Back to Journals » Cancer Management and Research » Volume 10
Quantitative assessment of aberrant P16INK4a methylation in ovarian cancer: a meta-analysis based on literature and TCGA datasets
Authors Ruan J, Xu P , Fan W, Deng Q, Yu M
Received 11 April 2018
Accepted for publication 14 June 2018
Published 29 August 2018 Volume 2018:10 Pages 3033—3046
DOI https://doi.org/10.2147/CMAR.S170818
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Harikrishna Nakshatri
Jie Ruan,1,* Peipei Xu,2,3,* Wei Fan,4 Qiaoling Deng,3 Mingxia Yu3
1Key Laboratory for Medical Molecular Diagnostics of Guangdong, Guangdong Medical University, Dongguan, Guangdong, 523808, China; 2Department of Clinical Laboratory, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, 450072, China; 3Department of Clinical Laboratory, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071, China; 4Department of Pathology, Zhongnan Hospital of Wuhan University, Wuhan, Hubei, 430071, China
*These authors contributed equally to this work
Abstract: Epigenetic alteration of P16INK4a is conventionally thought to induce the initiation of carcinoma. However, the role of P16INK4a methylation in ovarian cancer still remains controversial. Therefore, we performed a meta-analysis to further elucidate the relationship between P16INK4a promoter methylation and ovarian cancer. A total of 24 studies, including 20 on risk, 10 on clinicopathological features, and 3 on prognosis, were included in our meta-analysis. Our results indicated that the frequency of P16INK4a methylation in cancer tissues was significantly higher than normal tissues and low malignant potential tumor tissues (odds ratio [OR] =5.01, 95% CI=1.55–16.14; OR =1.88, 95% CI=1.10–3.19, respectively), but similar to benign tissues (OR =1.18, 95% CI=0.52–2.65). Furthermore, P16INK4a promoter methylation was not strongly correlated with age, clinical stage, tumor differentiation, or histological subtype in patients with ovarian cancer. Additionally, survival analysis showed that patients with P16INK4a promoter methylation had a shorter progression-free survival in univariate and multivariate Cox regression models (hazard ratio =1.68, 95% CI=1.26–2.24; hazard ratio =1.55, 95% CI=1.15–2.08; respectively). In The Cancer Genome Atlas datasets, the methylation levels of seven out of nine CpG sites were significantly increased in the ovarian tumor tissues compared with the normal tissues. In conclusion, the present meta-analysis suggests that P16INK4a promoter methylation may be useful in distinguishing malignant cancer from healthy ovarian tissues, and it may be a potential predictive marker for prognosis in patients with ovarian cancer.
Keywords: ovarian cancer, P16INK4a promoter methylation, TCGA datasets, meta-analysis
Introduction
Ovarian cancer is the fifth leading cause of cancer-related deaths in women. According to the GLOBOCAN 2012 database, the incidences of ovarian cancer are 9.1 per 1,00,100 in developed countries and 5.0 per 1,00,000 in developing countries.1 Thereinto, approximately 70% is high-grade serous carcinomas.2 Up to now, despite the effective treatments including radical resection, systemic chemotherapy, and targeted drugs for patients, the average 5-year survival is still only at 46%.3 Ovarian cancer is a multifactorial disease caused by the interaction of genetic and epigenetic factors.4,5 DNA methylation, as the most common epigenetic alteration, could occur at CpG island in the promoter region, 5′ or 3′ untranslated regions, and even in gene body of tumor suppressor genes (TSGs). Hypermethylation in the proximal promoter region often contributes to the transcriptional downregulation but methylation in exons is associated with active transcription.6,7 Recently, mounting evidences demonstrated that DNA methylation was involved in ovarian cancer.8–10 Therefore, identifying the role of TSG methylation in patients with ovarian cancer is of value.
