/jbc.273.27.16792
3. Allegretta G, Maurer CK, Eberhard J, et al. In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front Microbiol. 2017;8:924. doi:10.3389/fmicb.2017.00924
4. Holban AM, Bleotu C, Chifiriuc MC, Lazar V. Control of bacterial virulence by cell-to-cell signalling molecules Microbial pathogens and strategies for combating them: science, technology and education. Formatex. 2013;1:978–984.
5. Gholizadeh P, Aghazadeh M, Asgharzadeh M, Kafil H. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur J Clin Microbiol Infect Dis. 2017;36(11):2043–2051. doi:10.1007/s10096-017-3036-2
6. Narenji H, Gholizadeh P, Aghazadeh M, Rezaee MA, Asgharzadeh M, Kafil HS. Peptide nucleic acids (PNAs): currently potential bactericidal agents. Biomed Pharmacother. 2017;93:580–588. doi:10.1016/j.biopha.2017.06.092
7. Gholizadeh P, Ş K, Dao S, et al. How CRISPR-Cas system could be used to combat antimicrobial resistance. Infect Drug Resist. 2020;13:1111–1121. doi:10.2147/IDR.S247271
8. El‐Mowafy S, Shaaban M, Abd El Galil K. Sodium ascorbate as a quorum sensing inhibitor of P seudomonas aeruginosa. J Appl Microbiol. 2014;117(5):1388–1399. doi:10.1111/jam.12631
9. Abbas HA, Shaldam MA. Glyceryl trinitrate is a novel inhibitor of quorum sensing in Pseudomonas aeruginosa. Afr Health Sci. 2016;16(4):1109–1117. doi:10.4314/ahs.v16i4.29
10. Guendouze A, Plener L, Bzdrenga J, et al. Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front Microbiol. 2017;8:227. doi:10.3389/fmicb.2017.00227
11. Kordbacheh H, Eftekhar F, Ebrahimi S. Anti-quorum sensing activity of Pistacia atlantica against Pseudomonas aeruginosa PAO1 and identification of its bioactive compounds. Microb Pathog. 2017;110:390–398. doi:10.1016/j.micpath.2017.07.018
12. Starkey M, Lepine F, Maura D, et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10(8):e1004321. doi:10.1371/journal.ppat.1004321
13. Kalia VC. Quorum sensing inhibitors: an overview. Biotechnol Adv. 2013;31(2):224–245. doi:10.1016/j.biotechadv.2012.10.004
14. Dong Y-H, Zhang L-H. Quorum sensing and quorum-quenching enzymes. J Microbiol. 2005;43(1):101–109.
15. Sifri CD. Quorum sensing: bacteria talk sense. Clin Infect Dis. 2008;47(8):1070–1076. doi:10.1371/journal.pone.0134684
16. McDougald D, Rice SA, Kjelleberg S. Bacterial quorum sensing and interference by naturally occurring biomimics. Anal Bioanal Chem. 2007;387(2):445–453. doi:10.1007/s00216-006-0761-2
17. Dean SN, Chung M-C, van Hoek ML. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Appl Environ Microbiol. 2015;81(20):7057–7066. doi:10.1128/AEM.02165-15
18. LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77(1):73–111.
19. Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86(5):1267–1279. doi:10.1007/s00253-010-2521-7
20. Xavier KB, Bassler BL. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6(2):191–197. doi:10.1016/S1369-5274(03)00028-6
21. Depluverez S, Daled S, De Waele S, et al. Microfluidics-based LC-MS MRM approach for the relative quantification of Burkholderia cenocepacia secreted virulence factors. Rapid Commun Mass Spectrometry. 2018;32(6):469–479. doi:10.1002/rcm.8059
22. Subhadra B, Oh MH, Choi CH Quorum sensing in Acinetobacter: with special emphasis on antibiotic resistance, biofilm formation and quorum quenching. AIMS Microbiol. 2016;2(1):27–41. doi:10.3934/microbiol.2016.1.27
23. Haque S, Ahmad F, Dar SA, et al. Developments in strategies for Quorum Sensing virulence factor inhibition to combat bacterial drug resistance. Microb Pathog. 2018;121:293–302. doi:10.1016/j.micpath.2018.05.046
24. Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2(11):a012427. doi:10.1101/cshperspect.a012427
25. Li S, Chen S, Fan J, et al. Anti-biofilm effect of novel thiazole acid analogs against Pseudomonas aeruginosa through IQS pathways. Eur J Med Chem. 2018;145:64–73. doi:10.1016/j.ejmech.2017.12.076
26. Pérez-Pérez M, Jorge P, Pérez Rodríguez G, Pereira MO, Lourenço A. Quorum sensing inhibition in Pseudomonas aeruginosa biofilms: new insights through network mining. Biofouling. 2017;33(2):128–142. doi:10.1080/08927014.2016.1272104
27. Fong J, Yuan M, Jakobsen TH, et al. Disulfide bond-containing ajoene analogues as novel quorum sensing inhibitors of Pseudomonas aeruginosa. J Med Chem. 2016;60(1):215–227. doi:10.1021/acs.jmedchem.6b01025
28. Feltner JB, Wolter DJ, Pope CE, et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. MBio. 2016;7(5):e01513–e01516.
