Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Pyroptosis and Inflammasome-Related Genes-NLRP3, NLRC4 and NLRP7 Polymorphisms Were Associated with Risk of Lung Cancer
Authors Jing X, Yun Y, Ji X, Yang E, Li P
Received 4 June 2023
Accepted for publication 11 August 2023
Published 25 August 2023 Volume 2023:16 Pages 795—804
DOI https://doi.org/10.2147/PGPM.S424326
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Xin Jing,* Yuhui Yun,* Xiang Ji, Ende Yang, Pei Li
Department of Thoracic Surgery, Tangdu Hospital, The Fourth Military Medical University, Xi’an, Shaanxi, 710038, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Pei Li; Ende Yang, Department of Thoracic Surgery, Tangdu Hospital, The Fourth Military Medical University, Xi’an, Shaanxi, 710038, People’s Republic of China, Email [email protected]; [email protected]
Background: Cancer development and tumor immune microenvironment remodeling are closely linked to pyroptosis and inflammasome activation. However, little information is available in single nucleotide polymorphisms (SNPs) in pyroptosis and inflammasome-related genes in patients with lung cancer. This study aims to evaluate the associations between pyroptosis-related gene (NLRP3, NLRC4, and NLRP7) polymorphisms and the risk of lung cancer.
Methods: The MassARRAY platform was used to genotype six SNPs of the NLRP3, NLRC4, and NLRP7 genes in 660 lung cancer cases and 660 controls.
Results: Individuals with rs35829419-A, rs385076-C, and rs775882-T alleles exhibited a higher risk of lung cancer (p < 0.01), while rs212704-T appears protective (p = 0.006). The rs35829419-AA, rs385076-TC/CC, and rs775882-CT/TT genotypes were associated with various degrees of elevated risk of lung cancer (p< 0.02), whereas rs212704-TT was associated with a reduced risk of the disease (p=0.014). Genetic models analysis showed that rs35829419, rs385076, and rs775882 was associated with an increased risk of lung cancer, while rs212704 was related to a reduced risk in all three models (p < 0.05). The four SNPs remained significant in smoker and nonsmoker subgroups (p < 0.05). However, rs35829419 was correlated with risk of adenocarcinoma and small cell lung cancer, and rs212704 was only protective for squamous cell carcinoma. The rs385076 and rs775882 were associated with all three pathological types (p < 0.01).
Conclusion: Besides providing candidate markers for identification of high-risk populations and early prevention of the disease, our research also provided new insight into anti-tumor strategies targeting inflammasomes and pyroptosis.
Keywords: lung cancer, single nucleotide polymorphisms, pyroptosis, inflammasome, case–control study
Introduction
Lung cancer is the most common malignant tumor in China, and its incidence and mortality rank first among all tumors.1 Most patients with lung cancer were at an advanced stage by the time they noticed the symptoms of the disease.2 Currently, lung cancer is treated mainly with surgical resection, radiotherapy, and chemotherapy, adjuvant to emerging molecular targeted therapy.3 Although treatment has improved significantly, survival rates for most advanced cancer patients are still difficult to improve. According to statistics, only 15% of lung cancer patients survive 5 years after diagnosis.4 Therefore, it is important to strengthen basic research in the field of lung cancer and find specific immune markers for improving the survival rate of the patients. Single nucleotide polymorphisms (SNPs) genotyping has the characteristics of being simple, quick, and efficient, making it a promising method for identification of high-risk populations and avoiding the onset of the disease to the maximum extent.
Pyroptosis is a form of cell death closely related to inflammatory response.5 It is manifested by the continuous expansion of cells until the integrity of the cell membrane is lost, followed by the release of cell contents and damage-associated molecular patterns, finally leading to a strong inflammatory immune response in the body.6 In recent years, the importance of pyroptosis in various diseases and cancers has been gradually revealed, making it a research hotspot.7 Pyroptosis is mediated by the gasdermin (GSDM) family, including the caspase-1 classical pathway and the caspase4/5/11 and caspase3/8 nonclassical pathways.8 Inflammasomes are cytosolic polyprotein complexes that consist of NOD-like receptor (NLR), PYRIN domain, and HIN domain-containing family. It can mediate the lysis of a variety of inflammatory proteins such as GSDM family and are closely associated with pyroptosis.9 A NLRP3 inflammasome activation could affect pyroptosis or hyperactivity, resulting in cascade immunity or inflammatory response and affecting anti-tumor immunity.10 The formation and activation of NLRP3 inflammasomes has been linked to various tumors including colorectal, gastric, and lung cancers.11 Moreover, activation of NLRC4 can recruit the apoptosis speck protein and further activate caspase-1, which ultimately triggers cleavage of interleukin precursor and GSDMD, thereby causing inflammation and pyroptosis.12 In addition, NLRP7 is also a member of NLR family that participates in inflammatory responses and apoptosis. Both NLRC4 and NLRP7 have been recently identified as playing an essential role in tumor immunity and associated with prognosis of lung squamous cell carcinoma.13 However, few studies have focused on the SNPs of NLRP3, NLRC4, and NLRP7 in lung cancer.
