Back to Journals » Drug Design, Development and Therapy » Volume 18
Qifu Yixin Formula Improves Heart Failure by Enhancing β-Arrestin2 Mediated the SUMOylation of SERCA2a
Authors Wang X , Yang J, Lu C, Hu Y , Xu Z, Wan Q, Zhang M, Shi T, Liu Z, Liu Y
Received 13 November 2023
Accepted for publication 8 March 2024
Published 11 March 2024 Volume 2024:18 Pages 781—799
DOI https://doi.org/10.2147/DDDT.S446324
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Xinting Wang,1,* Jiahui Yang,1,* Cheng Lu,2 Yinqin Hu,1 Zhaohui Xu,1 Qiqi Wan,1 Meng Zhang,1 Tianyun Shi,1 Zhirui Liu,1 Yongming Liu1
1Department of Cardiology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 200021, People’s Republic of China; 2Department of Cardiology, Seventh People’s Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 200137, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongming Liu, Department of Cardiology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, No. 185, Pu’an Road, Huangpu District, Shanghai, 200021, People’s Republic of China, Email [email protected]
Purpose: This study aimed to elucidate the protective mechanism of Traditional Chinese Medicine (TCM) Qifu Yixin formula (QFYXF) to improve heart failure (HF) by promoting β-arrestin2 (β-arr2)-mediated SERCA2a SUMOylation.
Materials and Methods: The transverse aortic constriction (TAC)-induced HF mice were treated with QFYXF or carvedilol for 8 weeks. β-arr2-KO mice and their littermate wild-type (WT) mice were used as controls. Neonatal rat cardiomyocytes (NRCMs) were used in vitro. Cardiac function was evaluated by echocardiography and serum NT-proBNP. Myocardial hypertrophy and myocardial fibrosis were assessed by histological staining. β-arr2, SERCA2a, SUMO1, PLB and p-PLB expressions were detected by Western blotting, immunofluorescence and immunohistochemistry. SERCA2a SUMOylation was detected by Co-IP. The molecular docking method was used to predict the binding ability of the main active components of QFYXF to β-arr2, SERCA2a, and SUMO1, and the binding degree of SERCA2a to SUMO1 protein.
Results: The HF model was constructed 8 weeks after TAC. QFYXF ameliorated cardiac function, inhibiting myocardial hypertrophy and fibrosis. QFYXF promoted SERCA2a expression and SERCA2a SUMOylation. Further investigation showed that QFYXF promoted β-arr2 expression, whereas Barbadin (β-arr2 inhibitor) or β-arr2-KO reduced SERCA2a SUMOylation and attenuated the protective effect of QFYXF improved HF. Molecular docking showed that the main active components of QFYXF had good binding activities with β-arr2, SERCA2a, and SUMO1, and SERCA2a had a high binding degree with SUMO1 protein.
Conclusion: QFYXF improves HF by promoting β-arr2 mediated SERCA2a SUMOylation and increasing SERCA2a expression.
Keywords: heart failure, Qifu Yixin Formula, β-arrestin2, SERCA2a, SUMOylation
Introduction
The 2022 AHA/ACC/HFSA definition heart failure (HF) is a complex clinical syndrome with symptoms and signs that result from any structural or functional impairment of ventricular filling or ejection of blood.1 There are more than 64.3 million patients with HF worldwide,2 and the mortality rate of HF patients is as high as 50% within 5 years. HF has become the leading global public health problem due to its high incidence, high rate of rehospitalization, high mortality, poor quality of life, and high-cost treatment.3
Reduced expression and activity of sarco/endoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) is one of the hallmarks and key pathological mechanisms of HF.4 Sarcoplasmic reticulum is an organelle that stores Ca2+ and mediates the release and re-uptake of Ca2+ during contraction and relaxation. SERCA2a mediates the re-entry of Ca2+ in the cytoplasm into sarcoplasmic reticulum, regulating cardiac systolic and diastolic function. Gene therapy that increases the expression and activity of SERCA2a in the heart has been proven to be an effective strategy for treating HF. Although targeting SERCA2a has become a research hotspot in the treatment of HF, the efficacy of SERCA2a gene therapy in clinical trials of HF is not satisfactory.5 Therefore, it is of great clinical significance to find effective drugs to improve SERCA2a expression and activity to treat HF and to clarify the upstream regulatory mechanisms of SERCA2a.
SERCA2a expression is also regulated by post-translational modifications (PTMs). Small ubiquitin-like modifier (SUMO)ylation is a ubiquitin-like reversible PTM. Research studies have shown that the SUMO1 (not SUMO2/3) and SUMOylation of SERCA2a in HF patients are significantly reduced, thereby reducing SERCA2a expression.6 Adenovirus mediated overexpression of SUMO1 increased the expression of SERCA protein in the heart of transverse aortic constriction (TAC) induced HF mice and improved cardiac function. On the other hand, shRNA knockdown of SUMO1 expression leads to a decrease in SERCA expression and deterioration of cardiac function in HF mice. SUMOylation is a critical PTM that regulates SERCA2a function in HF.