29. Sun S, Zhou L, Jin K, Jiang H, He Y-W. Quorum sensing systems differentially regulate the production of phenazine-1-carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Sci Rep. 2016;6(1):30352. doi:10.1038/srep30352
30. Feltner J, Wolter D, Pope C, et al. Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa. MBio. 2016;7(5):e01513–e01516. doi:10.1128/mBio.01513-16
31. Liang H, Deng X, Li X, Ye Y, Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 2014;42(16):10307–10320. doi:10.1093/nar/gku586
32. Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6(1):26–41. doi:10.1007/s13238-014-0100-x
33. Qin X, Kräft T, Goycoolea FM. Chitosan encapsulation modulates the effect of trans-cinnamaldehyde on AHL-regulated quorum sensing activity. Colloids Surf B Biointerfaces. 2018;169:453–461. doi:10.1016/j.colsurfb.2018.05.054
34. Dembitsky VM, AAA Aq, Srebnik M. Natural and synthetic small boron-containing molecules as potential inhibitors of bacterial and fungal quorum sensing. Chem Rev. 2010;111(1):209–237. doi:10.1021/cr100093b
35. Kaufmann GF, Sartorio R, Lee S-H, et al. Antibody interference with N-acyl homoserine lactone-mediated bacterial quorum sensing. J Am Chem Soc. 2006;128(9):2802–2803. doi:10.1021/ja0578698
36. Jha B, Kavita K, Westphal J, Hartmann A, Schmitt-Kopplin P. Quorum sensing inhibition by Asparagopsis taxiformis, a marine macro alga: separation of the compound that interrupts bacterial communication. Mar Drugs. 2013;11(1):253–265. doi:10.3390/md11010253
37. Ditu L-M, Chifiriuc MC, Bezirtzoglou E, et al. Modulation of virulence and antibiotic susceptibility of enteropathogenic Escherichia coli strains by Enterococcus faecium probiotic strain culture fractions. Anaerobe. 2011;17(6):448–451. doi:10.1016/j.anaerobe.2011.05.019
38. Chifiriuc M-C, Diţu L, Banu O, et al. Subinhibitory concentrations of phenyl lactic acid interfere with the expression of virulence factors in Staphylococcus aureus and Pseudomonas aeruginosa clinical strains. Roum Arch Microbiol Immunol. 2009;68(1):27–33.
39. Li W, Lyte M, Freestone PP, Ajmal A, Colmer-Hamood JA, Hamood AN. Norepinephrine represses the expression of toxA and the siderophore genes in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;299(1):100–109. doi:10.1111/j.1574-6968.2009.01739.x
40. Memar MY, Raei P, Alizadeh N, Aghdam MA, Kafil HS. Carvacrol and thymol: strong antimicrobial agents against resistant isolates. Rev Med Microbiol. 2017;28(2):63–68. doi:10.1097/MRM.0000000000000100
41. Vadekeetil A, Alexandar V, Chhibber S, Harjai K. Adjuvant effect of cranberry proanthocyanidin active fraction on antivirulent property of ciprofloxacin against Pseudomonas aeruginosa. Microb Pathog. 2016;90:98–103. doi:10.1016/j.micpath.2015.11.024
42. Singh BR, Shoeb M, Sharma S, Naqvi A, Gupta VK, Singh BN. Scaffold of selenium nanovectors and honey phytochemicals for inhibition of Pseudomonas aeruginosa quorum sensing and biofilm formation. Front Cell Infect Microbiol. 2017;7:93.