P16INK4a (also known as CDKN2A), a classical TSG, is located on chromosome 9p21 and plays an important role in cell cycle regulation by decelerating cells progression from G1 to S phase.11,12 It has become clear that the expression of P16 is reduced by DNA methylation.13–15 Also, P16INK4a inactivation upregulates retinoblastoma (RB) protein by stimulating the cyclin-dependent kinases (CDKs) and RB pathway, which leads to dysfunction of cell proliferation and apoptosis, thereby further facilitating carcinogenesis.16 Indeed, several types of cancer, including ovarian cancer, exhibit a methylation phenotype of P16INK4a.17–19
To date, even though abundant studies have been conducted to explore the role of P16INK4a promoter methylation in ovarian cancer, the results are still inconclusive. Several studies reported that P16INK4a promoter methylation was associated with an increasing trend in ovarian cancer,20–23 while, other studies suggested that P16INK4a promoter methylation was not related to the occurrence of ovarian cancer.24–30 Interestingly, even the conclusions in two published meta-analyses were inconsistent. Xiao et al reported that aberrant methylation of P16INK4a was significantly associated with ovarian carcinogenesis,31 while Jiang et al suggested no association between P16INK4a methylation and epithelial ovarian cancer.32
Considering these conflicting conclusions on the role of methylated P16INK4a in ovarian cancer, we performed an adaptive synthesized analysis to quantitatively evaluate the occurrence frequency, clinicopathological features, and potential prognostic significance of P16INK4a promoter methylation in ovarian cancer. Moreover, we searched The Cancer Genome Atlas (TCGA) database, collecting hundreds of ovarian cancer samples with whole genome DNA methylation datasets to validate our meta-analysis.
Materials and methods
Search strategy and selection criteria
PubMed, Embase, Web of Science, and China National Knowledge Infrastructure were searched up to April 12, 2017, by the following keywords and search items: (P16 OR P16INK4a OR CDKN2A) AND (methylation OR hypermethylation OR demethylation) AND (ovarian OR ovary) AND (cancer OR carcinoma OR neoplasm). The search was limited to human studies, without language restriction. Moreover, a manual search of the relevant references was implemented to identify the potentially additional articles.
The following criteria were used for screening eligible studies: 1) case–control studies evaluating the association between P16INK4a promoter methylation and ovarian cancer risk, or case only studies evaluating the association of P16INK4a promoter methylation with clinicopathological features or prognosis in ovarian cancer; 2) articles providing sufficient information for calculating an odds ratio (OR) and corresponding 95% CI, or study offering hazard ratio (HR) and 95% CI directly; 3) sample types limited to tissues; and 4) studies with full-text articles. It is worth noting that when multiple reports were published from a same study population, only the most recent or complete information was included in this meta-analysis. Meanwhile, studies with Newcastle Ottawa Scale (NOS) scores greater than or equal to five were enrolled.
Data extraction and quality assessment
With a preformed unified form, data were extracted independently by two investigators, and disagreements were resolved by discussion till consensus was achieved. The following information was extracted from studies: the first author’s name, publication year, country, geographical location, sample size, age of patients in the case group, the frequencies of methylation in the case and control groups, methods for detecting methylation, methylation site, disease stage, tumor grade, histological subtype, and effects on survival outcomes.
The quality of eligible case–control studies was assessed according to the NOS criteria.33 The NOS criteria are based on three aspects: 1) subject selection: 0–4; 2) comparability of subject: 0–2; 3) clinical outcome: 0–3.
Statistical analysis
Statistical analysis was conducted with Review Manager 5.2 (Cochrane Collaboration, Oxford, UK) and the Stata 12.0 (Stata Corporation, College Station, TX, USA). ORs with corresponding 95% CIs were calculated to estimate the association between P16INK4a promoter methylation and ovarian cancer risk or clinicopathological features. Meanwhile, HRs and 95% CIs were used to assess the prognosis of P16INK4a promoter methylation on ovarian cancer. Inter-study heterogeneity was estimated with the Cochran’s Q statistic and I2 tests. P<0.05 or I2>50% indicated substantial heterogeneity, and then the random-effects model was applied. Otherwise, the fixed-effects model was selected. We also explored sources of heterogeneity using meta-regression and subgroup analyses by publication year, geographical location, method, and case sample size. Additionally, sensitivity analysis was performed to investigate the influence of individual study. Publication bias was evaluated by funnel plots and Begg’s test, and P<0.05 was considered statistically significant. It is worth mentioning that, for some trials containing no events in both case and control arms, as no information supplied about the likely magnitude of the effect, we excluded such trials when synthesizing data.34
TCGA datasets extraction and analysis
We collected DNA methylation datasets of 582 ovarian cancer cases and 12 ovarian normal tissues from TCGA (“TCGA-ovary [OV]” project) program.35 The methylation measurement was performed using Illumina HumanMethylation27 BeadChip. Beta value of each CpG site was extracted to assess the methylation level of CDKN2A gene. Beta value was calculated based on the intensities of the methylated (M) and unmethylated (U) bead types: beta value = M/(M+U).36 The difference of DNA methylation level of CpG sites between ovarian tumor tissues and normal ovarian tissues in TCGA database was analyzed by Student’s t-test on the means. P16INK4a gene expression value (fragments per kilobase of transcript per million mapped reads) in ovarian tumor tissues (TCGA, “TCGA-OV” project) was also extracted. Pearson’s product-moment correlation between P16INK4a gene expression levels and methylation of its CpG islands was evaluated. Data analysis was performed using R software (R i386 3.4.0). P-values were adjusted via Bonferroni correction.
