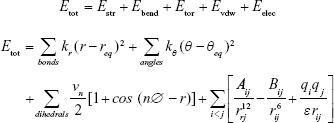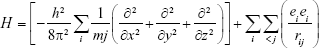Back to Journals » Drug Design, Development and Therapy » Volume 11
Quantum mechanics implementation in drug-design workflows: does it really help?
Authors Arodola OA , Soliman MES
Received 1 November 2016
Accepted for publication 22 December 2016
Published 31 August 2017 Volume 2017:11 Pages 2551—2564
DOI https://doi.org/10.2147/DDDT.S126344
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sukesh Voruganti
Olayide A Arodola,1 Mahmoud ES Soliman1,2
1Department of Pharmaceutical Chemistry, University of KwaZulu-Natal, Durban, South Africa; 2Department of Pharmaceutical Organic Chemistry, Faculty of Pharmacy, Zagazig University, Egypt
Abstract: The pharmaceutical industry is progressively operating in an era where development costs are constantly under pressure, higher percentages of drugs are demanded, and the drug-discovery process is a trial-and-error run. The profit that flows in with the discovery of new drugs has always been the motivation for the industry to keep up the pace and keep abreast with the endless demand for medicines. The process of finding a molecule that binds to the target protein using in silico tools has made computational chemistry a valuable tool in drug discovery in both academic research and pharmaceutical industry. However, the complexity of many protein–ligand interactions challenges the accuracy and efficiency of the commonly used empirical methods. The usefulness of quantum mechanics (QM) in drug–protein interaction cannot be overemphasized; however, this approach has little significance in some empirical methods. In this review, we discuss recent developments in, and application of, QM to medically relevant biomolecules. We critically discuss the different types of QM-based methods and their proposed application to incorporating them into drug-design and -discovery workflows while trying to answer a critical question: are QM-based methods of real help in drug-design and -discovery research and industry?
Keywords: quantum mechanics, drug discovery, drug design, molecular mechanics, molecular dynamics, in silico tools
Corrigendum for this paper has been published
Introduction
Drug discovery plays an important role in the growth of any pharmaceutical company and society, as newer and safer drugs are launched in the market with the sole objective of improving the therapeutic value and safety of drugs. The pharmaceutical industry has consistently shown that it can discover and develop innovative medicines for a wide range of diseases.1
Drug research, as it is called today, began when chemistry had reached the peak of its career, allowing chemical principles and theories to be applied to problems outside the scope of chemistry, and when pharmacology became an independent scientific discipline on its own. By 1870, some of the important foundations of chemistry theory had been laid.2,3 In the twentieth century, biochemistry had remarkable influence on drug research in numerous ways (Table 1). It was during this period that the concept of targeting enzymes and designing drugs as inhibitors came into existence.4 However, the current drug-discovery process is very time consuming and expensive and can take up to 12–16 years of exhaustive research, huge financial investment, and clinical trials before a molecule can be recognized as a drug (Figure 1).5
  | Table 1 Some of the important discoveries in medicine in the last two centuries |
  | Figure 1 Flowchart of drug-discovery and -development process. |
Despite the diverse research and development (R&D) approaches adopted by pharmaceutical companies, the attrition rate is inadmissibly high. One of the factors contributing to the high attrition rates is an active compound with unacceptable absorption, distribution, metabolism, excretion, and toxicity (ADMET) adverse effects that thus needs to be withdrawn from development. This factor represents approximately 50% of all costly failures in drug development,6 and it has become widely appreciated that these areas should be considered as early as possible in the drug-discovery process.7,8 It is evident that the pitfall in the current drug-discovery process urges an unconventional approach, which would not only truncate the R&D time but also reduce the cost involved.9
The process of finding a molecule that binds to the target protein has now moved from the laboratory to the computer.10 Years ago, drug design (DD) substantially extended its range of applications from target identification to clinical trials.11 Computer-generated models, which serve as good predictive models for the evaluation of biological activities, have had numerous successes predicting the possible structures of biological targets, thus reducing fruitless effort using nuclear magnetic resonance and spectroscopy-structure elucidations.12 With in silico tools, it is possible to accelerate the drug-discovery process by modeling the most relevant ADMET properties.12 A molecule could be too toxic, too quickly eliminated from the body, possess fast metabolic reaction, unstable, too challenging to synthesize in large volume, or too expensive to produce. Therefore, many promising compounds will regrettably have to be rejected once they are found to show unacceptable adverse effects in humans. Furthermore, compared to the status a decade ago, protein structure-based DD is swiftly gathering energy, and results have shown a remarkable increase in the structural knowledge of medically relevant proteins through various methods,13–18 as well as computer-aided programs. The large number of structural studies on medically relevant proteins suggests that the structure of a potential drug target is treasurable knowledge for any pharmaceutical company, not only for lead discovery and lead optimization but also in the later stages of drug development, where such concerns as toxicity, bioavailability, and binding modes of potential drug candidates to the target protein are extremely important.
A drug reveals its action when it binds to its biological target (enzyme, nucleic acid, or antibody), typically receptors. Receptors possess the active sites for the binding of a drug. Therefore, it is important to know the structure of the target, in order to design a good drug and identify an accurate binding site. In DD, predicting drug–receptor interactions involves the development of pharmacophore-based and molecular docking/scoring techniques. However, some biologically relevant biomolecules lack X-ray crystal structures. To resolve this, homology modeling has been implemented,19 and modeled proteins behave somewhat like the real proteins in their native biological environment when simulated.20 Recently, many computer-assisted models have been developed, and several thousand candidates are being screened for various activities using these models. The methods of choice for activity screening using these models are computer programs that superimpose molecules with flexible alignment to develop pharmacophoric patterns and/or quantitative structure–activity relationships (QSARs), dock molecules to the receptor or a pseudoreceptor, or construct new drugs within a predefined active site.21,22 Different molecular properties (eg, electrostatic, steric, and hydrophobic) and hydrogen bond-acceptor and donor fields have been used to achieve this purpose.23 Computational studies of biological targets allow the study of their structure, function, and dynamics at molecular and atomic levels. The entire process is about the simulation of the biological targets using quantum mechanics (QM) calculation, which is based on the principles of chemistry and physics.
In this review, we discuss how the implementation of QM methods in academic and pharmaceutical companies’ research can be a useful tool in the elucidation of drug–target interactions, which will help DD and drug development with respect to accuracy, time, and cost.
Computer-aided drug-design (CADD) approaches
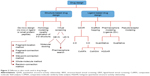
CADD is aimed at improving the development and efficacy of drugs using modern computational tools that are fast and cost-effective compared to conventional methods.24–26 The development of drugs that bind to specific targets has been recognized by the pharmaceutical industry as an important foundation that provides it with the necessary return on investment to invest in further R&D, leading to a discovery-and-development cycle.27 Broadly speaking, DD is divided into two areas: structure-based DD (SBDD) and ligand-based DD (Figure 2).
The CADD approach has been applied to various successful drugs, some of which are in use in the market. Examples include imatinib28 and nilotinib.29 Several other targets include ER,30,31 EGFR,32,33 PKCβ2,34 and BCR-Abl.35 A biomolecular system can be simulated using molecular mechanics (MM), QM, or a hybrid method (QM/MM), depending on the research problem to be answered.
Molecular mechanics
MM is commonly applied in large systems to calculate molecular structures and relative potential energies of a molecular conformation or atom arrangement.36–38 The electrons in the studied system are not explicitly considered, but instead each atom – specifically, the atomic nucleus and the associated electrons – is treated as a single particle. The exclusion of electrons in MM is justified on the basis of Born–Oppenheimer approximation,39 which states that electronic and nuclear motions can be uncoupled from each other and considered separately. Energy differences between conformations are significant in such calculations, rather than absolute values of potential energies.