43. Song Y, Cai ZH, Lao YM, et al. Antibiofilm activity substances derived from coral symbiotic bacterial extract inhibit biofouling by the model strain Pseudomonas aeruginosa PAO 1. Microb Biotechnol. 2018;11(6):1090–1105. doi:10.1111/1751-7915.13312
44. Kalia VC, Purohit HJ. Quenching the quorum sensing system: potential antibacterial drug targets. Crit Rev Microbiol. 2011;37(2):121–140. doi:10.3109/1040841X.2010.532479
45. Huma N, Shankar P, Kushwah J, et al. Diversity and polymorphism in AHL-lactonase gene (aiiA) of Bacillus. J Microbiol Biotechnol. 2011;21(10):1001–1011. doi:10.4014/jmb.1105.05056
46. Uroz S, Dessaux Y, Oger P. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. Chem Bio Chem. 2009;10(2):205–216. doi:10.1002/cbic.200800521
47. Romero M, A-B M-C, Roca-Rivada A, Cabello AM, Otero A. Quorum quenching in cultivable bacteria from dense marine coastal microbial communities. FEMS Microbiol Ecol. 2011;75(2):205–217. doi:10.1111/j.1574-6941.2010.01011.x
48. Kang BR, Lee JH, Ko SJ, et al. Degradation of acyl-homoserine lactone molecules by Acinetobacter sp. strain C1010. Can J Microbiol. 2004;50(11):935–941. doi:10.1139/w04-083
49. Park S-Y, Lee SJ, Oh T-K, et al. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology. 2003;149(6):1541–1550. doi:10.1099/mic.0.26269-0
50. Dong Y-H, Xu J-L, Li X-Z, Zhang L-H. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci. 2000;97(7):3526–3531. doi:10.1073/pnas.97.7.3526
51. Liu D, Momb J, Thomas PW, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures. Biochemistry. 2008;47(29):7706–7714. doi:10.1021/bi800368y
52. Momb J, Wang C, Liu D, et al. Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 2. Substrate modeling and active site mutations. Biochemistry. 2008;47(29):7715–7725. doi:10.1021/bi8003704
53. Reimmann C, Ginet N, Michel L, et al. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology. 2002;148(4):923–932. doi:10.1099/00221287-148-4-923
54. Dong W, Zhu J, Guo X, et al. Characterization of AiiK, an AHL lactonase, from Kurthia huakui LAM0618 T and its application in quorum quenching on Pseudomonas aeruginosa PAO1. Sci Rep. 2018;8(1):6013. doi:10.1038/s41598-018-24507-8
55. Chen F, Gao Y, Chen X, Yu Z, Li X. Quorum quenching enzymes and their application in degrading signal molecules to block quorum sensing-dependent infection. Int J Mol Sci. 2013;14(9):17477–17500. doi:10.3390/ijms140917477
56. Kusada H, Tamaki H, Kamagata Y, Hanada S, Kimura N. A novel quorum-quenching N-acylhomoserine lactone acylase from Acidovorax sp. strain MR-S7 mediates antibiotic resistance. Appl Environ Microbiol. 2017;83(13):e00080–e00017. doi:10.1128/AEM.00080-17
57. Utari PD, Vogel J, Quax WJ. Deciphering physiological functions of AHL quorum quenching acylases. Front Microbiol. 2017;8:1123. doi:10.3389/fmicb.2017.01123
58. Sio CF, Otten LG, Cool RH, et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74(3):1673–1682. doi:10.1128/IAI.74.3.1673-1682.2006
59. Park S-Y, Kang H-O, Jang H-S, Lee J-K, Koo B-T, Yum D-Y. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl Environ Microbiol. 2005;71(5):2632–2641. doi:10.1128/AEM.71.5.2632-2641.2005
60. Huang JJ, Han J-I, Zhang L-H, Leadbetter JR. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2003;69(10):5941–5949. doi:10.1128/AEM.69.10.5941-5949.2003
61. Romero M, Diggle SP, Heeb S, Camara M, Otero A. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol Lett. 2008;280(1):73–80. doi:10.1111/j.1574-6968.2007.01046.x
62. Lin YH, Xu JL, Hu J, et al. Acyl‐homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum‐quenching enzymes. Mol Microbiol. 2003;47(3):849–860. doi:10.1046/j.1365-2958.2003.03351.x
63. Shepherd RW, Lindow SE. Two dissimilar N-acyl-homoserine lactone acylases of Pseudomonas syringae influence colony and biofilm morphology. Appl Environ Microbiol. 2009;75(1):45–53. doi:10.1128/AEM.01723-08
64. Leadbetter JR, Greenberg E. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J Bacteriol. 2000;182(24):6921–6926. doi:10.1128/JB.182.24.6921-6926.2000
65. Huang JJ, Petersen A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72(2):1190–1197. doi:10.1128/AEM.72.2.1190-1197.2006
66. Uroz S, Chhabra SR, Camara M, Williams P, Oger P, Dessaux Y. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151(10):3313–3322. doi:10.1099/mic.0.27961-0
67. Tarkka MT, Sarniguet A, Frey-Klett P. Inter-kingdom encounters: recent advances in molecular bacterium–fungus interactions. Curr Genet. 2009;55(3):233–243. doi:10.1007/s00294-009-0241-2
68. Zhang LH, Dong YH. Quorum sensing and signal interference: diverse implications. Mol Microbiol. 2004;53(6):1563–1571. doi:10.1111/j.1365-2958.2004.04234.x
69. Paul D, Kim YS, Ponnusamy K, Kweon JH. Application of quorum quenching to inhibit biofilm formation. Environ Eng Sci. 2009;26(8):1319–1324. doi:10.1089/ees.2008.0392
70. Billecke S, Draganov D, Counsell R, et al. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab Disposition. 2000;28(11):1335–1342.