Results
Identification of relevant studies
The procedure of study selection is outlined in Figure 1. We identified 233 articles in the initial literature search. A total of 153 references remained after removing duplicates. After reading titles and abstracts, 84 records were identified for further full-text assessment, which further excluded 60 more articles. Finally, 24 studies from 1997 to 2015 were included in this meta-analysis.17,20,22–30,37–49
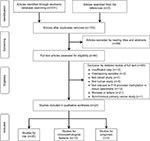  | Figure 1 Flow diagram of study selection. |
Baseline characteristics of included studies
Out of the 24 studies, 11 studies were conducted in Asia, 7 in Europe, 4 in America, 1 in Africa, and 1 in Oceania. The detection methods of methylation in 20 studies were methylation-specific PCR (MSP) and real-time quantitative MSP, while methylation-specific multiplex ligation-dependent probe amplification was used in two studies, MethyLight was used in one study, and Southern analysis was used in one study. Among the 24 articles, 20 studies17,20,22–30,37–40,42,45–47,49 addressed the risk of P16INK4a promoter methylation in ovarian cancer, 10 studies20,25,28,29,38,41,43,44,47,48 covered clinicopathological features, and 3 studies20,42,43 discussed prognosis. To explore the relationship between P16INK4a promoter methylation and ovarian cancer risk, three groups, that is, normal tissues, benign tissues, and low malignant potential or borderline tumor tissues (LMP), were compared. The NOS scores of all case–control studies were ≥5. The basic characteristics of all included studies are summarized in Tables 1 and 2.
Quantitative data synthesis
Association between P16INK4a promoter methylation and ovarian cancer risk
A total of 1,217 ovarian cancers, 116 LMP cancers, 271 benign patients, and 351 normal controls were quantitatively synthesized in this analysis. Results indicated that the frequency of P16INK4a promoter methylation in cancer tissues was significantly elevated than that in normal tissues (OR=5.01, 95% CI=1.55–16.14) and LMP tissues (OR =1.88, 95% CI=1.10–3.19), but similar to benign tissues (OR =1.18, 95% CI=0.52–2.65; Figure 2). Further analyses showed that the frequencies of P16INK4a promoter methylation in benign tissues and LMP tissues were not higher than those in normal tissues (OR =2.28, 95% CI=0.37–14.09; OR =2.28, 95% CI=0.15–34.73, respectively; Figure 3).
With large heterogeneity, meta-regression and subgroup analyses were conducted by the publication year, geographical location, method, and case sample size in the comparison of cancer tissues vs normal tissues. Meta-regression found that case sample size was significantly correlated with the inter-study heterogeneity (P=0.041) while other covariates were not (Table 3). Furthermore, as shown in Table 3, subgroup analyses revealed that the OR was 5.69 (95% CI=0.42–76.14) for the publication year ≤2005 and 4.71 (95% CI=1.30–17.07) for >2005 under the random-effects model. For geographical location, the OR was 7.85 (95% CI=1.33–46.32) in Asia, 2.31 (95% CI=0.24–22.01) in America, and 6.10 (95% CI=1.89–19.69) in Africa under random-effects model. For test method, the OR for MSP was 4.49 (95% CI=0.97–20.64) under random-effects model and 8.11 (95% CI=2.93–22.40) for other methods under fixed-effects model. In addition, the OR was 15.75 (95% CI=4.05–61.34) for sample size <50 in fixed-effects model and 2.21 (95% CI=1.33–3.67) for that ≥50 in random-effects model.