MM can simply be viewed as a ball-and-spring model of atoms and molecules with classical forces between them.40 Such forces are accounted by potential energy functions with respect to such structural features as bond length, bond angles, and torsional angles. Potential energy functions are equipped with parameters designed to reproduce experimental properties.37 The MM or rather the total potential energy of a molecule is described as the sum of bond-stretching energy (Estr), bond angle-bending energy (Ebend), torsion energy (Etor), and energy of interactions among unbound atoms (Enb). Energy contributions of the latter constitute van der Waals and electrostatic interactions:
|
where Etot is total potential energy, stretch terms refer to Estr, bend terms refer to bond angle-bending energy Ebend, torsional terms refer to Etor or twisting energy, and unbound interactions are van der Waals forces and electrostatic forces between atoms that are not chemically bonded. Energy contributions from special treatment of hydrogen bonding and stretch–bend coupling interactions may also be seen in MM.
Quantum mechanics
The QM method treats molecules as collections of nuclei and electrons without any reference to “chemical bonds”. QM is important in understanding the behavior of systems at the atomic level. QM methods apply the laws of QM to approximate the wave function and to solve the Schrödinger equation.36,41 The solution to the Schrödinger equation is in terms of the motions of electrons, which in turn lead directly to molecular structure and energy among other observables, as well as to information about bonding. However, the Schrödinger equation cannot actually be solved for any but a one-electron system (the hydrogen atom), and approximations need to be made. According to QM, an electron bound to an atom cannot possess any arbitrary energy or occupy any position in space. These characteristics can be determined by solving the time-independent Schrödinger equation:42,43
H = T + V |
where H is the Hamiltonian operator (sum of kinetic energy), T the potential energy, and V the operator. H can also be defined as:
|
QM methods include ab initio44 density functional theory (DFT)45–47 and semiempirical calculations.48–50 For more accurate QM calculations, electron correlation methods, namely, CCSDT and MP2, etc, are necessary.46 DFT methods conduct calculations by electron correlation approximation.46,47,51 These methods can be employed to calculate crucial properties of a system such as vibrational frequencies, equilibrium molecular structure, dipole moments and free energy of reaction, which cannot be achieved by experimental methods.36 They also help to identify the activated complex when applied to reacting chemical species and therefore in the identification of a reaction pathway. Since the Schrödinger equation cannot be solved for complex molecular systems, semiempirical ab initio DFT methods were developed to approximate the precise QM solution to the problem.36,46,52 QM models are the most accurate, but also the most expensive methods in terms of time and computational resources, and are thus applied on small systems.
Classical mechanics or QM – which to choose?
Classical mechanics, also called MM, is the alternative to QM when chemical reactions do not need to be considered in a simulation. MM does not start from an “exact theory” (the Schrödinger equation), but rather describes molecules in terms of “bonded atoms”, which have been distorted from some idealized geometry due to unbound van der Waals and Columbic interactions. Though MM does not solve the Schrödinger equation for electron motions, it requires an explicit description of chemical bonding and lots of information about the structures of molecules. It is the use and extent of this information that distinguishes different MM models.53–56 While many of the details of mechanical and biochemical interactions in enzymes are currently unclear, MM can rely on force fields with fixed parameters to provide better understanding of conformational analysis between conformers,57 mechanical deformation of DNA, RNA, and proteins, and changes in cellular structure, response, and function.58 This understanding can offer new prognoses of diseases, as MM calculations are used to provide qualitative descriptions of molecular interactions.
QM has been said to succeed outstandingly in the area where MM failed. In contrast to QM, MM ignores electrons, fails to illustrate reality, and also computes the energy of a system as only a function of the nuclear positions. Generally, QM incorporates four phenomena for which MM cannot justify. These include quantization of some physical properties, quantum entanglement, the principle of uncertainty, and wave–particle duality. QM is applied in the determination of interactions between possible drugs and enzyme active sites.59–65 It is slow but accurate with respect to DD. In spite of the advantages of MM, it has some setbacks, such as inappropriate parameterization, inability to predict chemical reactions, or explain bond breaking/formation.66–70
QM in CADD arena
In answering research questions, computational chemists have a vast selection of methodologies at their disposal. The key tools available belong to six all-encompassing classes: molecular dynamic (MD) simulation, MM, QM, ab initio calculations, DFT, and semiempirical calculations. MM can be used to study very large molecules, because other QM methods, such as semiempirical calculations, ab initio, and DFT are relatively slow and would exhaust computational resources. However, MM methods are unable to address interactions between the ligand and the receptor in metal-containing systems.
Such algorithms as hybrid QM/MM, which combine QM and MM, have been developed to limit issues brought about by the individual application of these methods.71–74 QM methods are the most accurate, but also computationally expensive and time-consuming calculations.75 QM calculations are employed in semiempirical methods (eg, AM1, PM3) only for valence electrons in the system, whereas for other electrons and atomic nuclei behavior of other atoms, approximations are made.75 Combined QM-MM methods provide the accuracy of a QM description with low computational cost of MM.71,76 Even though QM-MM may not be applicable in every SBDD project, the majority of important systems cannot be well addressed by any other computational methods. QM-MM is thus the crucial component in computational drug discovery.
Five key facets are imperative in planning a QM-MM calculation on an enzyme: choice of the QM method, choice of MM force field, segregation of the system into QM and MM regions, simulation type (eg, MD simulation or calculation of potential energy profiles), and whether advanced conformational sampling will be performed. The choice of QM method is crucial. A plethora of different QM methods exists, ranging from fast, semiempirical methods (eg, AM1, PM3, SCC-DFTB; low accuracy and maximum of 2,000 atoms) to more accurate but more computationally expensive Hartree–Fock and density-functional (eg, B3LYP; medium accuracy and maximum of 500 atoms), and molecular orbital ab initio (eg, MP2, coupled cluster; very high accuracy and maximum of 20 atoms) methods. Not all methods are applicable to all systems, for reasons of accuracy, practicality, or lack of parameters (eg, for semiempirical methods) (Table 2). Generally, but not always, improved accuracy comes at the price of increased calculation expenses.
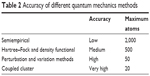  | Table 2 Accuracy of different quantum mechanics methods |
Typical applications of QM in DD include calculation of energies and structure optimization of ligand and/or protein–ligand complexes,77 especially for docking studies to obtain the correct binding mode of a ligand.78 QM-MM methods have shown promise for their accurate predictions when employed in the calculation of binding energies; however, this approach still requires further sampling of ligand–target conformations through MD simulations.79 QM methods have proved useful in the study of some target proteins, including HIV1 integrase,77 trypsin,78 West Nile virus NS3 serine protease, HIV1PR, and CDK2.79
The use of supercomputing to calculate QM has been attributed to expensive calculations for small systems. However, the use of Hadoop80 could make QM faster and more scalable and efficient. Hadoop could allow for better cluster utilization as well to accommodate larger jobs, which will help QM, as it needs more computational resources to run calculations for larger systems.
Recent QM developments in drug discovery
Tangible advances in the use of QM to solve relevant pharmaceutical problems have been seen in the last decade, eg, the use of the hybrid QM-MM approach to determine the free-energy landscape of the enzymatic reaction mechanism.81,82 The next step in the evolution of drug discovery is the routine use of QM in all levels of in silico DD.