β-arr2 overexpression in post-myocardial infarction HF mice can improve cardiac function, inhibit apoptosis, inflammation, and cardiac remodeling. Further research found that the interaction between β-arrestin2 (β-arr2) and SERCA2a increases the SUMOylation modification and expression of SERCA2a, thereby enhancing cardiac systolic function.7
SERCA2a activity is reversibly regulated by phospholamban (PLB/PLN). Non-phosphorylated PLB is an inhibitor of SERCA, while phosphorylated PLB can relieve the inhibition of SERCA2a and restore SERCA2a activity.8
Traditional Chinese medicine (TCM) has a long history of preventing and treating HF with significant therapeutic effects. Qifu Yixin formula (QFYXF) is composed of astragali radix (Huangqi), aconiti lateralis radix praeparata (Fuzi), ophiopogonis radix (Maidong) and curcumae rhizoma (Ezhu) (Table 1). QFYXF is a TCM with the main effect of invigorating Qi and warm Yang on improving HF. Clinical research shows that QFYXF significantly improves cardiac function, TCM syndromes, exercise tolerance and quality of life in patients with HF.9 Animal experiments showed that QFYXF inhibits TAC-induced myocardial fibrosis and ameliorated HF by down-regulating the expression of TGF-β1/Smad3 pathway.10 QFYXF is effective in the treatment of HF. However, the pharmacological mechanism by which QFYXF regulates SERCA expression to improve HF remains unclear.
 |
Table 1 The Components of Qifu Yixin Formula |
In this study, we constructed a TAC-induced HF model and demonstrated that QFYXF could improve HF by enhancing β-arr2 mediated the SUMOylation of SERCA2a. This study further elucidates the protective mechanism of QFYXF on HF and provides a scientific basis for the transformation of QFYXF.
Materials and Methods
Animals and Ethics
Male adult C57BL/6J mice aged 6 weeks were purchased from the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. After an acclimation period of 1 week, the mice were randomly assigned to four groups (n ≥ 10 per group) as follows: sham, TAC-induced HF (Model), TAC-induced HF + QFYXF (QFYXF), and TAC-induced HF + carvedilol (Carvedilol) groups.
β-arr2-KO CRISPR/Cas9 C57BL/6JGpt mice were purchased from GemPharmatech Co., Ltd. β-arr2-KO-tF1 (sequence: 5-GCTGTTGGAGGTTCACGAAATAG-3), β-arr2-KO-tR1 (sequence: 5-CCTCATCATGGAAACAGAGGAAC-3), β-arr2-WT-tF1 (sequence: 5-AGGCTGTGGGCTGTTTATCTCAG-3), β-arr2-WT-tR1 (sequence: 5-CAAGCCAGAGTTCTCAAAG-3). β-arr2-KO C57BL/6J mice raised in Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. All procedures in this study were approved by the Experimental Animal Welfare and Ethics Committee of Shanghai University of Traditional Chinese Medicine (Ethics No. PZSHUTCM2303140005). All animal experiments were conducted following the animal experimental guidelines set by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Animals were housed at 21 ± 1 °C with a 12 h light-dark cycle and provided with standard chow and water.
Drug and Reagent Preparation
QFYXF consists of astragali radix 30g, aconiti lateralis radix praeparata 10g, ophiopogonis radix 10g, and curcumae rhizoma 10g. TCM QFYXF granules were all produced by Tianjiang Pharmaceutical Group (Jiangyin, China) under the supervision of Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (batch number: 2304315), and the standard information of the corresponding nine components of QFYXF is shown in Table 1. QFYXF was prepared as a suspension at a 0.78g/mL concentration. Carvedilol was purchased from Qilu Pharmaceutical Co., LTD. 10mg/tablet. Preparation of the concentration of 0.20mg/mL of Carvedilol suspension liquid, sealed, avoid light in 4°C. Barbadin was purchased from MedChemExpress (product number, HY-119706) with a purity of 98.93%, dissolved in DMSO at a formulated concentration of 10 mM.
UHPLC-Q-Orbitrap High-Resolution Mass Spectrometry (HRMS) Analysis of QFYXF
The QFYXF extract quality was confirmed by qualitative and quantitative analyses of the major bioactive components through chromatography-quadrupole/electrostatic field orbitrap HRMS (UHPLC-Q/Exactive; Thermo Fisher Scientific, San Jose, CA, USA). Separations were performed using the Waters ACQUITY UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm) with gradient elution using methanol (A) and 0.1% formic acid water (B) as mobile phase at a flow rate of 0.3 mL/min. The injection volume of QFYXF decoction was 2 μL. The ultra-high performance liquid series quadrupole/electrostatic field orbital trap mass spectrometer was equipped with an electrospray ion source, and data were analyzed and acquired using Xcalibur 4.1. The ion source was in positive ion mode. The optimized MS parameters included a capillary temperature of 325°C; The flow rate of sheath gas was 45 arb. The auxiliary gas flow rate was 10 arb. The sweep gas flow rate was 0 arb. The spray voltage was 3.5 kV. Transmission voltage 50 V. The temperature of the auxiliary gas heater was 300 °C. Full scan mode: scan range 80–1200 m/z; Maximum injection time (IT): 200 ms; Scanning resolution 70,000 FWHM (m/z/s); Automatic gain control (AGC) target: 1.0e6. Calycosin-7-O-β-D-glucoside ([M + H]+, m/z 447.12857, characteristic component of astragali radix), benzoylmesaconine ([M + H]+, m/z 590.29597, characteristic component of aconiti lateralis radix praeparata), benzoylaconine ([M + H]+, m/z 604.31162, characteristic component of aconiti lateralis radix praeparata), benzoylhypaconine ([M + H]+, m/z 574.30106, characteristic component of aconiti lateralis radix praeparata), mesaconitine ([M + H]+, m/z 632.30654, characteristic component of aconiti lateralis radix praeparata), hypaconitine ([M + H]+, m/z 616.31162, characteristic component of aconiti lateralis radix praeparata), aconitine ([M + H]+, m/z 646.32219, characteristic component of aconiti lateralis radix praeparata), astragaloside IV ([M + H]+, m/z 785.46818, characteristic component of astragali radix) and ruscogenin ([M + H]+, m/z 431.31558, characteristic components of ophiopogonis Radix) were analyzed quantitatively and qualitatively.