  | Table 3 Meta-regression and subgroup analyses of P16INK4a methylation in comparison of cancer tissues vs normal tissues Abbreviations: MSP, methylation-specific PCR; OR, odds ratio. |
Association between P16INK4a promoter methylation and clinicopathological features in patients with ovarian cancer
Ten studies comprising 680 samples were enrolled to assess whether or not the abnormal P16INK4a promoter methylation was associated with ovarian cancer clinicopathological characteristics. As displayed in Figure 4, no statistically significant correlation was found between P16INK4a promoter methylation and age of patients (≥60 vs<60: OR =1.39, 95% CI=0.66–2.92), clinical stage (III–IV vs I–II: OR =1.21, 95% CI=0.81–1.82), grade (3 vs 1–2: OR=1.20, 95% CI=0.82–1.1.75) as well as histological subtype (serous vs non-serous: OR=1.09, 95% CI=0.76–1.55).
Prognostic value of P16INK4a promoter methylation in patients with ovarian cancer
Only two studies42,43 containing 464 patients evaluated the P16INK4a promoter methylation on progression-free survival (PFS) and three studies20,42,43 containing 600 patients on overall survival (OS). The combined results revealed P16INK4a promoter methylation was significantly associated with a poor PFS by univariate Cox proportional hazards regression model (HR=1.68, 95% CI=1.26–2.24; Figure 5A). After considering potential confounders by adjusting for age at diagnosis or surgery, disease stage, histological grade, and residual tumor size, the pooled HR was 1.55 (1.15–2.08; Figure 5B). Survival analysis also showed that P16INK4a promoter methylation reduced OS in univariate and multivariate Cox regression models (HR =1.28, 95% CI=0.97–1.68; HR =1.16, 95% CI=0.87–1.55, respectively; Figure 5C and D), but the differences were not statistically significant.
Sensitivity analysis and publication bias
As presented in Figure 6A–C, no single study significantly affected the pooled ORs in the sensitivity analysis, indicating our analysis was relatively stable and credible. Funnel plots and Begg’s test were used to evaluate the publication bias. The funnel plots were largely symmetric suggesting there were no publication biases in the meta-analysis of P16INK4a promoter methylation and ovarian cancer risk, which was confirmed by the Begg’s test (Figure 6D–F).
Methylation level of P16INK4a measured by TCGA program
To further explore the methylation level of P16INK4a in ovarian tumor tissues, we extracted DNA methylation data of P16INK4a CpG sites measured with Illumina HumanMethylation27 BeadChip from TCGA program. As shown in Table 4, the beta values of 582 ovarian tumor tissues and 12 normal ovarian tissues were extracted for analysis. Obviously, the methylation levels of seven out of nine CpG sites were significantly increased in the ovarian tumor tissues compared with the normal tissues (cg03079681, cg07752420, cg09099744, cg10895543, cg11653709, cg12840719, and cg26673943). Among these regions, methylation level of probe cg26673943 region (located at the promoter region of P16INK4a) was negatively associated with P16INK4a expression in ovarian cancer patients (adjusted P-value <0.000001). However, methylation levels of the rest six probes, which are located at non-promoter region tended to be positively associated with P16INK4a gene expression. Additionally, we found that methylation level of probe cg13479669 region was lower in tumor tissues compared with normal tissues, and negatively associated with P16INK4a gene expression in tumor tissues. These results suggest that hypermethylation of P16INK4a might be correlated with ovarian carcinogenesis and development. Nevertheless, it seems that the methylation at promoter region or non-promoter region has contrary effects on P16INK4a gene expression.