SBDD is an important factor in the drug-discovery process, and designs more potent molecules with few alterations made, ie, derivatives of “lead” molecules.83–86 In silico tools can be used to design molecules to investigate existing protein–ligand interactions, as well as explore the active site for any supplementary hydrophilic or hydrophobic interactions that can increase binding affinity.87–89 The use of in silico tools allows the testing of a theory in a short time frame, using high-throughput empirical methods. However, there are concerns regarding the accuracy of these methods, particularly in the area of docking and scoring90–93 and QSAR.94,95
The in silico approach is fast and environmentally friendly, but it does not replace experimentation. Regardless, failures encountered in the pharmaceutical industry at the drug-discovery stage can be attributed to a number of factors that are not limited to wrong force-field parameters, especially for metals,96 disregard for protein flexibility97,98 or domain of applicability,99 or nonrigorous validation of the QSAR model.100,101
QM, a method used to replicate an experimental work accurately, proffers a potential solution to the failures mentioned.88,102 Increasingly, QM-MM methods are being applied to enzymes that are drug targets, often with the aim of providing information for DD.103–115 Examples include the HIV1-replication enzymes: reverse transcriptase, protease, and integrase. The reaction mechanisms of these enzymes have been studied using the QM-MM approach.116–126 Other examples are G-protein-coupled receptors,127–129 5-HT4 receptors,130,131 design and evaluation of a novel class of FKBP12 ligands,132,133 and novel inhibitors of human DHFR.134–136 The number of accidental discoveries in drug history is also legion.83,137–141 Another development in DD research is the hybrid QM-MM method, developed to improve the accuracy of biomolecular simulations,71–75 QM docking, QM virtual screening, and QM-QSAR.142,143 The QM approach has been successfully applied in drug discovery in pharmaceutical companies, eg, in the combination of artificial intelligence and cloud computing to search molecular entities and aid in the design of novel drugs.144
QM in DD research: time vs accuracy
In spite of growing computational resources, simulations of complex biosystems at the atomic/molecular level remain a challenge. The application of QM is limited to relatively small systems (Table 2). On the other hand, MM methods can treat millions of atoms or more. The hybrid QM-MM method, which combines the accuracy of QM descriptions with the low computational cost of MM modeling, and other QM-based methods (QM docking, QM virtual screening, QM-QSAR) can thus offer a promising solution to the computational challenge in DD. In addition, a recent article implicitly explained that the need for suitable computational approaches or tools could enhance success rates in the drug-discovery process.145
QM/MM docking method
The process of docking involves the correct prediction of ligand conformation and orientation within a targeted binding site, while scoring predicts the binding free energy of a complex formation.146 There are numerous molecular docking programs,147 such as Dock,148 AutoDock,149 Gold,150 Flexx,151 Glide,152,153 ICM,147 PhDock,154 and Surflex.154 However, some problems have been reported in docking,155,156 which could be due to either posing or scoring. Each docking program is ideal for precise docking problems;157 however, combining different computational methods can improve the reliability and accuracy of results.158 QM is specifically useful in DD when the interaction of the drug involves a chemical reaction. As such, implementation of QM docking would systematically improve the accuracy of description of enzyme–ligand interactions, as well as binding affinity. Limitations in scoring functions are being increasingly exposed, particularly as more challenging and electronically complex pockets are being probed, eg, systems with metals.159 Cho et al160 obtained ligand atomic charges using a QM-MM calculation. Calculations indicated generally improved poses after docking. The ability of QM-MM docking has been further evaluated in other studies to predict the poses of metalloproteins.161,162 Another study163 employed full QM calculation rather than QM-MM calculations to obtain partial charges for the ligand and receptor. Therefore, there appears to be evidence that QM-based models provide scoring functions that can improve the quality of predicted docking poses for challenging receptors.
QM virtual screening method
Virtual screening has become a powerful tool in the drug-discovery process to search for novel compounds with desired properties.164,165 This method has found its application in screening of combinatorial chemistry, genomics, protein, and peptide libraries.166 Virtual screening involves the docking of selected lead molecules against a specific biological target. This is followed by a scoring function.167 Virtual screening, which can be ligand-based168,169 or structure-based,168,170,171 screens the library by applying Lipinski’s rule of five before further evaluation.172 Pharmaceutical companies rely on this method when searching for new novel compounds. Considering today’s computational resources, several million compounds can be screened in a few days on supercomputers, and QM could be used to evaluate binding for further drug development.
QM-QSAR method
QSAR is a mathematical representation that attempts to correlate a set of compounds with dependent variables (activity values, eg, Ki, EC50, ED50, IC50) and a set of independent variables called descriptors.173 There are various statistical models that are used to derive a QSAR equation,174–176 and a QSAR model can be 2-D, 3-D, or 4-D.177–179 Using the QSAR method, predicted chemical structures that possess good activity values need only be synthesized.180–182 QM-QSAR uses QM methods to develop quantum-based QSAR models.183 Studies have provided details of QM-based descriptors used in QSAR programs, such as Codessa-, AM1-, and DFT-based descriptors, to understand the relationship between physicochemical properties and their descriptors.184–186
QM implementation in the pharmaceutical industry: time vs accuracy
As the ADMET properties of a drug determine its activity, the development of a new drug with reasonable ADMET makes drug discovery a more difficult and challenging process in the pharmaceutical industry. The pharmaceutical industry is progressively operating in an era where development costs are constantly under pressure, higher percentages of drugs are demanded, and the drug-discovery process is a trial-and-error run. The profits that flow in with the discovery of new drugs have always been the motivation for the industry to keep up the pace and keep abreast with the endless demand for medicines.187,188
In recent years, the use of CADD to simulate drug–receptor interactions has made rational DD feasible and cost-effective. in silico tools, such as docking, virtual screening, QSAR, molecular simulation, MM, and QM, use their respective mathematical equation to predict rapidly the binding affinities of a large library of compounds, as well as analyze homolytic or heterolytic fission/fusion before undergoing chemical (synthesis) and biological (activity) evaluation as a novel compound.
However, more attention should be paid to the way pharmaceutical companies use in silico tools. While docking, virtual screening, QSAR, and MM manage computational resources and allow rapid scans of large libraries, the accuracy of the results is in question when it comes to experimental data correlation. Compared to those used in pharmaceutical companies, there are more efficient methods, but the cost with respect to computer time/resources is high when one has to scan a really large library of compounds. Therefore, using a combination of QM to parameterize the molecules and MM to describe and solvate the protein, a more accurate understanding of binding affinity and protein–molecule interaction could be gained (Figure 3). If this method was implemented in pharmaceutical companies’ R&D, it would give correct binding affinities and free binding energy using different ligand geometries in QM-MM energy calculations. Furthermore, using QSAR to predict the activity of an existent molecule may lead to remarkable savings with respect to development time and cost (Table 3).
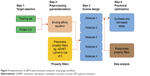  | Figure 3 Implementation of QM in pharmaceutical companies’ drug design workflow. |
Probably, most pharmaceutical companies today follow common technology processes for discovering drugs. These include cloning and expression of human receptors and enzymes using high-throughput, automated screening and the application of combinatorial chemistry.189 The field of combinatorial chemistry is in constant evolution. We believe that pharmaceutical companies rarely use accurate DD tools, eg, QM, in their DD, owing to the fast pace of their work. The use of QM is not limited to creating a computational model of a drug, but can also be applied to proteins, DNA, carbohydrates, and lipids, as well as solvent molecules that are involved in drug transportation, binding, and signaling.
Future perspectives
The application of QM-based approaches in guiding SBDD is not new. QM has featured in some medicinally relevant chemistry calculations in providing informative descriptors for QSAR and 3-D conformation for ligands. QM methods offer the ability to provide an accurate representation of ligands and proteins where MM parameterization struggles. QM approaches hold promise in addressing pharmacological problems on the time scale demanded by drug-discovery research. After ups and downs in the perception of CADD and perhaps some overhyping of its promises in drug development, it could be said that CADD is becoming a routinely used component of drug discovery.
Currently, sophisticated CADD tools are typically applied by modeling experts, but are increasingly spreading to the desktops of medicinal chemists as well. Ligand poses predicted from docking to receptors, such as metalloproteins, have been shown to resemble experiments more closely when partial charges are derived from QM or QM-MM calculations. The use of QM and QM-MM approaches in computation of protein–ligand binding affinities has met with mixed success. However, the QM-MM approach appears to be of most benefit for low-resolution X-ray structures, where an incorrectly assigned ligand structure due to its MM force field is more likely. Studies demonstrate that the use of accurate charges, in many cases, leads to improvement in docking accuracy in a wide range of Protein Data Bank complexes. The principal uncertainty at this point is whether this improved performance in docking can be noticed in other in silico methods.