Neonatal Rat Cardiomyocytes
Extract neonatal rat cardiomyocytes (NRCMs) from Wistar rats within 24 hours of birth. The hearts of Wistar neonatal rats were collected, finely crushed, and digested in digestive solution for 16h. The supernatant was discarded, and ADS was added to mix. The cardiomyocytes were purified by gradient centrifugation using Percoll concentration gradient. Preheated 37 °C medium containing 10% serum was added, cells were counted, and plates were laid, and cultured in a cell incubator for 24h before experiments were performed.
Transverse Aortic Constriction
The HF model was established using TAC. Mice were anesthetized via the intraperitoneal injection of 3% pentobarbital sodium (3mL/kg). After hair clipping, the mice were placed in the supine position. The surgical areas of the mice were disinfected with iodine fluoride. The anterior midline skin of the sternum was incised longitudinally, and the first and second ribs were incised along the left border of the sternum to strip the thymus and expose the aortic arch. TAC was created by taping around the aortic arch with a 7–0 silk suture and placing a pad between the silk suture and the aortic arch with a 27G blunt needle. After confirming ligation, the needle was withdrawn and the mediastinum was closed layer by layer. The mice were placed in a holding box after surgery and kept normally until they were awake. The sham group underwent the same surgical procedure without aortic arch ligation.
Echocardiography
Echocardiography was performed using a high-resolution ultrasound imaging system (VEVO 2100, VisualSonics, America) with a probe frequency of 30 MHz. Mouse prethoracic hair was depilated with depilatory cream 1 d before examination. M-mode recordings were taken from the parasternal left ventricular long axis slices. Left ventricular ejection fraction (LVEF), fractional shortening (FS), left ventricular internal diameter at end-diastole (LVIDd) and left ventricular mass (LV Mass) were measured.
Enzyme-Linked Immunosorbent Assay (ELISA) Analysis
Collect mouse blood samples and separate serum after high-speed centrifugation. According to the manufacturer’s instructions, the serum levels of N-terminal pro-B-type natriuretic peptide (NT-proBNP) were detected using the corresponding ELISA kits.
Histology, Immunohistochemistry and Immunofluorescence Staining
The heart tissue samples were washed with PBS at 4°C, fixed with 4% paraformaldehyde for 48h, and embedded in dehydrated paraffin. Next, 5μm-thick sections were cut for hematoxylin-eosin (HE) staining to observe the changes in heart size, and Masson trichrome staining and Sirius Red (SR) were performed to observe cardiac fibrosis. SERCA2a and β-arr2 were detected by immunohistochemistry staining. β-arr2, SERCA2a and SUMO1 were detected by immunofluorescence staining. After staining, all sections were scanned under an Olympus microscope. Collagen deposition based on Masson trichrome staining and SR staining was analyzed using ImageJ software.
Western Blotting
Western blotting assay was performed as previously described.11 The primary antibodies used were a glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phospholamban (PLB) rabbit mAb (Cell Signaling Technology, America), Phospho-Phospholamban (Ser16/Thr17) Antibody (Cell Signaling Technology, America), ATP2A2/SERCA2 rabbit mAb (Cell Signaling Technology, America), β-arrestin2 antibody (Santa Cruz, America) and SUMO-1 (C9H1) Rabbit mAb (Cell Signaling Technology, America).
Co-Immunoprecipitation
After the heart and cells were treated, the lysate was lysed according to the experimental steps of the Co-immunoprecipitation (Co-IP) kit, centrifuged, and quantified by BCA. The antibodies against target proteins SERCA2a and SUMO1 were added, and non-specific immune homologous antibodies were used as controls. 20uLProteinA/G was added and centrifuged after 3h at 4°C at 30 rpm/h to retain the precipitate. Use wash buffer cleaning precipitation, centrifugal, repeat 3 times. The precipitate was resuspended by adding SDS buffer, vortexed and shaken at 95°C for 5min, centrifuged, and detected by Western blotting.
Cell Counting Kit 8
NRCM was treated with 0(blank control),40, 80, 120, 160, 200, 400, and 500 μg/mLQFYXF for 24h, and Cell Counting Kit 8 (CCK8) was used to detect the absorbance value at 450nm and calculate the cell viability.
Molecular Docking Method
Using β-arrestin2, SERCA2a and SUMO1 as keywords, the protein structure was retrieved from UniProt, and the species of mouse were screened in the database. ARRB2_MOUSE (UniProt ID: Q91YI4), AT2A2_MOUSE (UniProt ID: O55143) and SUMO1_MOUSE (UniProt ID: P63166) full-length AlphaFold as target protein structure prediction structure file were selected. The SDF structure files of small-molecule ligand compounds were downloaded from PubChem database, input Chem 3D software to adjust them to 3D structure and optimize Minimize energy, and output as.pdb format files. The small-molecule compound structure and protein structure files were imported into AutoDock Tools 1.5.7 software for hydrogenization, calculation of charge number and other processing, and the files were converted into pdbqt format. AutoDock Vina 1.1.2 was used for molecular docking, and Pymol software was used for visualization. Binding energies of less than −4.25 kcal/mol, −5.0 kcal/mol, or −7.0 kcal/mol indicate certain, good, or strong-binding activity between the ligand and the receptor, respectively.