On the basis of literature, six SNPs were selected in NLRP3, NLRC4, and NLRP7 genes. The rs35829419 and rs10754558 in NLPR3 were closely tied to bladder cancer risk.14 The NLRC4-rs212704 was a protective variant of pulmonary aspergillosis,15 NLRC4-rs455060 was correlated with lipid and glucose metabolism,16 and NLRC4-rs385076 had effect on lung function in patients with tuberculosis.17 In addition, NLRP7-rs775882 has been investigated in infertility or recurrent pregnancy loss.18 However, little information has been found about these SNPs in lung cancer patients. Genotyping these SNPs in our study participants will hopefully lead to the discovery of novel markers and contribute to early detection and prevention of the disease.
Materials and Methods
Subjects
A case–control study was conducted with 660 lung cancer patients and 660 healthy controls. Each participant was a Chinese Han individual recruited from Tangdu Hospital. The patients were diagnosed with lung cancer by histopathological examination of biopsy specimens. Healthy individuals without a history of cancer were randomly selected for the control group. Consent was obtained from all participants in writing. This study was approved by the Ethics Committee of Tangdu Hospital and carried out in accordance with the World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects.
Genotyping
Five milliliters of whole blood was collected from each subject in tubes containing ethylenediamine tetraacetic acid. DNA was extracted using a QIAamp DNA Blood Midi Kit (QIAGEN, Germany). Spectrometry (DeNovix DS-11FX Ultramicro spectrophotometer, United States) was used to measure the DNA concentration. Primers were designed using Sequenom MassARRAY Assay Design 3.0 software. SNP genotyping was performed on a Mass ARRAY iPLEX platform (Sequenom, San Diego, CA, USA) according to the manufacturer’s instructions. Assay design and mass spectrometric genotyping were performed as previously described.19
Statistical Analysis
Statistical analysis was performed with SPSS package version 20.0 (SPSS, Chicago, IL, USA). The chi-square test was used to compare the gender and smoking status, and the Student’s t-test was used to compare the age between the cases and controls, respectively. A divergence from Hardy–Weinberg equilibrium was evaluated for Minor allele frequencies. SNPstats (https://www.snpstats.net/start.htm) was used to estimate the association between SNPs and lung cancer risk and expressed as odds ratios (ORs) and 95% confidence intervals (CIs) with adjustments for sex, age, and smoking status. Statistical significance was established when p < 0.05.
Results
The demographic characteristics of the participants are presented in Table 1. A total of 660 lung cancer cases and 660 healthy controls were included, and the sex and age were matched (p > 0.05). The distribution of smoking status was similar to sex, and the smoking status of the two groups did not differ significantly (p > 0.05). Among the lung cancer cases, 300 are adenocarcinomas, 213 are squamous cell carcinomas, 110 are small cell carcinomas, and 25 are rare types. In addition, 24.1% of the lung cancer cases were diagnosed with stage I or II, and 75.9% of the cases were stage III or IV.
 |
Table 1 The Demographic Characteristics of the Participants |
The location information of SNPs and their MAFs in lung cancer cases and healthy controls are listed in Table 2. The NLRP3-rs35829419 and NLRP7-rs775882 are missense variants, NLRC4-rs455060 is a synonymous variant, and other SNPs were in 3’UTR or intron region. HWE was confirmed for all SNPs (p > 0.05). Comparing MAFs from two groups, we observed that three SNPs, NLRP3-rs35829419, NLRC4-rs385076 and NLRP7-rs775882, exhibited a higher disease risk (rs35829419: OR=1.361, 95% CI: 1.078–1.718, p=0.009; rs385076: OR=1.568, 95% CI: 1.306–1.882, p<0.0001; rs775882: OR=2.007, 95% CI: 1.704–2.532, p<0.0001), while NLRC4-rs212704 showed a protective role for the disease (OR=0.807, 95% CI: 0.692–0.941, p=0.006).