In this review, we have discussed how the implementation of QM-based methods could help the drug-discovery and DD process in the pharmaceutical industry. This review outlines the major roles played by QM in the DD workflow and its importance in the drug-discovery process to avoid “dead-end” lead compounds. This method could have strong impact in future drug development, because of the endless demand for new drugs and the short time frame pharmaceutical companies have in developing them. Pharmaceutical companies have to reach a compromise between accuracy and productivity by applying QM in their research. The selection of the most appropriate method (MM, QM, or QM-MM) during drug development is of extreme importance. QM should be applied to “lead” compounds to provide insight into the free-energy landscape. Most importantly, before embarking on CADD, it is appropriate to evaluate the diversity and demand of accuracy of molecules to be designed in the project, which in turn dictates the most appropriate approach to select. It is also possible to reparameterize approximate methods in order to improve the accuracy of results in specific reactions that require numerous energy evaluations. A number of studies have sought to incorporate QM and QM-MM into their approaches for calculating ligand–receptor binding affinities. These approaches show promising results, but require further development to be broadly applicable. Finally, QM methods have proved valuable in quantitative analysis of the energetics of ligand deformation on binding. Although computation of binding energies remain a challenging and evolving area, current QM approaches could offer detailed information on the nature and relative strengths of complex active-site interaction, which is valuable in molecular design. It is likely that QM will become a more prominent tool in the repertoire of the computational medicinal chemist. Therefore, modern QM approaches will play a more direct role in informing and streamlining the drug-discovery process. The insight gained from this review could serve as a cornerstone for medicinal chemists, industry R&D and clinicians. This could provide better understanding of the in silico tools in drug design and development with improved ADMET, pharmacokinetics and the timely assessment of property profiles.
Acknowledgments
The authors acknowledge the College of Health Sciences, University of KwaZulu-Natal and the National Research Foundation (NRF) NRF-DST Centre of Excellence in HIV Prevention for financial support.
Disclosure
The authors report no conflicts of interest in this work.
References
Ratti E, Trist D. Continuing evolution of the drug discovery process in the pharmaceutical industry. Pure Appl Chem. 2011;73:67–75. | ||
Drews J. Drug discovery: a historical perspective. Science. 2000;287:1960–1964. | ||
Drews J. In Quest of Tomorrow’s Medicine. New York: Springer-Verlag; 1999. | ||
Meldrum NU, Roughton FJ. Carbonic anhydrase: its preparation and properties. J Physiol. 1933;80:113–142. | ||
Kuhlmann J. Drug research: from the idea to the product. Int J Clin Pharmacol Ther. 1997;35:541–552. | ||
Bharath EN, Manjula SN, Vijaychand A. In silico drug design: tool for overcoming the innovation deficit in the drug discovery process. Int J Pharm Pharm Sci. 2011;3:8–12. | ||
Parasuraman S. Toxicological screening. J Pharmacol Pharmacother. 2011;2:74–79. | ||
Alomar MJ. Factors affecting the development of adverse drug reactions (review article). Saudi Pharm J. 2014;22:83–94. | ||
Jochum C, Gasteiger J. Canonical numbering and constitutional symmetry. J Chem Inf Comput Sci. 1977;17:113–117. | ||
Balaban AT, Basak SC, Colburn T, Grunwald GD. Correlation between structure and normal boiling points of haloalkanes C1-C4 using neural networks. J Chem Inf Comput Sci. 1994;34:1118–1121. | ||
Rucker C, Meringer M, Kerber A. QSPR using MolGen-QSPR: the example of haloalkane boiling points. J Chem Inf Comput Sci. 2004;44:2070–2076. | ||
Augen J. The evolving role of information technology in the drug discovery process. Drug Discov Today. 2002;7:315–323. | ||
Schertler GF. Overproduction of membrane proteins. Curr Opin Struct Biol. 1992;2:534–544. | ||
[No authors listed]. Guide to protein purification. Methods Enzymol. 1990;182:1–818. | ||
Pardi A. Isotope labelling for NMR studies of biomolecules. Curr Opin Struct Biol. 1992;2:832–835. | ||
Pflugrath JW. Developments in X-ray detectors. Curr Opin Struct Biol. 1992;2:811–815. | ||
Ealick SE, Walter RL. Synchrotron beamlines for macromolecular crystallography. Curr Opin Struct Biol. 1993;3:725–736. | ||
Watenpaugh KD. Macromolecular crystallography at cryogenic temperatures. Curr Opin Struct Biol. 1991;1:1012–1015. | ||
Pitman MR, Menz RI. Methods for protein homology modelling. Bioinformatics. 2006;6:37–59. | ||
Taft C, Da Silva V, Da Silva C. Current topics in computer-aided drug design. J Pharm Sci. 2008;97:1089–1098. | ||
Klebe G. Toward a more efficient handling of conformational flexibility in computer-assisted modelling of drug molecules. Perspect Drug Discov Des. 1995;3:85–105. | ||
Kubinyi H. Combinatorial and computational approaches in structure-based drug design. Curr Opin Drug Discov Devel. 1998;1:16–27. | ||
Mestres J, Rohrer DC, Maggiora GM. MIMIC – a molecular-field matching program: exploiting applicability of molecular similarity approaches. J Comput Chem. 1997;18:934–954. | ||
Mandal S, Moudgil M, Mandal SK. Rational drug design. Eur J Pharmacol. 2009;625:90–100. | ||
Hao GF, Yang GF, Zhan CG. Structure-based methods for predicting target mutation-induced drug resistance and rational drug design to overcome the problem. Drug Discov Today. 2012;17:1121–1126. | ||
Hedvat M, Emdad L, Das SK, et al. Selected approaches for rational drug design and high throughput screening to identify anti-cancer molecules. Anticancer Agents Med Chem. 2012;12:1143–1155. | ||
Szymkowski DE. Creating the next generation of protein therapeutics through rational drug design. Curr Opin Drug Discov Devel. 2005;8:590–600. | ||
Druker BJ, Lydon NB. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest. 2000;105:3–7. | ||
Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. | ||
Bazer FW, Spencer TE, Johnson GA. Interferons and uterine receptivity. Semin Reprod Med. 2009;27:90–102. | ||
Gupta A, Mandal SK, Leblanc V, Descôteaux C, Asselin E, Bérubé G. Synthesis and cytotoxic activity of benzopyran-based platinum(II) complexes. Bioorg Med Chem Lett. 2008;18:3982–3987. | ||
Mandal S, Davie JR. An integrated analysis of genes and pathways exhibiting metabolic differences between estrogen receptor positive breast cancer cells. BMC Cancer. 2007;7:181. | ||
Sharma PS, Sharma R, Tyagi T. Receptor tyrosine kinase inhibitors as potent weapons in war against cancers. Curr Pharm Des. 2009;15:758–776. | ||
Grodsky N, Li Y, Bouzida D, et al. Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry. 2006;45:13970–13981. | ||