Using SERCA2a and SUMO1 as keywords, the protein structure was retrieved from UniProt, and the species of mouse were screened in the database. AT2A2_MOUSE (UniProt ID: O55143) and SUMO1_MOUSE (UniProt ID: P63166) full-length AlphaFold as target protein structure prediction structure file were selected. Protein–protein docking is based on the 3D structures of two known proteins and the near-native structure of the complex is predicted by molecular simulation methods. Docking is a process of sampling and scoring. HDOCK is a method to develop efficient molecular docking algorithms and accurate scoring functions for biomolecular interactions by combining physics-based and bioinformatic approaches. Appropriate docking parameters were set and protein–protein docking was performed using HDOCK. The results included two indicators: one was the Docking Score, with more negative docking scores indicating more likely binding models. Second, the confidence score, when the confidence score is higher than 0.7, the possibility of two molecules to combine is high.
Statistical Analysis
SPSS24.0 statistical software was used for statistical analysis, and GraphPad 9 and Photoshop software were used for figure production. Data were in accordance with normal distribution and homogeneity of variance and were expressed as mean ± standard error (SEM). The one-way ANOVA test was used to compare multiple groups, and the LSD test was used to compare groups. A non-parametric test was used to compare the data that did not meet the normal distribution or variance, and the Kruskal–Wallis test was used to compare the data between multiple groups. Statistical significance was set at P < 0.05.
Results
The Fingerprint Chromatograms of QFYXF
The QFYXF extract quality was confirmed by qualitative and quantitative analysis of the major bioactive components through chromatography-quadrupole/electrostatic field orbitrap HRMS. The contents of calycosin-7-O-β-D-glucoside, benzoylmesaconine, benzoylaconine, benzoylhypaconine, astragaloside IV, and ruscogenin in QFYXF were 6.92μg/mL, 2.95μg/mL, 0.91μg/mL, 2.97μg/mL, 0.92μg/mL, 0.0091μg/mL, respectively. The contents of mesaconitine, hypaconitine and aconitine were below the detection limit (Figure 1).
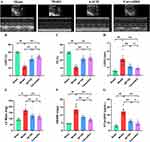
QFYXF Protects the Cardiac Function of TAC-Induced HF Mice
To evaluate the effect of QFYXF on cardiac function in HF mice, we successfully constructed an HF model induced 8 weeks after TAC surgery. First, we detected the LVEF, FS, LVIDd and LV Mass of mice in each group by echocardiography (Figure 2A–E), compared with sham group, LVEF and FS of model group mice were significantly decreased, and LVIDd and LV Mass were significantly increased. Compared with the model group, QFYXF and carvedilol groups significantly increased LVEF, FS, decreased LVIDd and LV Mass, and there was no significant difference between QFYXF and carvedilol groups. Secondly, heart weight/body weight (HW/BW) was used to evaluate heart function, and the higher the HW/BW ratio, the worse the heart function (Figure 2F), HW/BW of mice in Model group was significantly increased compared with Sham group. Compared with Model group, HW/BW of mice in QFYXF and carvedilol groups was significantly decreased, and there was no significant difference between QFYXF and carvedilol groups. Finally, the serum HF biomarker NT-proBNP was detected by Elisa (Figure 2G), compared with Sham group, serum NT-proBNP in Model group was significantly increased. Compared with Model group, serum NT-proBNP in QFYXF and carvedilol groups was significantly decreased, and there was no significant difference between QFYXF and carvedilol groups. The results of echocardiography, HW/BW and NT-proBNP showed that both QFYXF and carvedilol could significantly improve the cardiac function of mice with HF induced by pressure overload.
QFYXF Inhibits Cardiac Hypertrophy and Myocardial Fibrosis Induced in HF Mice
In order to study whether QFYXF can inhibit the cardiac remodeling of HF mice, the hearts of mice in each group were stained by HE, Masson and SR. 1mm scale HE staining showed that compared with the sham group, the model group had obvious myocardial hypertrophy and increased cardiac cross-sectional area. Compared with the model group, QFYXF and carvedilol significantly inhibited pressure overload-induced cardiac hypertrophy and reduced cardiac cross-sectional area. HE staining at 20μm scale showed that compared with Sham group, myocardial cells in Model group were disordered and interstitial was loose. Compared with Model group, cardiomyocytes in QFYXF and carvedilol groups were arranged orderly and interstitial was compact (Figure 3A and B). To further investigate whether QFYXF can inhibit myocardial fibrosis in HF mice, Masson and SR staining were performed. Masson staining is shown in Figure 3C and D. Compared with Sham group, the blue dye region of cardiac fibrosis in Model group was enlarged, and the collagen volume fraction was significantly increased. Compared with Model group, QFYXF and carvedilol group reduced the blue dye region of cardiac fibrosis and significantly decreased the collagen volume fraction. SR staining is shown in Figure 3E and 3F. Compared with Sham group, the red dye area of cardiac fibrosis in Model group was increased, and the percentage of fiber area was significantly increased. Compared with Model group, the red dye area of cardiac fibrosis in QFYXF and carvedilol groups was reduced, and the percentage of fiber area was significantly reduced. These results indicate that QFYXF can inhibit cardiac hypertrophy and myocardial fibrosis induced by pressure overload in HF mice and improve cardiac remodeling, with the same effect as carvedilol.