71. Teiber JF, Horke S, Haines DC, et al. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-L-homoserine lactone. Infect Immun. 2008;76(6):2512–2519. doi:10.1128/IAI.01606-07
72. Draganov DI, Teiber JF, Speelman A, Osawa Y, Sunahara R, La Du BN. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J Lipid Res. 2005;46(6):1239–1247. doi:10.1194/jlr.M400511-JLR200
73. Khersonsky O, Tawfik DS. Structure− reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44(16):6371–6382. doi:10.1021/bi047440d
74. Marin SDL, Xu Y, Meijler MM, Janda KD. Antibody catalyzed hydrolysis of a quorum sensing signal found in Gram-negative bacteria. Bioorg Med Chem Lett. 2007;17(6):1549–1552. doi:10.1016/j.bmcl.2006.12.118
75. Kaufmann GF, Park J, Mee JM, Ulevitch RJ, Janda KD. The quorum quenching antibody RS2-1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol Immunol. 2008;45(9):2710–2714. doi:10.1016/j.molimm.2008.01.010
76. D’Almeida R, Molina R, Viola C, et al. Comparison of seven structurally related coumarins on the inhibition of quorum sensing of Pseudomonas aeruginosa and Chromobacterium violaceum. Bioorg Chem. 2017;73:37–42. doi:10.1016/j.bioorg.2017.05.011
77. de Almeida FA, Vargas ELG, Carneiro DG, Pinto UM, Vanetti MCD. Virtual screening of plant compounds and nonsteroidal anti-inflammatory drugs for inhibition of quorum sensing and biofilm formation in Salmonella. Microb Pathog. 2018;121:369–388. doi:10.1016/j.micpath.2018.05.014
78. Teplitski M, Mathesius U, Rumbaugh KP. Perception and degradation of N-acyl homoserine lactone quorum sensing signals by mammalian and plant cells. Chem Rev. 2010;111(1):100–116. doi:10.1021/cr100045m
79. Vattem D, Mihalik K, Crixell S, McLean R. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia. 2007;78(4):302–310. doi:10.1016/j.fitote.2007.03.009
80. Teplitski M, Robinson JB, Bauer WD. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant Microbe Interact. 2000;13(6):637–648. doi:10.1094/MPMI.2000.13.6.637
81. Choo J, Rukayadi Y, Hwang JK. Inhibition of bacterial quorum sensing by vanilla extract. Lett Appl Microbiol. 2006;42(6):637–641. doi:10.1111/j.1472-765X.2006.01928.x
82. Teplitski M, Chen H, Rajamani S, et al. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 2004;134(1):137–146. doi:10.1104/pp.103.029918
83. Rasmussen TB, Skindersoe ME, Bjarnsholt T, et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology. 2005;151(5):1325–1340. doi:10.1099/mic.0.27715-0
84. Zhang M, Wang M, Zhu X, Yu W, Gong Q. Equisetin as potential quorum sensing inhibitor of Pseudomonas aeruginosa. Biotechnol Lett. 2018;40(5):865–870. doi:10.1007/s10529-018-2527-2
85. Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Del Rev. 2013;65(13–14):1803–1815. doi:10.1016/j.addr.2013.07.011
86. Thanh Nguyen H, Goycoolea F. Chitosan/Cyclodextrin/TPP nanoparticles loaded with quercetin as novel bacterial quorum sensing inhibitors. Molecules. 2017;22(11):1975. doi:10.3390/molecules22111975
87. Hoseinzadeh E, Makhdoumi P, Taha P, Hossini H, Stelling J, Amjad Kamal M. A review on nano-antimicrobials: metal nanoparticles, methods and mechanisms. Curr Drug Metab. 2017;18(2):120–128. doi:10.2174/1389200217666161201111146
88. Qais FA, Khan MS, Ahmad I. Nanoparticles as Quorum Sensing Inhibitor: Prospects and Limitations. Springer: Biotechnological Applications of Quorum Sensing Inhibitors; 2018:227–244.
89. Liu X, Ma L, Mao Z, Gao C. Chitosan-Based Biomaterials for Tissue Repair and Regeneration. Springer: Chitosan for Biomaterials II; 2011:81–127.
90. Gopu V, Meena C, Shetty P. Quercetin influences quorum sensing in food borne bacteria. In-Vitro and In-Silico Evidence. PLoS One. 2015;10:e0134684.
91. Natrajan D, Srinivasan S, Sundar K, Ravindran A. Formulation of essential oil-loaded chitosan–alginate nanocapsules. J Food Drug Anal. 2015;23(3):560–568. doi:10.1016/j.jfda.2015.01.001
92. Aranaz I, Harris R, Heras A. Chitosan amphiphilic derivatives. Chemistry and applications. Curr Org Chem. 2010;14(3):308–330. doi:10.2174/138527210790231919
93. Souza MP, Vaz AF, Correia MT, Cerqueira MA, Vicente AA, Carneiro-da-Cunha MG. Quercetin-loaded lecithin/chitosan nanoparticles for functional food applications. Food Bioproc Tech. 2014;7(4):1149–1159. doi:10.1007/s11947-013-1160-2
94. Ong S-Y, Wu J, Moochhala SM, Tan M-H LJ. Development of a chitosan-based wound dressing with improved hemostatic and antimicrobial properties. Biomaterials. 2008;29(32):4323–4332. doi:10.1016/j.biomaterials.2008.07.034
95. Ilk S, Sağlam N, Özgen M, Korkusuz F. Chitosan nanoparticles enhances the anti-quorum sensing activity of kaempferol. Int J Biol Macromol. 2017;94:653–662.