 |
Table 2 The MAF and HWE of Candidate SNPs Between Lung Cancer Cases and Healthy Controls |
Table 3 displays the genotype frequency of SNPs. Compared with CC genotype, the AA genotype of NLRP3-rs35829419 was tied to a 3.08-fold higher disease risk (95% CI: 1.20–7.89, p = 0.018). By analogy, the TC/CC genotypes of NLRC4-rs385076 and CT/TT genotypes of NLRP7-rs775882 were associated with a 1.50, 2.92, 2.05, 4.59-fold elevated risk of the disease, respectively (p<0.0001). Conversely, individuals carrying the NLRC4-rs212704-TT genotype were significantly less likely to develop lung cancer (OR=0.6, 95% CI: 0.43–0.85, p=0.014).
 |
Table 3 Genotype Frequency Distributions Between Lung Cancer Cases and Healthy Controls |
Table 4 shows three genetic models used to better evaluate SNPs’ associations with lung cancer risk. The results are consistent with allele and genotype models. The NLRP3-rs35829419, NLRC4-rs385076 and NLRP7-rs775882 were associated with various degrees of increased risk of lung cancer in all genetic models, except that NLRC4-rs212704 was associated with a reduced risk (p < 0.05).
 |
Table 4 Association Between SNPs and Lung Cancer Risk in Genetic Models |
Considering the effect of smoking on lung cancer risk and the differences of pathogenesis, we divided the participants into smoker and nonsmoker subgroups (Table 5) and evaluated the associations in each pathological type (Table 6). We observed that the four SNPs remained significant in smoker and nonsmoker subgroups (p < 0.05). However, in different pathological type groups, NLRP3-rs35829419 was correlated with risk of adenocarcinoma and small cell lung cancer, but not squamous cell carcinoma; and NLRC4-rs212704 was only protective for squamous cell carcinoma. The NLRC4-rs385076 and NLRP7-rs775882 were associated with all three pathological types (p < 0.01).
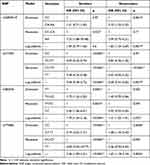 |
Table 5 Associations of Candidate SNPs with Lung Cancer Risk in Smokers and Nonsmokers |
 |
Table 6 Association Between Candidate SNPs and Risk of Adenocarcinoma, Squamous Cell Carcinoma, and Small Cell Lung Cancer |
Discussion
Inflammasomes have a dual effect on tumorigenesis, and their effects on the tumor development changed with the type of tumor.20 Genetic mutations in the inflammasome component may increase the body’s susceptibility to tumor, and anti-tumor immune strategy targeting inflammasomes has become a research hotspot in the field of oncology.21 In the present study, we demonstrated that NLRP3-rs35829419, NLRC4-rs385076 and NLRP7-rs775882 were associated with different levels of increased lung cancer risk, while NLRC4 -rs212704 exhibited a reduced risk. Our data provide further insight into the effects of pyroptosis and inflammasomes in lung cancer.
NLRP3 inflammasome is the most characteristic and deeply investigated inflammasome, which is composed of NLRP3 protein, apoptosis-associated spot-like protein containing a CARD (ASC) and pro-caspase-1.22 Studies have shown that tumor cells can escape from pyroptosis when their inflammasome components are silenced by epigenetic modifications.23 For example, the caspase-1 protein was downregulated in prostate tumor cells,24 and methylation suppresses ASC expression in many cancers.25 Also, knock out of NLRP3, ASC, and Caspase-1 promoted tumor growth in colorectal cancer mice model, indicating that dysfunction of NLRP3 inflammasome increased tumor burden.26 In addition, according to Yuan et al, Cucurbitacin B can directly activate the NLRP3 inflammasome through binding to Toll-like receptor 4, and trigger pyroptosis to inhibit development of lung cancer.27 Liang et al found that lycorine can inhibit NLRP3 activation through interference with its interaction with ASC and finally ameliorate pulmonary inflammation and fibrosis.28 In brief, NLRP3 inflammasome establishes the link between immunity and anti-tumor by inducing immune cells and tumor cells to undergo pyroptosis or hyperactivity, and plays a nonnegligible role in the process of anti-tumor immunity. According to our results, A NLRP3-rs35829419 mutation increased the risk of lung cancer, especially in adenocarcinoma and small cell lung cancer. Considering rs35829419 is a missense variant, we speculated that rs35829419 might exert an effect on development of the disease by changing the conformation of NLRP3 and influencing the activation of NLRP3 inflammasome and further pyroptosis combined with other factors.