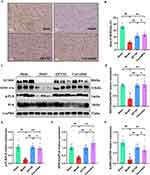
QFYXF Enhanced the SUMOylation of SERCA2a in Failing Myocardium
In order to investigate whether QFYXF can affect SERCA2a activity and SUMOylation in HF mice, SERCA2a was detected by immunohistochemistry and Western-Blot. Compared with Sham group, SERCA2a protein abundance was significantly decreased in Model group. Compared with Model group, QFYXF and carvedilol significantly increased SERCA2a protein abundance in HF mice, and there was no significant difference between QFYXF and carvedilol groups (Figure 4A–D). Secondly, in order to study SERCA2a activity, we further examined the protein expression of PLB and p-PLB. Compared with Sham group, cardiac p-PLB protein expression and SERCA2a/PLB in Model group were significantly decreased. Compared with Model group, QFYXF and carvedilol significantly increased p-PLB protein expression and SERCA2a/PLB in HF mice, and there was no significant difference between QFYXF and carvedilol groups. QFYXF enhanced p-PLB expression in failing myocardium, relieved PLB inhibition on SERCA2a, and increased SERCA2a expression. The effect was similar to carvedilol (Figure 4C–F). Finally, in order to investigate the SUMOylation of SERCA2a, we further examined the protein expression of SUMO1. Compared with Sham group, SUMO1 protein expression in the heart of Model group was significantly decreased. Compared with Model group, QFYXF and carvedilol significantly increased the expression of heart SUMO1 protein in HF mice (Figure 4C–G). Therefore, QFYXF can enhance SUMOylation of SERCA2a in failing myocardium.
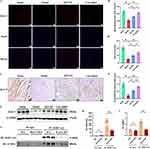
QFYXF Promotes β-arrestin2-Mediated the SUMOylation of SERCA2a in Failing Myocardium
To further investigate whether the SUMOylation of SERCA2a in failing myocardium enhanced by QFYXF is mediated by β-arr2, we tested cardiac β-arr2. Firstly, by Western-Blot, immunofluorescence and immunohistochemistry, it was found that the heart β-arr2 protein abundance of HF mice was significantly decreased. Both QFYXF and carvedilol significantly increased β-arr2 protein abundance in failing myocardium (Figure 5A–F). Based on this, we hypothesize that QFYXF promotes β-arr2-mediated the SUMOylation of SERCA2a in failing myocardium, thereby ameliorating HF. To verify this hypothesis, we performed CO-IP detection of SERCA2a and SUMO1 in the hearts of β-arr2-KO mice (Figure 5G–I). Compared with WT mice, the expression of SERCA2a and SUMO1 protein, SUMOylation indicators of SERCA2a in the heart of β-arr2-KO mice were significantly down-regulated. Therefore, we concluded that β-arr2 can mediate the SUMOylation of SERCA2a, and QFYXF promotes β-arr2-mediated the SUMOylation of SERCA2a in failing myocardium, thereby ameliorating HF.
QFYXF Promotes β-arrestin2-Mediated the SUMOylation of SERCA2a in Hypertrophic Cardiomyocytes
To further investigate the protective effect of QFYXF on HF by enhancing β-arr2 mediated SUMOylation of SERCA2a, primary NRCMs were cultured in vitro. CCK8 assay showed that QFYXF at concentrations between 40 and 500ug/mL for 24h incubation had no toxic effect on NRCMs (Figure 6A). First, the cytoskeleton protein β-actin was used to mark the cross-sectional area of cardiomyocytes by immunofluorescence. NRCMs were incubated with 1μM AngII for 24h to induce hypertrophy, and then treated with 40μg/mL (low dose), 80μg/mL (medium dose), and 120μg/mL (high dose) QFYXF. QFYXF at 40μg/mL, 80μg/mL, and 120μg/mL significantly inhibited AngII-induced NRCMs hypertrophy, and there was no significant difference among the low, medium, and high QFYXF concentrations (Figure 6B and C). We used a medium dose of 80μg/mL QFYXF for subsequent studies. As shown in Figure 6D–I, NRCMs treated with QFYXF tended to increase β-arr2 compared with the blank control group. Compared with the blank control group, AngII significantly reduced the expression of β-arr2, SERCA2a, SUMO1, p-PLB and SERCA2a/PLB. Compared with AngII intervention group, QFYXF treatment significantly enhanced the expression of β-arr2, SERCA2a, SUMO1, p-PLB, and SERCA2a/PLB in hypertrophic NRCMs. To this end, we adopted Co-IP further testing, compared with blank control group, Ang II intervention to reduce SERCA2a SUMOylation. Addition of QFYXF restored the SUMOylation of SERCA2a (Figure 6J–O). Consistent with the results in vivo, on the one hand, QFYXF can enhance p-PLB expression in hypertrophic NRCMs, remove PLB inhibition of SERCA2a, improve SERCA2a expression. On the other hand, QFYXF could enhance the expression of β-arr2, SERCA2a, and SUMO1 in hypertrophic NRCMs.
Barbadin, a β-arr2 Inhibitor, Blocked QFYXF-Promoted SERCA2a SUMOylation in Hypertrophic Cardiomyocytes
To further determine whether the promotion of SERCA2a SUMOylation by QFYXF was mediated by β-arr2, Barbadin, a β-arr2 inhibitor, was used for verification. As shown in Figure 7A–D, Barbadin 50μm inhibited β-arr2 expression compared with the blank control, together with the inhibition of SRECA2a and SUMO1 protein expression. Barbadin blocked the effect of QFYXF, which upregulated SERCA2a and SUMO1 expression in AngII-induced hypertrophic NRCMs. In Figure 7E–J, we further observed the fluorescence intensity of the target proteins (β-arr2, SERCA2a, and SUMO1) in NRCMs. AngII increased the morphology of NRCMs and decreased the fluorescence intensity of β-arr2, SERCA2a, and SUMO1. However, QFYXF inhibited the AngII-induced morphological enlargement of NRCMs and enhanced the fluorescence intensity of β-arr2, SERCA2a, and SUMO1. Barbadin inhibited the fluorescence intensity of β-arr2, SERCA2a, and SUMO1 in NRCMs without affecting NRCMs morphology. Barbadin blocked the effect of QFYXF on enhancing the fluorescence intensity of β-arr2, SERCA2a, and SUMO1 in AngII-induced hypertrophic cardiomyocytes, but not the effect of QFYXF on inhibiting NRCMs hypertrophy. Thus, the enhancement of SERCA2a SUMOylation by QFYXF against HF is mediated through upstream β-arr2.