Patwardhan B, Bodeker G. Ayurvedic genomics: establishing a genetic basis for mind-body typologies. J Altern Complement Med. 2008;14:571–576. | ||
Atkins P, de Paula J. Atkins’ Physical Chemistry. 8th ed. New York: Macmillan; 2006. | ||
Rogers DW. Computational Chemistry Using the PC. 3rd ed. Hoboken (NJ): Wiley; 2003. | ||
Lewars, EG. Computational Chemistry: Introduction to the Theory and Applications of Molecular and Quantum Mechanics. 2nd ed. Alphen aan den Rijn, Netherlands; Kluwer; 2004. | ||
Tully JC. Perspective on “Zur Quantentheorie der Molekeln”. Theor Chem Acc. 2000;103:173–176. | ||
Hotokka M. Calculation of vibrational frequencies by molecular mechanics. In: Chalmers JM, Griffiths PR, editors. Handbook of Vibrational Spectroscopy. Hoboken (NJ): Wiley; 2006. | ||
Tannor DJ. Introduction to Quantum Mechanics: A Time-Dependent Perspective. Sausalito (CA): University Science Books; 2008. | ||
Mortimer R. Physical Chemistry. 3rd ed. Cambridge (MA): Academic Press; 2008. | ||
Lewars EG. Computational Chemistry: Introduction to the Theory and Applications of Molecular and Quantum Mechanics. Heidelberg: Springer; 2011. | ||
Jensen F. Introduction to Computational Chemistry. 2nd ed. Hoboken (NJ): Wiley; 1999. | ||
Perdew JP, Ruzsinszky A. Fourteen easy lessons in density functional theory. Int J Quantum Chem. 2010;110:2801–2807. | ||
Burke KJ. Perspective on density functional theory. J Chem Phys. 2012;136:150901. | ||
Grimme S, Antony J, Schwabe T, Mück-Lichtenfeld C. Density functional theory with dispersion corrections for supramolecular structures, aggregates, and complexes of (bio)organic molecules. Org Biomol Chem. 2007;5:741–758. | ||
Hehre WJ. A Guide to Molecular Mechanics and Quantum Chemical Calculations. Irvine (CA): Wavefunction; 2003. | ||
Dewar MJ, Zoebisch EG, Healy EF, Stewart JJ. AM1: a new general purpose quantum mechanical molecular model. J Am Chem Soc. 1985;107:3902–3909. | ||
Zerner M. Reviews of Computational Chemistry. Weinheim, Germany: Wiley-VCH; 1991. | ||
Car R. Introduction to density-functional theory and ab initio molecular dynamics. QSAR Comb Sci. 2002;21:97–104. | ||
Atkins PW, Friedman RS. Molecular Quantum Mechanics. 4th ed. Oxford: Oxford University Press; 2005. | ||
Atkins PW, Friedman RS. Molecular Quantum Mechanics. 3rd ed. Oxford: Oxford University Press; 2005. | ||
Levine IN. Quantum Chemistry. 5th ed. Upper Saddle River (NJ): Prentice Hall; 1999. | ||
Roothaan CC. New developments in molecular orbital theory. Rev Mod Phys. 1951;23:69–89. | ||
Hehre WJ, Radom L, Schleyer P, Pople J. Ab Initio Molecular Orbital Theory. Hoboken (NJ): Wiley; 1986. | ||
Lewars EG. Computational Chemistry. Heidelberg: Springer; 2011. | ||
Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2:715–725. | ||
Lipkowitz KB. Abuses of molecular mechanics: pitfalls to avoid. J Chem Educ. 1995;72:1070–1075. | ||
Nagase S. Polyhedral compounds of the heavier group 14 elements: silicon, germanium, tin, and lead. Acc Chem Res. 1995;28:469–476. | ||
Sella A, Basch H, Hoz S. Reactivity of strained compounds: is ground state destabilization the major cause for rate enhancement? J Am Chem Soc. 1996;118:416–420. | ||
Balaji V, Michl J. New strained organic molecules: theory guides experiment. Pure Appl Chem. 1988;60:189–194. | ||
Chakraborty S, Saha C. The Curtin-Hammett principle: a qualitative understanding. Reson J Sci Educ. 2016;21:151–171. | ||
Morgan J, Greenberg A. Curtin-Hammett principle: application to benzene oxide-oxepin tautomers. Struct Chem. 2013;24:1945–1956. | ||
Seeman JI. The Curtin-Hammett principle and the Winstein-Holness equation: new definition and recent extensions to classical concepts. J Chem Educ. 1986;63:42–48. | ||
Clark M, Cramer RD, Van Opdenbosch N. Validation of the general purpose Tripos 5.2 force field. J Comput Chem. 1989;10:982–1012. | ||
Burkert U, Allinger NL. Molecular Mechanics. Washington: American Chemical Society; 1982. | ||
Rappé AK, Casewit CJ. Molecular Mechanics Across Chemistry. Sausalito (CA): University Science Books; 1999. | ||
Halgren TA. Merck molecular force field – I: basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. | ||
Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl. 1996;48:1198–1229. | ||
Lin H, Truhlar DG. QM/MM: what have we learned, where are we, and where do we go from here? Theor Chem Acc. 2007;117:185–199. | ||
Garcia-Vela A, Gerber RB. Hybrid quantum/semiclassical wave packet method for molecular dynamics: application to photolysis of Ar⋯HCl. J Chem Phys. 1993;98:427–436. | ||
Sauer J, Sierka M. Combining quantum mechanics and interatomic potential functions in ab initio studies of extended systems, J Comput Chem. 2000;21:1470–1493. | ||
Monard G, Prat-Resina X, Gonzalez-Lafont A, Lluch JM. Determination of enzymatic reaction pathways using QM/MM methods. Int J Quantum Chem. 2003;93:229–244. | ||
Honarparvar B, Kruger HG, Maguire GE, Govender T, Soliman ME. Integrated approach to structure-based enzymatic drug design: molecular modeling, spectroscopy, and experimental bioactivity. Chem Rev. 2014;114(1):493–537. | ||
Menikarachchi LC, Gascon JA. QM/MM approaches in medicinal chemistry research. Curr Top Med Chem. 2010;10:46–54. | ||
Liao C, Nicklaus MC. Tautomerism and magnesium chelation of HIV-1 integrase inhibitors: a theoretical study. ChemMedChem. 2010;5:1053–1066. | ||
Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–931. | ||
Zhou T, Huang D, Caflisch A. Is quantum mechanics necessary for predicting binding free energy? J Med Chem. 2008;51:4280–4288. | ||
Cloudera. The Hadoop ecosystem. 2016. Available from: http://www.cloudera.com/training/library/hadoop-ecosystem.html. Accessed November 28, 2016. | ||
Chu WT, Wang J. Energy landscape topography reveals the underlying link between binding specificity and activity of enzymes. Sci Rep. 2016;6:27808. | ||
Ribeiro AJ, Santos-Martins D, Russo N, Ramos MJ, Fernandes PA. Enzymatic flexibility and reaction rate: a QM/MM study of HIV-1 protease. ACS Catal. 2015;5:5617–5626. | ||
Greer J, Erickson JW, Baldwin JJ, Varney MD. Application of the 3-dimensional structures of protein target molecules in structure-based drug design. J Med Chem. 1994;37:1035–1054. | ||
Kubinyi H. Structure-based design of enzyme inhibitors and receptor ligands. Curr Opin Drug Discov Devel. 1998;1:4–15. | ||
Babine RE, Bender SL. Molecular recognition of protein-ligand complexes: applications to drug design. Chem Rev. 1997;97:1359–1472. | ||
Anderson AC. The process of structure-based drug design. Chem Biol. 2003;10:787–797. | ||
Marrone TJ, Briggs JM, McCammon JA. Structure-based drug design: computational advances. Annu Rev Pharmacol Toxicol. 1997;37:71–90. | ||
Raha K, Peters MB, Wang B, et al. The role of quantum mechanics in structure-based drug design. Drug Discov Today. 2007;12:725–731. | ||
Davis AM, Teague SJ, Kleywegt GJ. Application and limitations of X-ray crystallographic data in structure-based ligand and drug design. Angew Chem Int Ed Engl. 2003;42:2718–2736. | ||
Tame JR. Scoring functions: a view from the bench. J Comput Aided Mol Des. 1999;13:99–108. | ||