Similar to other NLRs, NLRC4 has three structure domains: N-terminal isotype, central nucleotide-binding domain, and C-terminal leucine-rich repeat (LRR). In the absence of activation signals, NLRC4 is not activated by inhibiting self-polymerization; and when pathogenic microorganism infection, stress, or injury occurs, the LRRs domain recognizes the corresponding ligand, resulting in conformational changes, depolymerization, and activation of inflammasome.12 The expression and function of NLRC is inconsistent in different tumor type, or even in the same type.29 Hu et al demonstrated that NLRC4 inflammasome exerts influences on the process of colonic inflammation to tumor by regulating the epithelial cell response to injury.30 Janowski et al proved that NLRC4 could independently inhibit tumor growth without changed expression of ASC and caspase-1 or activation of inflammasome in melanoma.31 Kolb et al reported that obesity could induce the activation of NLRC4 and IL-1β, further change the VEGFA expression, and finally promote progression of breast cancer.32 In addition, Koichiro et al observed that NLRC4 could drive M2 polarization of tumor‐associated macrophages and upregulate IL‐1β and VEGF levels, resulting in liver metastasis of colon cancer.33 However, it is not fully understood how NLRC4 contributes to lung cancer. Here, we genotyped three SNPs in NLRC4 and identified that NLRC4-rs212704 was a risk-lowering variant for lung squamous cell carcinoma, while NLRC4-rs385076 was correlated with an increased risk of three types of the disease. Despite being located in the intron region, these two SNPs may influence the progression of lung cancer by changing motifs. In addition to providing new insight into the role of NLRC4, these results lay the groundwork for further functional studies on the protein.
NLRP7 is also a member of NLRP family and has a similar composition with other members and is the essential domain to function as an inflammasome sensor. However, the role of NLRP7 on inflammation in controversial. Kinoshita et al recognized that NLRP7 (also known as PYPAF3) could negatively regulate the secretion of IL-1β in caspase-1-dependent inflammasome signalling.34 Massaed et al further demonstrated that an inhibitory role of NLRP7 in formation of pro-IL-1β could be abolished by site-specific mutations after the Pyrin domain,35 indicating the anti- inflammatory role of NLRP7. On the contrary, Khare et al described that NLRP7 activation is necessary for activation of caspase-1 and release of IL-1β and IL-18 but not for secretion of IL-6 and TNF-α, in response to Mycoplasma infection.36 Zhou et al reported that NLRP7 inflammasome could promote IL-1β secretion and induce pyroptosis in THP-1 macrophages in response to M. bovis infection.37 Collectively, the anti-inflammatory or pro-inflammatory role of NLRP7 was observed under specific conditions. Previous studies on genetic polymorphisms of NLRP7 mainly focused on reproductive diseases, such as placental development, early pregnancy, and hydatidiform mole.38 We firstly established a link between NLRP7-rs775882 and risk of lung cancer. As rs775882 is a missense variant that may lead to the conformation change of NLRP7, we hypothesized that rs775882 may exert an effect on risk of the disease through disturbing the activation of NLRP7 inflammasome and pyroptosis in human body. In recent years, the latest association studies brought new ideas and inspiration for identification of susceptible genes for lung cancer. Yin et al reported that rs3136558 in pro-inflammatory gene IL1B has interactions with PPP1R13L and POLR1G in relation to lung cancer.39 Wang’s Lab identified BRCA2 that affect risk of lung cancer in a very large sample size including almost 30,000 individuals.40 Ji’s group reported a relationship between rs1948915 in long noncoding RNA (lncRNA) CCAT1 and risk of lung adenocarcinoma in the Chinese northeast population.41 In addition, Yu et al introduced a novel concept, RegQTL, to association study, and found that rs3768617 might have an influence on lung cancer risk by regulating the expression of miRNA-548b-3p-LAMC1 axis.42 These studies suggested that we could explore the interactions of genes and genes, polymorphisms of miRNA and lncRNA, or some novel regQTL-SNPs that related to lung cancer in a large sample size in further studies.
This study has some limitations. Firstly, a very long period of time was spent to recruit the subjects, we did not detect the serum levels of IL-1β and IL-18 of participants, therefore, we could not evaluate the effects of SNPs on activation of inflammasome pyroptosis. Secondly, the participants recruited in our hospital are mainly from northwest China, these results could only be interpreted as the associations between SNPs and risk of the disease in a Chinese Han population from northwest China. Thirdly, our results need to be validated by cell experiment and animal models.