Molecular Docking Method Showed That the Main Active Components of QFYXF Had Good Binding Activity with the Main Targets β-arr2, SERCA2a and SUMO1
The fingerprint chromatograms of QFYXF mainly detected calycosin-7-O-β-D-glucoside and astragaloside IV in astragali radix, benzoylmesaconine in aconiti lateralis radix praeparata, and ruscogenin in ophiopogonis radix. We used the MDM to predict the binding ability of calycosin-7-O-β-D-glucoside, astragaloside IV, benzoylmesaconine and ruscogenin to β-arr2, SERCA2a and SUMO1 proteins, respectively. Binding energies of less than −4.25 kcal/mol, −5.0 kcal/mol, or −7.0 kcal/mol indicate certain, good, or strong binding activity between the ligand and the receptor, respectively. The results showed that calycosin-7-O-β-D-glucoside, astragaloside IV, benzoylmesaconine and ruscogenin all had good binding ability with β-arr2, SERCA2a and SUMO1 proteins (Figure 8A and B). Among them, benzoylmesaconine and ruscogenin have strong binding ability to SERCA2a. In addition, the active ingredient curcumin in curcumae rhizoma was docked with β-arr2, SERCA2a and SUMO1 proteins. The results showed that curcumin had a good binding ability with β-arr2 and SERCA2a, and a certain binding ability with SUMO1 (Figure 8A–B).
On the other hand, we performed protein–protein docking analysis for SERCA2a and SUMO1; a smaller docking score indicates a better binding model, and a confidence score higher than 0.7 indicates a high probability of molecular binding. As shown in Figure 9A and B, the binding model with the best results, model_1 with docking score of −260.62 and confidence score of 0.9014, shows the binding process of the two proteins in the form of Surface as Figure 9C. The binding interface of the protein–protein complex was comprehensively described and systematically analyzed, and the interaction-related details were supplemented by PyMOL to form a 3D interaction diagram as shown in Figure 9D. There are a total of nine pairs of hydrophobic interactions, eight pairs of hydrogen bonds, one pair of salt Bridges, and one pair of π-π stacking between SERCA2a and SUMO1. For example, the PHE-92 amino acid residue of the protein SERCA2a interacts hydrophobic with the THR-10 amino acid residue of the protein SUMO1. In summary, on the one hand, SERCA2a and SUMO1 proteins bind to each other, which was confirmed by CO-IP experiments. On the other hand, the main active ingredients of QFYXF have good binding ability with β-arr2, SERCA2a, and SUMO1 proteins. In this study, QFYXF promoted β-arr2 mediated SUMOylation of SERCA2a in HF treatment.
 |
Figure 9 Protein interaction of SERCA2a and SUMO1. (A) SERCA2a and SUMO1 binding model; (B) Summary of Top 10 Models; (C) binding process of SERCA2a and SUMO1; (D) 3D interaction diagram. |
Discussions
TCM QFYXF improved HF by promoting β-arr2-mediated SERCA2a SUMOylation and enhancing SERCA2a expression. In addition, QFYXF enhanced PLB phosphorylation modification in failing myocardium, relieved PLB inhibition on SERCA2a, increased SERCA2a activity. The pharmacological mechanism of QFYXF is fully demonstrated (Figure 10).
 |
Figure 10 The pharmacological mechanism of QFYXF. |
TAC induced-pressure overload leads to the deterioration of cardiac function and ultimately HF.12 In this study, we successfully constructed an HF model induced by cardiac pressure overload 8 weeks after TAC surgery. β-receptor blocker is a classic drug used to treat HF, and studies have shown that carvedilol can prolong the survival of HF patients compared to metoprolol.13 Carvedilol can improve the severity of HF and reduce the risk of clinical deterioration and hospitalization.14 More importantly, carvedilol can activate the cardiac β-arr2 mediated the SUMOylation of SERCA2a, increase SERCA2a expression, convert into positive inotropic force, promote cardiac contraction, and improve HF.15 Therefore, this study selected carvedilol as the positive control drug for QFYXF.
The decrease of SERCA2a expression and activity has become one of the main pathological mechanisms of HF.4 The experimental study of HF mice model showed that the protein expression level of SERCA2a was significantly reduced.16 Heart specific SERCA2a-KO mice exhibit an HF phenotype.17 The results showed that QFYXF significantly increased SERCA2a expression in TAC-induced HF mice and AngII-induced hypertrophic cardiomyocytes. There are studies on QFYXF components to increase SERCA2a expression in the treatment of heart failure as follows: astragali radix and its active component astragaloside IV,18,19 aconite water-soluble alkaloid extract20 can upregulate SERCA2a expression and effectively prevent and treat HF. Astragaloside IV activates peroxisome proliferator-activated receptors α (PPARα) to stimulate fatty acids β-oxidize and increase cardiac energy production, promote cardiac excitatory-contraction coupling and SERCA2a expression, and improve HF.21
On the one hand, SERCA2a protein level in the heart of β-arr2-KO mice was significantly lower than that of WT mice, and β-arr2 overexpression improved HF after myocardial infarction through SERCA2a-dependent positive inotropy, and gene therapy targeting β-arr2 may be a new strategy for the treatment of HF.7 On the other hand, increasing the SUMOylation of SECA2a can restore the contractile function of failing myocardium, and N106, a small molecule targeting SERCA2a SUMOylation, is effective in the treatment of HF.22 The present study further verified that SERCA2a SUMOylation was significantly reduced in the hearts of β-arr2-KO mice compared with WT mice. TCM QFYXF in this study can promote the β-arr2 mediated SERCA2a SUMOylation and effectively improve HF. However, Barbadin, which inhibited upstream β-arr2 expression, blocked the mechanism by which QFYXF promoted β-arr2-mediated SERCA2a SUMOylation to improve HF.