Discussion
Ovarian cancer is one of the leading causes of cancer-related deaths in women.50 Identification of early disease indicators for diagnosis and prognosis is of clinical value. P16INK4a, which resembles classic TSGs such as P53, is an important negative regulator of cell growth and proliferation.16 It has been synthetically evaluated for aberrant P16INK4a methylation in numerous cancers,51–54 including ovarian cancer.31,32 Considering the conflicting conclusions in two meta-analyses and the lack of comprehensive assessment on the role of methylated P16INK4a in ovarian cancer, we performed an adaptive synthesized analysis to investigate the relationships between P16INK4a promoter methylation and ovarian cancer risk, as well as clinicopathological features and prognostic value in ovarian cancer. Meanwhile, we searched TCGA data to validate our meta-analysis.
Our meta-analysis demonstrated that P16INK4a promoter methylation in cancer tissues was significantly higher than that in normal tissues (P<0.05), but not much increased than that in benign tissues. Compared with normal tissues, the frequency of P16INK4a promoter methylation was 2.28-fold higher in both benign tissues and LMP tissues (P>0.05), but the differences were not statistically significant. The reason for this phenomenon may be that the transformation of normal cells to cancer cells is a long-term, gradual, and multiphase process.55 Although not establishing a strong correlation between P16INK4a promoter methylation and cancer progression, the above results do suggest a possibility that epigenetic alteration of P16INK4a promoter methylation might play a certain role in ovarian carcinogenesis and might be useful in distinguishing malignant tumor from healthy ovarian tissues. Considering the evident heterogeneity, we conducted subgroup analyses based on probable covariates in the comparison of cancer tissues vs normal tissues. For geographical location, P16INK4a promoter methylation is a risk factor in Asia and Africa, but not in America. The divergence may be underscored in a large part to a combination of differences in allele frequencies and complex epistasis or gene–environment interactions.56 A review also outlined that some factors such as distinct physical appearance, behavior, and response to environmental agents and drugs between human populations could have contributed to the epigenetic variations.57 Similar findings appeared in the subgroup analyses of different methods and publication year. Kurdyukov and Bullock58 suggested that it was essential to choose an appropriate method in a suitable region to answer a particular biological question in studies of DNA methylation. Additionally, the 95% CI was large in the group of small sample size while relatively small in the group of large sample size, implying the conclusion may not be reliable unless studies should be conducted using a sufficient number of samples. Previous studies also demonstrated that the methylation status in blood samples or fluids might be different from that in tissues.59,60 Thus, our results should be interpreted with caution because sample types were limited to tissues in studies included in this meta-analysis.
Previous studies indicated that P16INK4a promoter methylation was associated with poorly differentiated tumors and was different in histological subtype in ovarian cancer.22,43 However, we could not establish any significant correlations between P16INK4a promoter methylation and clinicopathological features, including age, clinical stage, tumor differentiation or histological subtype in this study. Therefore, it might not be essential to predict the invasion and metastasis of ovarian cancer.
Katsaros et al43 and Wiley et al42 reported association of P16INK4a promoter methylation with PFS and OS in ovarian cancer, while Bhagat et al20 found no significant value in predicting prognosis. In the present study, we discovered that P16INK4a promoter methylation represented a risk factor for PFS. For OS, patients with P16INK4a promoter methylation also had a slightly elevated risk, though the differences are not statistically significant. This trend was also observed in other types of cancer.51,54 However, its statistical confirmation requires large studies. The data from TCGA also indicated that methylation level of probe cg26673943 region (located at the promoter region of P16INK4a) in the ovarian tumor tissues was higher than normal ovarian tissues. Increased methylation of CpG island at the promoter region was negatively associated with P16INK4a gene expression, while methylation of CpG islands at non-promoter regions was positively associated with P16INK4a expression.
Compared with previous meta-analyses,31,32 our meta-analysis had several improvements. First, the development of ovarian cancer is a multistep procedure involving normal tissues, benign disease, LMP or borderline tumor, and malignant tumor.20 We compared malignant ovarian cancer with LMP tumors, benign disease, and normal samples to give more rigorously to the analysis. Second, with 1,217 malignant ovarian cancer patients, 116 LMP, 271 benign patients, and 351 normal samples, the sample size in our study is much larger than that of all previous meta-analyses. Finally, we included the clinicopathological features and prognostic significance of P16INK4a promoter methylation in ovarian cancer for more comprehensive understanding of the underlying pathogenesis of ovarian cancer. These strengths make our study a useful effort in seeking better understanding of the P16INK4a promoter methylation in ovarian cancer.