96. Akyuz L, Kaya M, Mujtaba M, et al. Supplementing capsaicin with chitosan-based films enhanced the anti-quorum sensing, antimicrobial, antioxidant, transparency, elasticity and hydrophobicity. Int J Biol Macromol. 2018;115:438–446. doi:10.1016/j.ijbiomac.2018.04.040
97. Ma Z, Garrido-Maestu A, Jeong KC. Application, mode of action, and in vivo activity of chitosan and its micro-and nanoparticles as antimicrobial agents: A review. Carbohyd Polym. 2017;176:257–265. doi:10.1016/j.carbpol.2017.08.082
98. Sahariah P, Gaware V, Lieder R, et al. The effect of substituent, degree of acetylation and positioning of the cationic charge on the antibacterial activity of quaternary chitosan derivatives. Mar Drugs. 2014;12(8):4635–4658. doi:10.3390/md12084635
99. O’Callaghan KA, Kerry JP. Preparation of low-and medium-molecular weight chitosan nanoparticles and their antimicrobial evaluation against a panel of microorganisms, including cheese-derived cultures. Food Control. 2016;69:256–261. doi:10.1016/j.foodcont.2016.05.005
100. Muslim SN, Kadmy IMA, Ali ANM, et al. Chitosan extracted from Aspergillus flavus shows synergistic effect, eases quorum sensing mediated virulence factors and biofilm against nosocomial pathogen Pseudomonas aeruginosa. Int J Biol Macromol. 2018;107:52–58. doi:10.1016/j.ijbiomac.2017.08.146
101. Vadekeetil A, Chhibber S, Harjai K. Efficacy of intravesical targeting of novel quorum sensing inhibitor nanoparticles against Pseudomonas aeruginosa biofilm-associated murine pyelonephritis. J Drug Target. 2019;1–9.
102. Lee J-H, Kim Y-G, Cho MH, Lee J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol Res. 2014;169(12):888–896. doi:10.1016/j.micres.2014.05.005
103. Zhang L, Jiang Y, Ding Y, et al. Mechanistic investigation into antibacterial behaviour of suspensions of ZnO nanoparticles against E. coli. J Nanopart Res. 2010;12(5):1625–1636. doi:10.1007/s11051-009-9711-1
104. Zhang L, Jiang Y, Ding Y, Povey M, York D. Investigation into the antibacterial behaviour of suspensions of ZnO nanoparticles (ZnO nanofluids). J Nanopart Res. 2007;9(3):479–489. doi:10.1007/s11051-006-9150-1
105. García‐Lara B, Saucedo‐Mora M, Roldán‐Sánchez J, et al. Inhibition of quorum‐sensing‐dependent virulence factors and biofilm formation of clinical and environmental P seudomonas aeruginosa strains by ZnO nanoparticles. Lett Appl Microbiol. 2015;61(3):299–305. doi:10.1111/lam.12456
106. Mouneyrac C, Buffet P-E, Poirier L, et al. Fate and effects of metal-based nanoparticles in two marine invertebrates, the bivalve mollusc Scrobicularia plana and the annelid polychaete Hediste diversicolor. Environ Sci Pollut Res. 2014;21(13):7899–7912. doi:10.1007/s11356-014-2745-7
107. Kim JS, Kuk E, Yu KN, et al. Antimicrobial effects of silver nanoparticles. Nanomed Nanotechnol Biol Med. 2007;3(1):95–101. doi:10.1016/j.nano.2006.12.001
108. Eom H-J CJ, Choi J. p38 MAPK activation, DNA damage, cell cycle arrest and apoptosis as mechanisms of toxicity of silver nanoparticles in Jurkat T cells. Environ Sci Technol. 2010;44(21):8337–8342. doi:10.1021/es1020668
109. Li N, Wang L, Yan H, et al. Effects of low-level engineered nanoparticles on the quorum sensing of Pseudomonas aeruginosa PAO1. Environ Sci Pollut Res. 2018;25(7):7049–7058. doi:10.1007/s11356-017-0947-5
110. Mohanty A, Tan CH, Cao B. Impacts of nanomaterials on bacterial quorum sensing: differential effects on different signals. Environ Sci. 2016;3(2):351–356.