Warren GL, Andrews CW, Capelli AM, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. | ||
Enyedy IJ, Egan WJ. Can we use docking and scoring for hit-to-lead optimization? J Comput Aided Mol Des. 2008;22:161–168. | ||
Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J Comput Chem. 2008;29:1316–1331. | ||
Johnson SR. The trouble with QSAR (or How I learned to stop worrying and embrace fallacy). J Chem Inf Model. 2008;48:25–26. | ||
Hawkins DM. The problem of overfitting. J Chem Inf Comput Sci. 2004;44:1–12. | ||
Halgren TA, Damm W. Polarizable force fields. Curr Opin Struct Biol. 2001;11:236–242. | ||
Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003;2:527–541. | ||
Davis AM, Teague SJ. Hydrogen bonding, hydrophobic interactions, and failure of the rigid receptor hypothesis. Angew Chem Int Ed Engl. 1999;38:737–749. | ||
Weaver S, Gleeson NP. The importance of the domain of applicability in QSAR modeling. J Mol Graph Model. 2008;26:1315–1326. | ||
Stouch TR, Kenyon JR, Johnson SR, Chen XQ, Doweyko A, Li Y. In silico ADME/Tox: why models fail. J Comput Aided Mol Des. 2003;17:83–92. | ||
Barril X, Hubbard RE, Morley SD. Virtual screening in structure-based drug discovery. Mini Rev Med Chem. 2004;4:779–791. | ||
Peters MB, Raha K, Merz KM. Quantum mechanics in structure-based drug design. Curr Opin Drug Discov Devel. 2006;9:370–379. | ||
Zhang, YK, Kua, J, McCammon, JA. Role of the catalytic triad and oxyanion hole in acetylcholinesterase catalysis: an ab initio QM/MM study. J Am Chem Soc. 2002;124:10572–10577. | ||
Zhou Y, Wang S, Zhang Y. Catalytic reaction mechanism of acetylcholinesterase determined by Born-Oppenheimer ab initio QM/MM molecular dynamics simulations. J Phys Chem B. 2010;114:8817–8825. | ||
Topf M, Varnai P, Richards WG. Ab initio QM/MM dynamics simulation of the tetrahedral intermediate of serine proteases: insights into the active site hydrogen-bonding network. J Am Chem Soc. 2002;124:14780–14788. | ||
Ishida T. Low-barrier hydrogen bond hypothesis in the catalytic triad residue of serine proteases: correlation between structural rearrangement and chemical shifts in the acylation process. Biochemistry. 2006;45:5413–5420. | ||
Schutz CN, Warshel A. The low barrier hydrogen bond (LBHB) proposal revisited: the case of the Asp⋯His pair in serine proteases. Proteins. 2004;55:711–723. | ||
Chen X, Fang L, Liu J, Zhan CG. Reaction pathway and free energy profile for butyrylcholinesterase-catalyzed hydrolysis of acetylcholine. J Phys Chem B. 2011;115:1315–1322. | ||
Boeck AT, Schopfer LM, Lockridge O. DNA sequence of butyrylcholinesterase from the rat: expression of the protein and characterization of the properties of rat butyrylcholinesterase. Biochem Pharmacol. 2002;63:2101–2110. | ||
Chen X, Fang L, Liu J, Zhan CG. Reaction pathway and free energy profiles for butyrylcholinesterase-catalyzed hydrolysis of acetylthiocholine. Biochemistry. 2012;51:1297–1305. | ||
Bourne Y, Kolb HC, Radic´ Z, Sharpless KB, Taylor P, Marchot P. Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation. Proc Natl Acad Sci U S A. 2004;101:1449–1454. | ||
Senapati S, Cheng Y, McCammon JA. In-situ synthesis of a tacrine-triazole-based inhibitor of acetylcholinesterase: configurational selection imposed by steric interactions. J Med Chem. 2006;49:6222–6230. | ||
Cheng Y, Cheng X, Radic´ Z, McCammon JA. Acetylcholinesterase: mechanisms of covalent inhibition of wild-type and H447I mutant determined by computational analyses. J Am Chem Soc. 2007;129:6562–6570. | ||
Liu J, Zhang Y, Zhan CG. Reaction pathway and free-energy barrier for reactivation of dimethylphosphoryl-inhibited human acetylcholinesterase. J Phys Chem B. 2009;113:16226–16236. | ||
Kwasnieski O, Verdier L, Malacria M, Derat E. Fixation of the two tabun isomers in acetylcholinesterase: a QM/MM study. J Phys Chem B. 2009;113:10001–10007. | ||
Piana S, Bucher D, Carloni P, Rothlisberger U. Reaction mechanism of HIV-1 protease by hybrid Car-Parrinello/classical MD simulations. J Phys Chem B. 2004;108:11139–11149. | ||
Rungrotmongkol T, Hannongbua S, Mulholland A. Mechanistic study of HIV-1 reverse transcriptase at the active site based on QM/MM method. J Theor Comput Chem. 2004;3:491–500. | ||
Rungrotmongkol T, Mulholland AJ, Hannongbua S. Active site dynamics and combined quantum mechanics/molecular mechanics (QM/MM) modelling of a HIV-1 reverse transcriptase/DNA/dTTP complex. J Mol Graph Model. 2007;26:1–13. | ||
Carnevale V, Raugei S, Piana S, Carloni P. On the nature of the reaction intermediate in the HIV-1 protease: a quantum chemical study. Comput Phys Commun. 2008;179:120–123. | ||
Ruiz-Pernia JJ, Alves CN, Moliner V, Silla E, Tunon I. A QM/MM study of the reaction mechanism for the 3′-processing step catalyzed by HIV-1 integrase. Theochem. 2009;898:115–120. | ||
Das A, Mahale S, Prashar V, Bihani S, Ferrer JL, Hosur MV. X-ray snapshot of HIV-1 protease in action: observation of tetrahedral intermediate and short ionic hydrogen bond SIHB with catalytic aspartate. J Am Chem Soc. 2010;132:6366–6373. | ||
Garrec J, Sautet P, Fleurat-Lessard P. Understanding the HIV-1 protease reactivity with DFT: what do we gain from recent functionals? J Phys Chem B. 2011;115:8545–8558. | ||
Suresh CH, Vargheese AM, Vijayalakshmi KP, Mohan N, Koga N. Role of structural water molecule in HIV protease-inhibitor complexes: a QM/MM study. J Comput Chem. 2008;29:1840–1849. | ||
Hensen C, Hermann JC, Nam KH, Ma SH, Gao JL, Holtje HD. A combined QM/MM approach to protein-ligand interactions: polarization effects of the HIV-1 protease on selected high affinity inhibitors. J Med Chem. 2004;47:6673–6680. | ||
Garrec J, Cascella M, Rothlisberger U, Fleurat-Lessard P. Low inhibiting power of N⋯CO based peptidomimetic compounds against HIV-1 protease: insights from a QM/MM study. J Chem Theory Comput. 2010;6:1369–1379. | ||
Gautier A, Pitrat D, Hasserodt J. An unusual functional group interaction and its potential to reproduce steric and electrostatic features of the transition states of peptidolysis. Bioorg Med Chem. 2006;14:3835–3847. | ||
Yeagle PL, Choi G, Albert AD. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry. 2001;40:11932–11937. | ||
Grossfield A. Recent progress in the study of G protein-coupled receptors with molecular dynamics computer simulations. Biochim Biophys Acta. 2011;1808:1868–1878. | ||
Tikhonova IG, Sum CS, Neumann S, et al. A bi-directional, iterative approach to the structural delineation of the functional “chemoprint” in GPR40 for agonist recognition. J Med Chem. 2007;50:2981–2989. | ||
Tack J, Camilleri M, Chang L, et al. Systematic review: cardiovascular safety profile of 5-HT4 agonists developed for gastrointestinal disorders. Aliment Pharmacol Ther. 2012;35:745–767. | ||
Sun H, Scott DO. Metabolism of 4-aminopiperidine drugs by cytochrome P450s: molecular and quantum mechanical insights into drug design. ACS Med Chem Lett. 2011;2:638–643. | ||