Limitations
Several potential limitations in our current study should be noted. First, the heterogeneity was still large after subgroup analyses in the assessment of the association between P16INK4a promoter methylation and ovarian cancer risk, which may affect the statistical power. Second, as a retrospective study, a potential unidentified confounding information and selection bias may exist in our meta-analysis. We could not eliminate the possibility of publication bias, where positive results are likely published than negative results. Third, the total sample size was still relatively small for reliably assessing the prognostic value of P16INK4a promoter methylation in ovarian cancer. Fourth, none of the studies included in our meta-analysis defined the region considered as promoter or provided specific methylation sites. Therefore, we are unable to establish whether or not they focused on the same sequence of P16INK4a gene. However, the impact of methylation on transcriptional potential depends on the density of the methylated CpG islands and their location relative to the transcription start site. This highlights the importance of a uniform and full-scale reporting of study designs and outcomes. Additionally, previous researches showed that the occurrence of P16INK4a promoter methylation may depend on the histological subtype.41,48,61 However, we are unable to extract sufficient data to analyze the association between P16INK4a promoter methylation and high-grade serous carcinomas because no detailed information of P16INK4a promoter methylation in high-grade serous carcinomas was provided in the eligible articles.
Although with certain limitations, our study is a comprehensive meta-analysis focusing on the correlation of aberrant P16INK4a promoter methylation with the initiation, development, and prognosis of ovarian cancer to provide a new insight into the pathogenesis of ovarian cancer.
Conclusion
In conclusion, our meta-analysis suggests that aberrant methylation of P16INK4a promoter may be essential to the initiation of ovarian cancer and in distinguishing malignant from healthy ovarian tissues. Besides, P16INK4a promoter methylation is a potential predictive factor for poor prognosis in ovarian cancer. This study indicates the need for multicenter large-scale studies to confirm the role of P16INK4a promoter methylation in ovarian cancer.
Acknowledgments
This study was supported by National Natural Science Funds (numbers 81472033 and 30901308), the National Science Foundation of Hubei Province (numbers 2013CFB233 and 2013CFB235), the Scientific and Technological Project of Wuhan City (number 2014060101010045), Hubei Province Health and Family Planning Scientific Research Project (WJ2015Q021), and Training Program of the Science and Technology Innovation from Zhongnan Hospital of Wuhan University (cxpy20160054). We thank the Beijing Circle & Dot Technology Co., Ltd. for assisting in analyzing the TCGA database.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23(Suppl 10):x111–x117. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Bai H, Cao D, Yang J, Li M, Zhang Z, Shen K. Genetic and epigenetic heterogeneity of epithelial ovarian cancer and the clinical implications for molecular targeted therapy. J Cell Mol Med. 2016;20(4):581–593. | ||
You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22(1):9–20. | ||
Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic silencing of tumor suppressor genes: paradigms, puzzles, and potential. Biochim Biophys Acta. 2016;1865(2):275–288. | ||
Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. | ||
Dong A, Lu Y, Lu B. Genomic/Epigenomic alterations in ovarian carcinoma: translational insight into clinical practice. J Cancer. 2016;7(11):1441–1451. | ||
Koukoura O, Spandidos DA, Daponte A, Sifakis S. DNA methylation profiles in ovarian cancer: implication in diagnosis and therapy (Review). Mol Med Rep. 2014;10(1):3–9. | ||