111. Singh BR, Singh BN, Singh A, Khan W, Naqvi AH, Singh HB. Mycofabricated biosilver nanoparticles interrupt Pseudomonas aeruginosa quorum sensing systems. Sci Rep. 2015;5(1):13719. doi:10.1038/srep13719
112. Wagh MS, Patil RH, Thombre DK, Kulkarni MV, Gade WN, Kale BB. Evaluation of anti-quorum sensing activity of silver nanowires. Appl Microbiol Biotechnol. 2013;97(8):3593–3601. doi:10.1007/s00253-012-4603-1
113. Gholamrezazadeh M, Shakibaie MR, Monirzadeh F, Masoumi S, Hashemizadeh Z. Effect of nano-silver, nano-copper, deconex and benzalkonium chloride on biofilm formation and expression of transcription regulatory quorum sensing gene (rh1R) in drug-resistance Pseudomonas aeruginosa burn isolates. Burns. 2018;44(3):700–708. doi:10.1016/j.burns.2017.10.021
114. Prateeksha S, Shoeb M, Sharma S, Naqvi A, Gupta V, Singh B. Scaffold of selenium nanovectors and honey phytochemicals for inhibition of Pseudomonas aeruginosa quorum sensing and biofilm formation. Front Cell Infect Microbiol. 2017;7:93. doi:10.3389/fcimb.2017.00093
115. Nafee N, Husari A, Maurer CK, et al. Antibiotic-free nanotherapeutics: ultra-small, mucus-penetrating solid lipid nanoparticles enhance the pulmonary delivery and anti-virulence efficacy of novel quorum sensing inhibitors. J Control Release. 2014;192:131–140. doi:10.1016/j.jconrel.2014.06.055
116. Davies J. Microbes have the last word. EMBO Rep. 2007;8(7):616–621. doi:10.1038/sj.embor.7401022
117. Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9(5):445–453. doi:10.1016/j.mib.2006.08.006
118. Yim G, Huimi Wang H, Davies Frs J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1195–1200. doi:10.1098/rstb.2007.2044
119. Kafil HS, Mobarez AM, Moghadam MF, Sadat Hashemi Z, Yousefi M. Gentamicin induces efaA expression and biofilm formation in Enterococcus faecalis. Microb Pathog. 2016;92:30–35. doi:10.1016/j.micpath.2015.12.008
120. Skindersoe ME, Alhede M, Phipps R, et al. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(10):3648–3663. doi:10.1128/AAC.01230-07
121. Bala A, Kumar R, Harjai K. Inhibition of quorum sensing in Pseudomonas aeruginosa by azithromycin and its effectiveness in urinary tract infections. J Med Microbiol. 2011;60(3):300–306. doi:10.1099/jmm.0.025387-0
122. Babić F, Venturi V, Maravić-Vlahoviček G. Tobramycin at subinhibitory concentration inhibits the RhlI/R quorum sensing system in a Pseudomonas aeruginosa environmental isolate. BMC Infect Dis. 2010;10(1):148. doi:10.1186/1471-2334-10-148
123. Sofer D, Gilboa-Garber N, Belz A, Garber NC. ‘Subinhibitory’erythromycin represses production of Pseudomonas aeruginosa lectins, autoinducer and virulence factors. Chemotherapy. 1999;45(5):335–341. doi:10.1159/000007224
124. Tateda K, Comte R, Pechere J-C, Köhler T, Yamaguchi K, Van Delden C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001;45(6):1930–1933. doi:10.1128/AAC.45.6.1930-1933.2001
125. El-Mowafy SA, Kha EG, Habib E-SE, Shaaban MI. Quorum sensing inhibitory activity of sub-inhibitory concentrations of β-lactams. Afr Health Sci. 2017;17(1):199–207. doi:10.4314/ahs.v17i1.25
126. Goh E-B, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci. 2002;99(26):17025–17030. doi:10.1073/pnas.252607699
127. Linares JF, Gustafsson I, Baquero F, Martinez J. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci. 2006;103(51):19484–19489. doi:10.1073/pnas.0608949103
128. El-Mowafy SA, Abd E, Galil KH, et al. Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa. Microb Pathog. 2014;74:25–32. doi:10.1016/j.micpath.2014.07.008
129. Soheili V, Bazzaz BSF, Abdollahpour N, Hadizadeh F. Investigation of Pseudomonas aeruginosa quorum-sensing signaling system for identifying multiple inhibitors using molecular docking and structural analysis methodology. Microb Pathog. 2015;89:73–78. doi:10.1016/j.micpath.2015.08.017
130. Chanda S, Rakholiya K. Combination therapy: synergism between natural plant extracts and antibiotics against infectious diseases. Microbiol Book Series. 2011;5:520–529.