HF belongs to the categories of “heart water”, “heart obstruction”, and “chest obstruction and heartache” in TCM. The heart dominates the blood vessels and blood circulation. When the heart Yang Qi is sufficient, it promotes the normal operation of the human blood and nourishes various organs. TCM believes that the pathogenesis of HF is heart Qi deficiency and inability to promote blood circulation, which causes blood stasis. Over time, both Yin and Yang of the heart are deficient, accelerating cardiac dysfunction. Therefore, invigorating heart Qi is the basic principle of HF treatment. QFYXF takes astragali radix as the monarch drug, which has the effect of invigorating heart Qi and helping blood circulation; curcumae rhizoma can activate Qi and blood circulation, aconiti lateralis radix praeparata can warm the heart Yang, ophiopogonis radix can nourish the heart Yin, and help Yin to calm the Yang. All three are minister drugs. QFYXF plays the function of “invigorating Qi and activating blood circulation, warming Yang and nourishing Yin”. QFYXF invigorates heart Qi and activates blood circulation, warms the heart Yang Qi and nourishes the heart Yin Qi, promoting the circulation of Qi, blood, and body fluid and improving the progress of HF.
In addition, in terms of the mechanism, Non-phosphorylated PLB formed a PLB-SERCA2a complex with SERCA2a to exert the inhibitory effect, whereas the dissociation of phosphorylated PLB from the PLB-SERCA2a complex abolished the inhibitory effect of PLB on SERCA2a.23 Therefore, dephosphorylated PLB is an inhibitor of SERCA and phosphorylation of PLB relieves this inhibition,24 and the radio of SERCA2a/PLB can represent SERCA2a activity. QFYXF improves HF by increasing PLB phosphorylation, relieving PLB inhibition of SERCA2a, and increasing SERCA2a expression and activity. Astragaloside, an active component of astragali radix in QFYXF, can up regulate the expression of p-PLB and SERCA2a in failing myocardium, relieve the inhibition of PLB on SERCA2a, promote the normalization of myocardial excitation-contraction coupling, and improve HF.25
Appropriate protein SUMOylation modification plays a crucial role in the development and metabolism of the heart.26 β-arresin2 regulates protein SUMOylation to increase protein stability, which is also applicable in the heart.27 Promoting cardiac β-arr2 expression directly increases positive inotropy is by stimulating the SUMOylation of SERCA2a, which in turn increases SERCA2a expression and activity.7 Although targeting β-arresin2 can improve HF by promoting cardiac contractility and inhibiting adverse cardiac remodeling.
TCM QFYXF can significantly increase β-arr2-mediated SUMOylation of SERCA2a in this study and effectively treat HF. However, the study has some limitations. TCM QFYXF improves HF through multi-target and multi-pathway effects. QFYXF has many active components, but this study did not identify the components that promote β-arr2 mediated SERCA2a SUMOylation to improve HF. In addition, the upstream mechanism by which QFYXF promotes β-arr2 expression remains unclear and needs further investigation. In the future, we will conduct research on the above limitations to provide a high-quality, evidence-based medical basis for treating HF with TCM QFYXF.
Conclusions
TCM QFYXF improved HF by promoting β-arr2-mediated SERCA2a SUMOylation and enhancing SERCA2a expression, whereas Barbadin blocked the protective effect of QFYXF on SERCA2a SUMOylation by inhibiting upstream β-arr2 expression. Meanwhile, β-arr2-KO significantly reduced cardiac SERCA2a SUMOylation.
Author Contributions
Xinting Wang and Jiahui Yang are the first authors. Yongming Liu is the corresponding author.
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (grant number 81973611, 81102671), Shanghai Municipal Health Commission (grant number 202340158), and the Shanghai Sailing Program (grant number 20YF1436800).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Heidenreich PA, Bozkurt B, Aguilar D, et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: a Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2022;145(18):e895–e1032. doi:10.1161/CIR.0000000000001063
2. Bragazzi NL, Zhong W, Shu J, et al. Burden of heart failure and underlying causes in 195 countries and territories from 1990 to 2017. Eur J Prev Cardiol. 2021;28(15):1682–1690. doi:10.1093/eurjpc/zwaa147
3. Becher PM, Lund LH, Coats A, Savarese G. An update on global epidemiology in heart failure. Eur Heart J. 2022;43(32):3005–3007. doi:10.1093/eurheartj/ehac248
4. Zhihao L, Jingyu N, Lan L, et al. SERCA2a: a key protein in the Ca(2+) cycle of the heart failure. Heart Fail Rev. 2020;25(3):523–535. doi:10.1007/s10741-019-09873-3
5. Yla-Herttuala S. Gene Therapy for Heart Failure: back to the Bench. Mol Ther. 2015;23(10):1551–1552. doi:10.1038/mt.2015.158
6. Kho C, Lee A, Jeong D, et al. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477(7366):601–605. doi:10.1038/nature10407
7. McCrink KA, Maning J, Vu A, et al. beta-Arrestin2 improves post-myocardial infarction heart failure via Sarco(endo)plasmic Reticulum Ca(2+)-ATPase-dependent positive inotropy in cardiomyocytes. Hypertension. 2017;70(5):972–981. doi:10.1161/HYPERTENSIONAHA.117.09817
8. James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature and site of phospholamban regulation of the Ca2+ pump of sarcoplasmic reticulum. Nature. 1989;342(6245):90–92. doi:10.1038/342090a0
9. Zhaohui X, Yihang Z, Tianyun S, et al. Clinical effect of qifu yixin prescription on chronic heart failure in patients with syndrome of heart qi deficiency. Zhongguo Shi Yan Fang Ji Xue Za Zhi. 2023;29(23):98–105.