Gloss BS, Samimi G. Epigenetic biomarkers in epithelial ovarian cancer. Cancer Lett. 2014;342(2):257–263. | ||
Sharpless NE, Depinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. | ||
Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. | ||
Qin Y, Liu JY, Li B, Sun ZL, Sun ZF. Association of low p16INK4a and p15INK4b mRNAs expression with their CpG islands methylation with human hepatocellular carcinogenesis. World J Gastroenterol. 2004;10(9):1276–1280. | ||
Kim BN, Yamamoto H, Ikeda K, et al. Methylation and expression of p16INK4 tumor suppressor gene in primary colorectal cancer tissues. Int J Oncol. 2005;26(5):1217–1226. | ||
Gao SJ, Zhang GF, Zhang RP. High CpG island methylation of p16 gene and loss of p16 protein expression associate with the development and progression of tetralogy of Fallot. J Genet. 2016;95(4):831–837. | ||
Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576(1–2):22–38. | ||
Moselhy SS, Kumosani TA, Kamal IH, Jalal JA, Jabaar HS, Dalol A. Hypermethylation of P15, P16, and E-cadherin genes in ovarian cancer. Toxicol Ind Health. 2015;31(10):924–930. | ||
di Vinci A, Perdelli L, Banelli B, et al. p16(INK4a) promoter methylation and protein expression in breast fibroadenoma and carcinoma. Int J Cancer. 2005;114(3):414–421. | ||
Shima K, Nosho K, Baba Y, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: cohort study and literature review. Int J Cancer. 2011;128(5):1080–1094. | ||
Bhagat R, Kumar SS, Vaderhobli S, et al. Epigenetic alteration of p16 and retinoic acid receptor beta genes in the development of epithelial ovarian carcinoma. Tumour Biol. 2014;35(9):9069–9078. | ||
Bammidi LS, Neerukonda GN, Murthy S, Kanapuram RD. p16 gene alterations in human ovarian cancers: comparison between tissue and blood samples. Int J Gynecol Cancer. 2012;22(4):553–560. | ||
Abou-Zeid AA, Azzam AZ, Kamel NA. Methylation status of the gene promoter of cyclin-dependent kinase inhibitor 2A (CDKN2A) in ovarian cancer. Scand J Clin Lab Invest. 2011;71(7):542–547. | ||
Dhillon VS, Aslam M, Husain SA. The contribution of genetic and epigenetic changes in granulosa cell tumors of ovarian origin. Clin Cancer Res. 2004;10(16):5537–5545. | ||
Ozdemir F, Altinisik J, Karateke A, Coksuer H, Buyru N. Methylation of tumor suppressor genes in ovarian cancer. Exp Ther Med. 2012;4(6):1092–1096. | ||
Shen WJ, Dai DQ, Guo KJ, Xm L. Promoter hypermethylation of RASSF1A, BRCA1 and p16 gene in epithelial ovarian cancer and its clinical significance. Chin J Cancer Prev Treat. 2008;15(7):530–533. | ||
Tam KF, Liu VW, Liu SS, et al. Methylation profile in benign, borderline and malignant ovarian tumors. J Cancer Res Clin Oncol. 2007;133(5):331–341. | ||
Li M, Huang ZJ, Dong WH, et al. Disfigurement of p16INK4A gene expression in development of ovarian cancer and the mechanism. Zhonghua Fu Chan Ke Za Zhi. 2006;41(6):408–412. | ||
Liu Z, Wang LE, Wang L, et al. Methylation and messenger RNA expression of p15INK4b but not p16INK4a are independent risk factors for ovarian cancer. Clin Cancer Res. 2005;11(13):4968–4976. | ||
Makarla PB, Saboorian MH, Ashfaq R, et al. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin Cancer Res. 2005;11(15):5365–5369. | ||
Rathi A, Virmani AK, Schorge JO, et al. Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin Cancer Res. 2002;8(11):3324–3331. | ||
Xiao X, Cai F, Niu X, Shi H, Zhong Y. Association between P16INK4a promoter methylation and ovarian cancer: a meta-analysis of 12 published studies. PLoS One. 2016;11(9):e0163257. | ||
Jiang Y, Yan F, Liang L, Wan Y, Liu J, Cheng W. Meta-analysis demonstrates no association between P16 INK4A promoter methylation and epithelial ovarian cancer. Arch Gynecol Obstet. 2017;295(3):697–704. | ||
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. | ||
Bradburn MJ, Deeks JJ, Berlin JA, Russell Localio A. Much ado about nothing: a comparison of the performance of meta-analytical methods with rare events. Stat Med. 2007;26(1):53–77. | ||