131. Roudashti S, Zeighami H, Mirshahabi H, Bahari S, Soltani A, Haghi F. Synergistic activity of sub-inhibitory concentrations of curcumin with ceftazidime and ciprofloxacin against Pseudomonas aeruginosa quorum sensing related genes and virulence traits. World J Microbiol Biotechnol. 2017;33(3):50. doi:10.1007/s11274-016-2195-0
132. Bahari S, Zeighami H, Mirshahabi H, Roudashti S, Haghi F. Inhibition of Pseudomonas aeruginosa quorum sensing by subinhibitory concentrations of curcumin with gentamicin and azithromycin. J Glob Antimicrob Resist. 2017;10:21–28. doi:10.1016/j.jgar.2017.03.006
133. Li Y, Huang J, Li L, Liu L. Synergistic activity of berberine with azithromycin against Pseudomonas aeruginosa isolated from patients with cystic fibrosis of lung in vitro and in vivo. Cell Physiol Biochem. 2017;42(4):1657–1669. doi:10.1159/000479411
134. Chanda W, Joseph TP, Padhiar AA, et al. Combined effect of linolenic acid and tobramycin on Pseudomonas aeruginosa biofilm formation and quorum sensing. Exp Ther Med. 2017;14(5):4328–4338. doi:10.3892/etm.2017.5110
135. Zhou J-W, Chen -T-T, Tan X-J, Sheng J-Y, Jia A-Q. Can the quorum sensing inhibitor resveratrol function as an aminoglycoside antibiotic accelerant against Pseudomonas aeruginosa? Int J Antimicrob Agents. 2018;52(1):35–41. doi:10.1016/j.ijantimicag.2018.03.002
136. Defoirdt T, Boon N, Bossier P. Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 2010;6(7):e1000989. doi:10.1371/journal.ppat.1000989
137. García-Contreras R, Maeda T, Wood TK. Resistance to quorum-quenching compounds. Appl Environ Microbiol. 2013;79(22):6840–6846. doi:10.1128/AEM.02378-13
138. García-Contreras R, Nunez-Lopez L, Jasso-Chávez R, et al. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 2015;9(1):115. doi:10.1038/ismej.2014.98
139. Maeda T, García-Contreras R, Pu M, et al. Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 2012;6(3):493. doi:10.1038/ismej.2011.122
140. García-Contreras R, Martínez-Vázquez M, Velázquez Guadarrama N, et al. Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Pathog Dis. 2013;68(1):8–11. doi:10.1111/2049-632X.12039
141. Song Z, Kong K, Wu H, et al. Panax ginseng has anti-infective activity against opportunistic pathogen Pseudomonas aeruginosa by inhibiting quorum sensing, a bacterial communication process critical for establishing infection. Phytomedicine. 2010;17(13):1040–1046. doi:10.1016/j.phymed.2010.03.015
142. Luo J, Dong B, Wang K, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS One. 2017;12(4):e0176883. doi:10.1371/journal.pone.0176883
143. Paczkowski JE, Mukherjee S, McCready AR, et al. Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J Biol Chem. 2017;292(10):4064–4076. doi:10.1074/jbc.M116.770552
144. Jakobsen TH, van Gennip M, Phipps RK, et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob Agents Chemother. 2012;56(5):2314–2325. doi:10.1128/AAC.05919-11
145. Ahmed SA, Rudden M, Smyth TJ, Dooley JS, Marchant R, Banat IM. Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl Microbiol Biotechnol. 2019;103(8):3521–3535. doi:10.1007/s00253-019-09618-0
146. Zhou S, Zhang A, Chu W. Phillyrin is an effective inhibitor of quorum sensing with potential as an anti-Pseudomonas aeruginosa infection therapy. J Vet Med Sci. 2019;81(3):473–479. doi:10.1292/jvms.18-0523
147. Hançer Aydemir D, Çifci G, Aviyente V, Boşgelmez‐Tinaz G. Quorum‐sensing inhibitor potential of trans‐anethole aganist Pseudomonas aeruginosa. J Appl Microbiol. 2018;125(3):731–739. doi:10.1111/jam.13892
148. Rajkumari J, Borkotoky S, Murali A, Suchiang K, Mohanty SK, Busi S. Cinnamic acid attenuates quorum sensing associated virulence factors and biofilm formation in Pseudomonas aeruginosa PAO1. Biotechnol Lett. 2018;40(7):1087–1100. doi:10.1007/s10529-018-2557-9
149. Zhou J-W, Luo H-Z, Jiang H, Jian T-K, Chen Z-Q, Jia A-Q. Hordenine: a novel quorum sensing inhibitor and antibiofilm agent against Pseudomonas aeruginosa. J Agric Food Chem. 2018;66(7):1620–1628. doi:10.1021/acs.jafc.7b05035
150. Zhou L, Zheng H, Tang Y, Yu W, Gong Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol Lett. 2013;35(4):631–637. doi:10.1007/s10529-012-1126-x
151. Musthafa KS, Ravi AV, Annapoorani A, Packiavathy ISV, Pandian SK. Evaluation of anti-quorum-sensing activity of edible plants and fruits through inhibition of the N-acyl-homoserine lactone system in Chromobacterium violaceum and Pseudomonas aeruginosa. Chemotherapy. 2010;56(4):333–339. doi:10.1159/000320185
152. Priya K, Yin W-F, Chan K-G. Anti-quorum sensing activity of the traditional Chinese herb, Phyllanthus amarus. Sensors. 2013;13(11):14558–14569. doi:10.3390/s131114558
153. Kumar L, Chhibber S, Kumar R, Kumar M, Harjai K. Zingerone silences quorum sensing and attenuates virulence of Pseudomonas aeruginosa. Fitoterapia. 2015;102:84–95. doi:10.1016/j.fitote.2015.02.002
154. Vandeputte OM, Kiendrebeogo M, Rajaonson S, et al. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2010;76(1):243–253. doi:10.1128/AEM.01059-09
155. Rasmussen TB, Bjarnsholt T, Skindersoe ME, et al. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol. 2005;187(5):1799–1814. doi:10.1128/JB.187.5.1799-1814.2005
156. Bjarnsholt T, PØ J, Rasmussen TB, et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology. 2005;151(12):3873–3880. doi:10.1099/mic.0.27955-0
157. Vasavi HS, Arun AB, Rekha PD. Anti‐quorum sensing activity of Psidium guajava L. flavonoids against Chromobacterium violaceum and Pseudomonas aeruginosa PAO1. Microbiol Immunol. 2014;58(5):286–293. doi:10.1111/1348-0421.12150
158. Husain FM, Ahmad I, Asif M, Tahseen Q. Influence of clove oil on certain quorum-sensing-regulated functions and biofilm of Pseudomonas aeruginosa and Aeromonas hydrophila. J Biosci. 2013;38(5):835–844. doi:10.1007/s12038-013-9385-9
159. Husain FM, Ahmad I, Khan MS, Al-Shabib NA. Trigonella foenum-graceum (Seed) extract interferes with quorum sensing regulated traits and biofilm formation in the strains of Pseudomonas aeruginosa and Aeromonas hydrophila. Evid Based Complement Alternat Med. 2015;2015.