Wu X, Wang L, Han Y, et al. Creating diverse target-binding surfaces on FKBP12: synthesis and evaluation of a rapamycin analogue library. ACS Comb Sci. 2011;13:486–495. | ||
Olivieri L, Gardebien F. Structure-affinity properties of a high-affinity ligand of FKBP12 studied by molecular simulations of a binding intermediate. PLoS One. 2014;9:e114610. | ||
Tosso RD, Andujar SA, Gutierrez L, et al. Molecular modeling study of dihydrofolate reductase inhibitors: molecular dynamics simulations, quantum mechanical calculations, and experimental corroboration. J Chem Inf Model. 2013;53:2018–2032. | ||
Oefner C, D’Arcy A, Winkler FK. Crystal structure of human dihydrofolate reductase complexed with folate. Eur J Biochem. 1988;174:377–385. | ||
Alves CN, Marti S, Castillo R, et al. A quantum mechanic/molecular mechanic study of the wild-type and N155S mutant HIV-1 integrase complexed with diketo acid. Biophys J. 2008;94:2443–2451. | ||
Kubinyi H. Chance favors the prepared mind: from serendipity to rational drug design. J Recept Signal Transduct Res. 1999;19:15–39. | ||
von Itzstein M, Wu WY, Kok GB, et al. Rational design of potent sialidase-based inhibitors of influenza-virus replication. Nature. 1993;363:418–423. | ||
Vacca JP, Condra JH. Clinically effective HIV-1 protease inhibitors. Drug Discov Today. 1997;2:261–272. | ||
Sternweis PC, Gilman AG. Aluminum: a requirement for activation of the regulatory component of adenylate-cyclase by fluoride. Proc Natl Acad Sci U S A. 1982;79:4888–4891. | ||
Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-Å crystal structure of transducin α·GDP·AIF-4. Nature. 1994;372:276–279. | ||
Gleeson MP, Gleeson D. QM/MM calculations in drug discovery: a useful method for studying binding phenomena? J Chem Inf Model. 2009;49:670–677. | ||
Zhang X, Zhao Y, Lu G. Recent development in quantum mechanics/molecular mechanics modeling for materials. Int J Multiscale Comput Eng. 2012;10:65–82. | ||
Cloud Pharmaceuticals. Technology. 2016. Available from: http://www.cloudpharmaceuticals.com/technology.html. Accessed November 28, 2016. | ||
Nantasenamat C, Prachayasittikul V. Maximizing computational tools for successful drug discovery. Expert Opin Drug Discov. 2015;10:321–329. | ||
Leach AR, Shoichet BK, Peishoff CE. Prediction of protein-ligand interactions: docking and scoring – successes and gaps. J Med Chem. 2006;49:5851–5855. | ||
Cross JB, Thompson DC, Rai BK, et al. Comparison of several molecular docking programs: pose prediction and virtual screening accuracy. J Chem Inf Model. 2009;49:1455–1474. | ||
Ewing TJ, Makino S, Skillman AG, Kuntz ID. Dock 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–428. | ||
Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des. 1996;10:293–304. | ||
Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using Gold. Proteins. 2003;52:609–623. | ||
Warren GL, Andrews CW, Capelli AM, et al. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49:5912–5931. | ||
Friesner RA, Banks JL, Murphy RB, et al. Glide: a new approach for rapid, accurate docking and scoring – 1: method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. | ||
Halgren TA, Murphy RB, Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring – 2: enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. | ||
Joseph-McCarthy D, Alvarez JC. Automated generation of MCSS-derived pharmacophoric Dock site points for searching multiconformation databases. Proteins. 2003;51:189–202. | ||
Caballero NA, Melendez FJ, Nino A, Munoz-Caro C. Molecular docking study of the binding of aminopyridines within the K+ channel. J Mol Model. 2007;13:579–586. | ||
Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012;279:532–542. | ||
Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem. 1992;13:505–524. | ||
Vajda S, Kozakov D. Convergence and combination of methods in protein-protein docking. Curr Opin Struct Biol. 2009;19:164–170. | ||
Seebeck B, Reulecke I, Kamper A, Rarey M. Modeling of metal interaction geometries for protein-ligand docking. Proteins. 2008;71:1237–1254. | ||
Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 200526:915–931. | ||
Cho AE, Rinaldo D. Extension of QM/MM docking and its applications to metalloproteins. J Comput Chem. 2009;30:2609–2616. | ||
Fong P, McNamara JP, Hillier IH, Bryce RA. Assessment of QM/MM scoring functions for molecular docking to HIV-1 protease. J Chem Inf Model. 2009;49:913–924. | ||
Fischer B, Fukuzawa K, Wenzel W. Receptor-specific scoring functions derived from quantum chemical models improve affinity estimates for in-silico drug discovery. Proteins. 2008;70:1264–1273. | ||
Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today. 2006;11:1046–1053. | ||
Heikamp K, Bajorath J. The future of virtual compound screening. Chem Biol Drug Des. 2013;81:33–40. | ||
Martis EA, Radhakrishnan R, Badye RR. High-throughput screening: the hits and leads of drug discovery – an overview. J Appl Pharm Sci. 2011;1:2–10. | ||
Rostami-Hodjegan A, Tucker G. ‘In silico’ simulations to assess the ‘in vivo’ consequences of ‘in vitro’ metabolic drug-drug interactions. Drug Discov Today. 2004;1:441–448. | ||
Bajorath J. Integration of virtual and high-throughput screening. Nat Rev Drug Discov. 2002;1:882–894. | ||
Eckert H, Bojorath J. Molecular similarity analysis in virtual screening: foundations, limitations and novel approaches. Drug Discov Today. 2007;12:225–233. | ||
Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004;3:935–949. | ||
Cheng T, Li Q, Zhou Z, Wang Y, Bryant SH. Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J. 2012;14:133–141. | ||
Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44:235–249. | ||
Hansch C, Maloney PP, Fujita T. Correlation of biological activity of phenoxyacetic acids with Hammett substituent constants and partition coefficients. Nature. 1962;194:178–180. | ||
Mevik BH, Cederkvist HR. Mean squared error of prediction (MSEP) estimates for principal component regression (PCR) and partial least squares regression (PLSR). J Chemom. 2004;18:422–429. | ||
Estienne F, Vander Heyden Y, Massart DL. Chemometrics and modeling. Chimia. 2001;55:70–80. | ||
Nantasenamat C, Isarankura-Na-Ayudhya C, Naenna T, Prachayasittikul V. A practical overview of quantitative structure-activity relationship. EXCLI J. 2009;8:74–88. | ||
Suh ME, Park SY, Lee HJ. Comparison of QSAR methods (CoMFA, CoMSIA, HQSAR) of anticancer 1-N-substituted imidazoquinoiine-4,9-dione derivatives. Bull Korean Chem Soc. 2002;23:417–422. | ||
Myint KZ, Xie XQ. Recent advances in fragment-based QSAR and multi-dimensional QSAR methods. Int J Mol Sci. 2010;11:3846–3866. | ||
Melani F, Gratteri P, Adamo M, Bonaccini C. Field interaction and geometrical overlap: a new simplex and experimental design based computational procedure for superposing small ligand molecules. J Med Chem. 2003;46:1359–1371. | ||
Kubinyi H. QSAR and 3D QSAR in drug design – 2: applications and problems. Drug Discov Today. 1997;2:538–546. | ||
Kubinyi H. QSAR and 3D QSAR in drug design – 1: methodology. Drug Discov Today. 1997;2:457–467. | ||