Conclusion
In conclusion, we identified four SNPs associated with risk of lung cancer: NLRP3-rs35829419, NLRC4-rs385076 and NLRP7-rs775882 were risk variants for lung cancer, while NLRC4-rs212704 was a protective factor for the disease. Our results not only provide candidate markers for identification of high-risk population and early prevention of the disease but also provide new insight for anti-tumor strategy targeting inflammasome and pyroptosis.
Disclosure
Xin Jing and Yuhui Yun are co-first authors. The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40–46. doi:10.1097/CCO.0000000000000703
3. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
4. Akhtar-Danseh GG, Akhtar-Danesh N, Finley C. Uptake and survival effects of minimally invasive surgery for lung cancer: a population-based study. Eur J Surg Oncol. 2021;47(7):1791–1796. doi:10.1016/j.ejso.2021.01.002
5. D’Souza CA, Heitman J. Dismantling the Cryptococcus coat. Trends Microbiol. 2001;9(3):112–113. doi:10.1016/S0966-842X(00)01945-4
6. Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol. 2016;16(1):7–21. doi:10.1038/nri.2015.7
7. Wei X, Xie F, Zhou X, et al. Role of pyroptosis in inflammation and cancer. Cell Mol Immunol. 2022;19(9):971–992. doi:10.1038/s41423-022-00905-x
8. Yu P, Zhang X, Liu N, et al. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6(1):128. doi:10.1038/s41392-021-00507-5
9. Pandey A, Shen C, Feng S, et al. Cell biology of inflammasome activation. Trends Cell Biol. 2021;31(11):924–939. doi:10.1016/j.tcb.2021.06.010
10. Brydges SD, Broderick L, McGeough MD, et al. Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. J Clin Invest. 2013;123(11):4695–4705. doi:10.1172/JCI71543
11. Sharma BR, Kanneganti TD. NLRP3 inflammasome in cancer and metabolic diseases. Nat Immunol. 2021;22(5):550–559. doi:10.1038/s41590-021-00886-5
12. Hu Z, Yan C, Liu P, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341(6142):172–175. doi:10.1126/science.1236381
13. Li T, Liu H, Dong C, et al. Prognostic implications of pyroptosis-related gene signatures in lung squamous cell carcinoma. Front Pharmacol. 2022;13:806995. doi:10.3389/fphar.2022.806995
14. Xu G, Huang R, Xia W, et al. Associations between inflammasome-related gene NLRP3 Polymorphisms (rs10754558 and rs35829419) and risk of bladder cancer in a Chinese population. J Clin Lab Anal. 2021;35(11):e23973. doi:10.1002/jcla.23973
15. Zhong J, Liu L, Lu Y, et al. NLRP3, NLRC4 and NLRC5 gene polymorphisms associate with susceptibility of pulmonary aspergillosis in non-neutropenic patients. J Clin Med. 2022;11(7):1870. doi:10.3390/jcm11071870
16. Gomes Torres A, Leite N, Tureck LV, et al. Association between Toll-like receptors (TLR) and NOD-like receptor (NLR) polymorphisms and lipid and glucose metabolism. Gene. 2019;685:211–221. doi:10.1016/j.gene.2018.11.065
17. Ravimohan S, Maenetje P, Auld SC, et al. A common NLRC4 gene variant associates with inflammation and pulmonary function in human immunodeficiency virus and tuberculosis. Clin Infect Dis. 2020;71(4):924–932. doi:10.1093/cid/ciz898
18. Aghajanova L, Mahadevan S, Altmae S, et al. No evidence for mutations in NLRP7, NLRP2 or KHDC3L in women with unexplained recurrent pregnancy loss or infertility. Hum Reprod. 2015;30(1):232–238. doi:10.1093/humrep/deu296
19. Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60(1). doi:10.1002/0471142905.hg0212s60
20. Karki R, Man SM, Kanneganti TD. Inflammasomes and Cancer. Cancer Immunol Res. 2017;5(2):94–99. doi:10.1158/2326-6066.CIR-16-0269
21. Mishra SR, Mahapatra KK, Behera BP, et al. Inflammasomes in cancer: effect of epigenetic and autophagic modulations. Semin Cancer Biol. 2022;83:399–412. doi:10.1016/j.semcancer.2020.09.013
22. Kelley N, Jeltema D, Duan Y, et al. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20(13):3328. doi:10.3390/ijms20133328
23. Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-“host”? Cell Death Dis. 2019;10(9):650. doi:10.1038/s41419-019-1883-8
24. Winter RN, Rhee JG, Kyprianou N. Caspase-1 enhances the apoptotic response of prostate cancer cells to ionizing radiation. Anticancer Res. 2004;24(3a):1377–1386.