160. Okusa PN, Rasamiravaka T, Vandeputte O, Stévigny C, El Jaziri M, Duez P. Extracts of Cordia gilletii de wild (Boraginaceae) quench the quorum sensing of Pseudomonas aeruginosa PAO1. J Intercult Ethnopharmacol. 2014;3(4):138. doi:10.5455/jice.20140710031312
161. Ilic-Tomic T, Sokovic M, Vojnovic S, et al. Diarylheptanoids from Alnus viridis ssp. viridis and Alnus glutinosa: modulation of Quorum Sensing activity in Pseudomonas aeruginosa. Planta Med. 2017;83(01/02):117–125. doi:10.1055/s-0042-107674
162. Kim H-S, Lee S-H, Byun Y, Park H-D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci Rep. 2015;5(1):8656. doi:10.1038/srep08656
163. Pejin B, Ciric A, Glamoclija J, Nikolic M, Sokovic M. In vitro anti-quorum sensing activity of phytol. Nat Prod Res. 2015;29(4):374–377. doi:10.1080/14786419.2014.945088
164. Yang Y-X, Xu Z-H, Zhang Y-Q, Tian J, Weng L-X, Wang L-H. A new quorum-sensing inhibitor attenuates virulence and decreases antibiotic resistance in Pseudomonas aeruginosa. J Microbiol. 2012;50(6):987–993. doi:10.1007/s12275-012-2149-7
165. Stacy DM, Le Quement ST, Hansen CL, et al. Synthesis and biological evaluation of triazole-containing N-acyl homoserine lactones as quorum sensing modulators. Org Biomol Chem. 2013;11(6):938–954. doi:10.1039/C2OB27155A
166. Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J Am Chem Soc. 2005;127(37):12762–12763. doi:10.1021/ja0530321
167. Persson T, Hansen TH, Rasmussen TB, Skindersø ME, Givskov M, Nielsen J. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org Biomol Chem. 2005;3(2):253–262. doi:10.1039/B415761C
168. Nizalapur S, Ö K, Biswas NN, et al. Design, synthesis and evaluation of N-aryl-glyoxamide derivatives as structurally novel bacterial quorum sensing inhibitors. Org Biomol Chem. 2016;14(2):680–693. doi:10.1039/C5OB01973G
169. Geske GD, O’Neill JC, Miller DM, et al. Comparative analyses of N‐acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. Chem Bio Chem. 2008;9(3):389–400. doi:10.1002/cbic.200700551
170. Miandji A, Ulusoy S, Dündar Y, et al. Synthesis and biological activities of some 1, 3-benzoxazol-2 (3H)-one derivatives as anti-quorum sensing agents. Arzneimittelforschung. 2012;62(07):330–334. doi:10.1055/s-0032-1312590
171. Heidari A, Noshiranzadeh N, Haghi F, Bikas R. Inhibition of quorum sensing related virulence factors of Pseudomonas aeruginosa by pyridoxal lactohydrazone. Microb Pathog. 2017;112:103–110. doi:10.1016/j.micpath.2017.09.043
172. Lee LY, Hupfield T, Nicholson RL, et al. 2-Methoxycyclopentyl analogues of a Pseudomonas aeruginosa quorum sensing modulator. Mol Biosyst. 2008;4(6):505–507. doi:10.1039/b801563e
173. Heidari A, Haghi F, Noshiranzadeh N, Bikas R. (S, E)-2-hydroxy-N-(2-hydroxy-5-nitrobenzylidene) propane hydrazide as a quorum sensing inhibitor of Pseudomonas aeruginosa. Med Chem Res. 2017;26(9):1947–1955. doi:10.1007/s00044-017-1908-8
174. O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci. 2013;110(44):17981–17986. doi:10.1073/pnas.1316981110
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.