Polanski J, Gieleciak R, Bak A. Probability issues in molecular design: predictive and modeling ability in 3D-QSAR schemes. Comb Chem High Throughput Screen. 2004;7:793–807. | ||
Dixon S, Merz KM, Lauri G, Ianni JC. QMQSAR: utilization of a semiempirical probe potential in a field-based QSAR method. J Comput Chem. 2005;26:23–34. | ||
Karelson M, Lobanov VS, Katritzky AR. Quantum-chemical descriptors in QSAR/QSPR studies. Chem Rev. 1996;96:1027–1043. | ||
Brustle M, Beck B, Schindler T, King W, Mitchell T, Clark T. Descriptors, physical properties, drug-likeness. J Med Chem. 2002;45:3345–3355. | ||
Wan J, Zhang L, Yang GF, Zhan CG. Quantitative structure-activity relationship for cyclic imide derivatives of protoporphyrinogen oxidase inhibitors: a study of quantum chemical descriptors from density functional theory. J Chem Inf Comput Sci. 2004;44:2099–2105. | ||
Halliday RG, Walker SR, Lumley CE. R&D philosophy and management in the world’s leading pharmaceutical companies. J Pharm Med. 1992;2:139–154. | ||
Dimasi JA, Hansen RW, Grabowski HG, Lasagna L. Research-and-development costs for new drugs by therapeutic category: a study of the United States pharmaceutical industry. Pharmacoeconomics. 1995;7:152–169. | ||
Kenakin T, Angus JA. The Pharmacology of Functional, Biochemical, and Recombinant Receptor Systems. Heidelberg: Springer; 2000. | ||
Gonçalves AS, França TC, Figueroa-Villar JD, Pascutti PG. Molecular dynamics simulations and QM/MM studies of the reactivation by 2-PAM of tabun inhibited human acethylcolinesterase. J Braz Chem Soc. 2011;22:155–165. | ||
Vidossich P, Magistrato A. QM/MM molecular dynamics studies of metal binding proteins. Biomolecules. 2014;4:616–645. | ||
Chen J, Wang J, Zhang Q, Chen K, Zhu W. A comparative study of trypsin specificity based on QM/MM molecular dynamics simulation and QM/MM GBSA calculation. J Biomol Struct Dyn. 2015;33:2606–2618. | ||
Makatini MM, Petzold K, Sriharsha SN, et al. Synthesis and structural studies of pentacycloundecane-based HIV-1 PR inhibitors: a hybrid 2D NMR and docking/QM/MM/MD approach. Eur J Med Chem. 2011;46:3976–3985. | ||
van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52:2708–2728. | ||
Woods CJ, Manby FR, Mulholland AJ. An efficient method for the calculation of quantum mechanics/molecular mechanics free energies. J Chem Phys. 2008;128:014109. | ||
Sproviero EM, Newcomer MB, Gascon JA, Batista ER, Brudvig GW, Batista VS. The MoD-QM/MM methodology for structural refinement of photosystem II and other biological macromolecules. Photosynth Res. 2009;102:455–470. | ||
Barone V, Biczysko M, Brancato G. Extending the range of computational spectroscopy by QM/MM approaches: time-dependent and time-independent routes. In: Sabin JR, Brandas E, Canuto S, editors. Combining Quantum Mechanics and Molecular Mechanics: Some Recent Progresses [sic] in QM/MM Methods. Oxford: Academic Press; 2010:17–57. | ||
Jensen JH, Li H, Robertson AD, Molina PA. Prediction and rationalization of protein pKa values using QM and QM/MM methods. J Phys Chem A. 2005;109:6634–6643. | ||
Ranaghan KE, Mulholland AJ. Investigations of enzyme-catalysed reactions with combined quantum mechanics/molecular mechanics (QM/MM) methods. Int Rev Phys Chem. 2010;29:65–133. | ||
Senn HM, Thiel W. QM/MM methods for biological systems. In: Reiher M, editor. Atomistic Approaches in Modern Biology: From Quantum Chemistry to Molecular Simulations. Heidelberg: Springer; 2007:173–290. | ||
Morozov AV, Misura KM, Tsemekhman K, Baker D. Comparison of quantum mechanics and molecular mechanics dimerization energy landscapes for pairs of ring-containing amino acids in proteins. J Phys Chem B. 2004;108:8489–8496. | ||
Park K, Goetz AW, Walker RC, Paesani F. Application of adaptive QM/MM methods to molecular dynamics simulations of aqueous systems. J Chem Theory Comput. 2012;8:2868–2877. | ||
Lian P, Guo HB, Riccardi D, et al. X-ray structure of a Hg2+ complex of mercuric reductase (MerA) and quantum mechanical/molecular mechanical study of Hg2+ transfer between the C-terminal and buried catalytic site cysteine pairs. Biochemistry. 2014;53:7211–7222. | ||
Akash K, Viera L, Dogan C, Daniel MK, Soumyendu R, Stefan B. A combination of docking, QM/MM methods, and MD simulation for binding affinity estimation of metalloprotein ligands. J Med Chem. 2005;48:5437–5447. | ||
Cho AE, Chung JY, Kim M, Park K. Quantum mechanical scoring for protein docking. J Chem Phys. 2009;131:134108. | ||
Burger SK, Thompson DC, Ayers PW. Quantum mechanics/molecular mechanics strategies for docking pose refinement: distinguishing between binders and decoys in cytochrome C peroxidase. J Chem Inf Model. 2011;51:93–101. | ||
Nisha C, Kotni MK, Chetan S, Manga V, Jitander KK, Pawan KS. QM/MM docking strategy and prime/MM-GBSA calculation of celecoxib analogues as N-myristoyltransferase inhibitors. Virol Mycol. 2015;4:141. | ||
Chaskar P, Zoete V, Roehrig UF. Toward on-the-fly quantum mechanical/molecular mechanical (QM/MM) docking: development and benchmark of a scoring function. J Chem Inf Model. 2014;54:3137–3152. | ||
Chung JY, Hah JM, Cho AE. Correlation between performance of QM/MM docking and simple classification of binding sites. J Chem Inf Model. 2009;49:2382–2387. | ||
Xu M, Lill MA. Induced fit docking, and the use of QM/MM methods in docking. Drug Discov Today Technol. 2013;10:e411–e418. | ||
Tan J, Tian F, Lv Y, et al. Integration of QSAR modelling and QM/MM analysis to investigate functional food peptides with antihypertensive activity. Mol Simul. 2013;39:1000–1006. | ||
Braga RC, Andrade CH. QSAR and QM/MM approaches applied to drug metabolism prediction. Mini Rev Med Chem. 2012;12:573–582. | ||
Chung JY, Chung HW, Cho SJ, Hah JM, Cho AE. QM/MM based 3D QSAR models for potent B-Raf inhibitors. J Comput Aided Mol Des. 2010;24:385–397. | ||
Lodola A, Rivara S, Mor M. Application of computational methods to the design of fatty acid amide hydrolase (FAAH) inhibitors based on a carbamic template structure. In: Christov C, editor. Computational Chemistry Methods in Structural Biology. Oxford: Academic Press; 2011:1–26. | ||
Reddy KK, Singh SK, Tripathi SK, Selvaraj C. Identification of potential HIV-1 integrase strand transfer inhibitors: in silico virtual screening and QM/MM docking studies. SAR QSAR Environ Res. 2013;24:581–595. | ||
Zheng F, Yang W, Ko MC, et al. Most efficient cocaine hydrolase designed by virtual screening of transition states. J Am Chem Soc. 2008;130:12148–12155. | ||
Wichapong K, Rohe A, Platzer C, et al. Application of docking and QM/MM-GBSA rescoring to screen for novel Myt1 kinase inhibitors, J Chem Inf Model. 2014;54:881–893. | ||
Ilatovskiy AV, Abagyan R, Kufareva I. Quantum mechanics approaches to drug research in the era of structural chemogenomics. Int J Quantum Chem. 2013;113:1669–1675. | ||
Waszkowycz B. Towards improving compound selection in structure-based virtual screening. Drug Discov Today. 2008;13:219–226. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.