25. Wang Q, Wang Y, Ding J, et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579(7799):421–426. doi:10.1038/s41586-020-2079-1
26. Zaki MH, Vogel P, Body-Malapel M, et al. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185(8):4912–4920. doi:10.4049/jimmunol.1002046
27. Yuan R, Zhao W, Wang -Q-Q, et al. Cucurbitacin B inhibits non-small cell lung cancer in vivo and in vitro by triggering TLR4/NLRP3/GSDMD-dependent pyroptosis. Pharmacol Res. 2021;170:105748. doi:10.1016/j.phrs.2021.105748
28. Liang Q, Cai W, Zhao Y, et al. Lycorine ameliorates bleomycin-induced pulmonary fibrosis via inhibiting NLRP3 inflammasome activation and pyroptosis. Pharmacol Res. 2020;158:104884. doi:10.1016/j.phrs.2020.104884
29. Wen J, Xuan B, Liu Y, et al. Updating the NLRC4 inflammasome: from bacterial infections to autoimmunity and cancer. Front Immunol. 2021;12:702527. doi:10.3389/fimmu.2021.702527
30. Hu B, Elinav E, Huber S, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci U S A. 2010;107(50):21635–21640. doi:10.1073/pnas.1016814108
31. Janowski AM, Colegio OR, Hornick EE, et al. NLRC4 suppresses melanoma tumor progression independently of inflammasome activation. J Clin Invest. 2016;126(10):3917–3928. doi:10.1172/JCI86953
32. Kolb R, Phan L, Borcherding N, et al. Obesity-associated NLRC4 inflammasome activation drives breast cancer progression. Nat Commun. 2016;7(1):13007. doi:10.1038/ncomms13007
33. Ohashi K, Wang Z, Yang YM, et al. NOD-like receptor C4 inflammasome regulates the growth of colon cancer liver metastasis in NAFLD. Hepatology. 2019;70(5):1582–1599.
34. Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J Biol Chem. 2005;280(23):21720–21725. doi:10.1074/jbc.M410057200
35. Messaed C, Akoury E, Djuric U, et al. NLRP7, a nucleotide oligomerization domain-like receptor protein, is required for normal cytokine secretion and co-localizes with Golgi and the microtubule-organizing center. J Biol Chem. 2011;286(50):43313–43323. doi:10.1074/jbc.M111.306191
36. Khare S, Dorfleutner A, Bryan N, et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity. 2012;36(3):464–476. doi:10.1016/j.immuni.2012.02.001
37. Zhou Y, Shah SZA, Yang L, et al. Virulent mycobacterium bovis Beijing strain activates the NLRP7 inflammasome in THP-1 macrophages. PLoS One. 2016;11(4):e0152853. doi:10.1371/journal.pone.0152853
38. Carriere J, Dorfleutner A, Stehlik C. NLRP7: from inflammasome regulation to human disease. Immunology. 2021;163(4):363–376. doi:10.1111/imm.13372
39. Yin J, Wang C, Vogel U, et al. Common variants of pro-inflammatory gene IL1B and interactions with PPP1R13L and POLR1G in relation to lung cancer among Northeast Chinese. Sci Rep. 2023;13(1):7352. doi:10.1038/s41598-023-34069-z
40. Wang C, Dai J, Qin N, et al. Analyses of rare predisposing variants of lung cancer in 6004 whole genomes in Chinese. Cancer Cell. 2022;40(10):1223–1239.e6. doi:10.1016/j.ccell.2022.08.013
41. Ji Y, Yang Y, Yin Z. Polymorphisms in lncRNA CCAT1 on the susceptibility of lung cancer in a Chinese northeast population: a case-control study. Cancer Med. 2023;12(1):500–512. doi:10.1002/cam4.4902
42. Yu Y, Mao L, Cheng Z, et al. A novel regQTL-SNP and the risk of lung cancer: a multi-dimensional study. Arch Toxicol. 2021;95(12):3815–3827. doi:10.1007/s00204-021-03170-5
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