National Institutes of Health; National Cancer Institute; National Human Genome Resarch Institute. The Cancer Genome Atlas. Available from: https://cancergenome.nih.gov/. Accessed August 19, 2018. | ||
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. | ||
Ho CM, Huang CJ, Huang CY, Wu YY, Chang SF, Cheng WF. Promoter methylation status of HIN-1 associated with outcomes of ovarian clear cell adenocarcinoma. Mol Cancer. 2012;11:53. | ||
Cul’bová M, Lasabova Z, Stanclova A, et al. Metylácia vybraných tumor-supresorických génov v benígnych a malígnych ovariálnych nádoroch [Methylation of selected tumor-suppressor genes in benign and malignant ovarian tumors]. Ceska /Gynekol. 2011;76(4):274–279. Czech. | ||
Gu XH, Lu Y, Ma D, Liu XS, Guo SW. Model of aberrant DNA methylation patterns and its applications in epithelial ovarian cancer. Zhonghua Fu Chan Ke Za Zhi. 2009;44(10):754–759. | ||
Wu Q, Lothe RA, Ahlquist T, et al. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. | ||
Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer. 2006;6:212. | ||
Wiley A, Katsaros D, Chen H, et al. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107(2):299–308. | ||
Katsaros D, Cho W, Singal R, et al. Methylation of tumor suppressor gene p16 and prognosis of epithelial ovarian cancer. Gynecol Oncol. 2004;94(3):685–692. | ||
Hashiguchi Y, Tsuda H, Yamamoto K, Inoue T, Ishiko O, Ogita S. Combined analysis of p53 and RB pathways in epithelial ovarian cancer. Hum Pathol. 2001;32(9):988–996. | ||
Brown I, Milner B, Rooney P, Haites N. Inactivation of the p16INK4A gene by methylation is not a frequent event in sporadic ovarian carcinoma. Oncol Rep. 2001;8(6):1359–1362. | ||
Strathdee G, Appleton K, Illand M, et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158(3):1121–1127. | ||
Mccluskey LL, Chen C, Delgadillo E, Felix JC, Muderspach LI, Dubeau L. Differences in p16 gene methylation and expression in benign and malignant ovarian tumors. Gynecol Oncol. 1999;72(1):87–92. | ||
Milde-Langosch K, Ocon E, Becker G, Löning T. p16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hyper-methylation or mutation in endometrioid and mucinous tumors. Int J Cancer. 1998;79(1):61–65. | ||
Shih YC, Kerr J, Liu J, et al. Rare mutations and no hypermethylation at the CDKN2A locus in epithelial ovarian tumours. Int J Cancer. 1997;70(5):508–511. | ||
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. | ||
Xing X, Cai W, Shi H, et al. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. Br J Cancer. 2013;108(12):2542–2548. | ||
Li J, Zhou C, Zhou H, et al. The association between methylated CDKN2A and cervical carcinogenesis, and its diagnostic value in cervical cancer: a meta-analysis. Ther Clin Risk Manag. 2016;12:1249–1260. | ||
Wang X, Zhu YB, Cui HP, Yu TT, Tt Y. Aberrant promoter methylation of p15 (INK4b) and p16 (INK4a) genes may contribute to the pathogenesis of multiple myeloma: a meta-analysis. Tumour Biol. 2014;35(9):9035–9043. | ||
Tang B, Li Y, Qi G, et al. Clinicopathological significance of CDKN2A promoter hypermethylation frequency with pancreatic cancer. Sci Rep. 2015;5:13563. | ||
Hogan C. Impact of interactions between normal and transformed epithelial cells and the relevance to cancer. Cell Mol Life Sci. 2012;69(2):203–213. | ||
Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13(2):R8. | ||
Kader F, Ghai M. DNA methylation-based variation between human populations. Mol Genet Genomics. 2017;292(1):5–35. | ||
Kurdyukov S, Bullock M. DNA methylation analysis: choosing the right method. Biology. 2016;5(1):3. | ||
Chang H, Yi B, Li L, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol. 2008;85(2):96–100. | ||
Zhu W, Qin W, Hewett JE, Sauter ER. Quantitative evaluation of DNA hypermethylation in malignant and benign breast tissue and fluids. Int J Cancer. 2010;126(2):474–482. | ||
Niederacher D, Yan HY, An HX, Bender HG, Beckmann MW. CDKN2A gene inactivation in epithelial sporadic ovarian cancer. Br J Cancer.1999;80(12):1920–1926. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.