10. Xinting W, Cheng L, Lei S, et al. Study on mechanism of AACO prescription for regulating TGF-β1/Smad3 signaling pathway to protect myocardial fibrosis in mice with chronic heart failure. Zhongguo Zhongyiyao Xinxi Zazhi. 2023;30(01):103–108.
11. Wang Q, Liu Y, Fu Q, et al. Inhibiting insulin-mediated beta2-adrenergic receptor activation prevents diabetes-associated cardiac dysfunction. Circulation. 2017;135(1):73–88. doi:10.1161/CIRCULATIONAHA.116.022281
12. Zhuang L, Jia K, Chen C, et al. DYRK1B-STAT3 drives cardiac hypertrophy and heart failure by impairing mitochondrial bioenergetics. Circulation. 2022;145(11):829–846. doi:10.1161/CIRCULATIONAHA.121.055727
13. Poole-Wilson PA, Swedberg K, Cleland JG, et al. Comparison of carvedilol and metoprolol on clinical outcomes in patients with chronic heart failure in the Carvedilol Or Metoprolol European Trial (COMET): randomised controlled trial. Lancet. 2003;362(9377):7–13. doi:10.1016/S0140-6736(03)13800-7
14. Packer M, Fowler MB, Roecker EB, et al. Effect of carvedilol on the morbidity of patients with severe chronic heart failure: results of the carvedilol prospective randomized cumulative survival (COPERNICUS) study. Circulation. 2002;106(17):2194–2199. doi:10.1161/01.CIR.0000035653.72855.BF
15. Maning J, Desimine VL, Pollard CM, Ghandour J, Lymperopoulos A. Carvedilol selectively stimulates betaArrestin2-dependent SERCA2a activity in cardiomyocytes to augment contractility. Int J Mol Sci. 2022;23(19). doi:10.3390/ijms231911315
16. Ye B, Zhou H, Chen Y, et al. USP25 ameliorates pathological cardiac hypertrophy by stabilizing SERCA2a in cardiomyocytes. Circ Res. 2023;132(4):465–480. doi:10.1161/CIRCRESAHA.122.321849
17. Louch WE, Hougen K, Mork HK, et al. Sodium accumulation promotes diastolic dysfunction in end-stage heart failure following Serca2 knockout. J Physiol. 2010;588(Pt 3):465–478. doi:10.1113/jphysiol.2009.183517
18. Xu XL, Ji H, Gu SY, Shao Q, Huang QJ, Cheng YP. Modification of alterations in cardiac function and sarcoplasmic reticulum by astragaloside IV in myocardial injury in vivo. Eur J Pharmacol. 2007;568(1–3):203–212. doi:10.1016/j.ejphar.2007.04.007
19. Su D, Li HY, Yan HR, Liu PF, Zhang L, Cheng JH. Astragalus improved cardiac function of adriamycin-injured rat hearts by upregulation of SERCA2a expression. Am J Chin Med. 2009;37(3):519–529. doi:10.1142/S0192415X09007028
20. Xu X, Xie X, Zhang H, et al. Water-soluble alkaloids extracted from Aconiti Radix lateralis praeparata protect against chronic heart failure in rats via a calcium signaling pathway. Biomed Pharmacother. 2021;135:111184. doi:10.1016/j.biopha.2020.111184
21. Dong Z, Zhao P, Xu M, et al. Astragaloside IV alleviates heart failure via activating PPARalpha to switch glycolysis to fatty acid beta-oxidation. Sci Rep. 2017;7(1):2691. doi:10.1038/s41598-017-02360-5
22. Kho C, Lee A, Jeong D, et al. Small-molecule activation of SERCA2a SUMOylation for the treatment of heart failure. Nat Commun. 2015;6(1):7229. doi:10.1038/ncomms8229
23. MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4(7):566–577. doi:10.1038/nrm1151
24. Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res. 2012;110(12):1646–1660. doi:10.1161/CIRCRESAHA.111.259754
25. Wang Y, Ji Y, Xing Y, Li X, Gao X. Astragalosides rescue both cardiac function and sarcoplasmic reticulum Ca(2)(+) transport in rats with chronic heart failure. Phytother Res. 2012;26(2):231–238. doi:10.1002/ptr.3492
26. Mendler L, Braun T, Muller S. The ubiquitin-like SUMO system and heart function: from development to disease. Circ Res. 2016;118(1):132–144. doi:10.1161/CIRCRESAHA.115.307730
27. Wyatt D, Malik R, Vesecky AC, Marchese A. Small ubiquitin-like modifier modification of arrestin-3 regulates receptor trafficking. J Biol Chem. 2011;286(5):3884–3893. doi:10.1074/jbc.M110.152116
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.