Back to Journals » Infection and Drug Resistance » Volume 15
Pulmonary and Disseminated Mycobacterium avium Complex Cases Confirmed by Tissue-Direct Polymerase Chain Reaction-Based Nucleic Acid Lateral Flow Immunoassay of Formalin-Fixed Paraffin-Embedded Tissues
Authors Fukushi D, Murakami K, Watanabe Y , Sugimoto N, Uehara H, Seki M
Received 19 January 2022
Accepted for publication 9 March 2022
Published 14 March 2022 Volume 2022:15 Pages 1049—1054
DOI https://doi.org/10.2147/IDR.S358112
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Daisuke Fukushi,1 Keigo Murakami,2 Yuji Watanabe,3 Norihiko Sugimoto,4 Hirotsugu Uehara,4 Masafumi Seki5
1Division of Gastroenterology, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan; 2Division of Pathology, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan; 3Laboratory for Clinical Microbiology, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan; 4Fuso Pharmaceutical Industry, Osaka City, Osaka, Japan; 5Division of Infectious Diseases and Infection Control, Tohoku Medical and Pharmaceutical University Hospital, Sendai City, Miyagi, Japan
Correspondence: Masafumi Seki, Division of Infectious Diseases and Infection Control, Tohoku Medical and Pharmaceutical University, 1-15-1 Fukumuro, Miyagino-ku, Sendai City, Miyagi, 983-8536, Japan, Tel +81-22-983-1221, Fax +81-22-290-8959, Email [email protected]; [email protected]
Background: Detection of Mycobacterium avium complex (MAC) in tissue is essential for the diagnosis of MAC infections when the Mycobacterium is not isolated from sputum. However, detection of MAC in paraffin-embedded sections has not been established.
Methods: We encountered two patients with suspected MAC infections after surgery: patient 1 had a pulmonary nodule that was initially suspected to be lung cancer and was excised under video-assisted thoracoscopic surgery (VATS). Patient 2, who was under treatment with steroids and anti-IL-6 inhibitors for rheumatoid arthritis, was suspected to have disseminated ileocecal cancer with metastasis to the lung and skin. In both cases, we postoperatively detected MAC genes in paraffin-embedded tissue sections using the novel mycobacterial nucleic acid identification test, ie tissue-direct polymerase chain reaction (tdPCR)-based nucleic acid lateral flow immunoassay (NALFIA). Both patients showed granulomatous lesions with hematoxylin-eosin staining, and mycobacteria by Ziehl–Neelsen staining in tissue sections from the lung and skin, respectively, although MAC were not isolated from the sections. MAC genes were finally detected by tdPCR-NALFIA in both cases.
Conclusion: Although Ziehl–Neelsen staining and culture tests are the gold standard in identifying causative mycobacteria, the rapid results of tdPCR-NALFIA performed simultaneously with sputum and/or tissue culture may make it an important auxiliary diagnostic tool for identifying mycobacterial infection, leading to improvement in the management of MAC patients.
Keywords: Mycobacterium avium complex, MAC, Ziehl–Neelsen staining, rapid diagnosis
Background
Mycobacterium avium complex (MAC) is the most commonly isolated non-tuberculous Mycobacterium (NTM) in Japan which leads to lung diseases that manifest as fibro-cavitary (FC) radiographic changes or bronchiectasis with nodular and reticulonodular radiographic changes, termed the nodular bronchiectatic (NB) type.1
Since the presentations are similar to those of pulmonary tuberculosis and lung cancers, a definitive diagnosis, such as by microbial identification and isolation of MAC from sputum, bronchial lavage fluid and dissected tissue sections, obtained by surgery and/or bronchoscopic biopsy, are important for determining the appropriate therapeutic agents, such as clarithromycin (CAM), rifampicin (RIP), and ethambutol (EB).2,3
Usually, diagnosis of NTM are recommended more than one positive sputum culture microbiologically because NTM can be isolated from respiratory specimens due to environmental contamination and some patients who have an NTM isolated from their respiratory tract do not show evidence of progressive disease.1 Therefore, two or more sputum culture positive are required, however, since the detection rate of MAC from sputum culture alone is relatively low, mycobacterial gene analysis is sometimes required for detecting MAC in tissue sections that have already been embedded in paraffin.4,5
In this report, we present two cases of MAC that were clinically diagnosed as cancers before operation, although mycobacterial infections were later suspected because granulomas and mycobacteria were found with hematoxylin-eosin (HE) and Ziehl–Neelsen (ZN) staining of surgical specimens, respectively. Gene analysis for MAC was not performed before surgery, although a novel mycobacterial nucleic acid identification test using a tissue-direct polymerase chain reaction (tdPCR)-based nucleic acid lateral flow immunoassay (NALFIA) later confirmed the MAC genes in paraffin-embedded tissue sections.
This study was approved by the Committee for Clinical Scientific Research of Tohoku Medical and Pharmaceutical University Hospital on July 08, October 09, 2015 (Nos. ID2015-2-008 and ID2015-2-025, as Trial cdPCR-NALFIA for sepsis and mycobacterium, respectively: https://www.hosp.tohoku-mpu.ac.jp/department/infection.html. The patients whose specimens were used provided written informed consent to have their case details and any accompanying images published.
Case Reports
Case 1
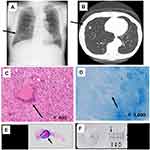
A 79-year-old male patient was found to have a small nodular shadow in his right middle lung lobe during an annual health examination that was suspected to be lung cancer (Figure 1A and B), and received the surgery because the malignant cells and some pathogenic bacteria including mycobacteria were not collected and isolated from his sputum. He was healthy and no history of the other drugs use and immunocompromised status, including HIV and diabetes. Although he underwent partial lobectomy, malignant cells were not found in the intraoperative frozen section. Subsequently, granulomatous lesions were found by hematoxylin-eosin (H&E) staining (Figure 1C and E) and mycobacteria were identified by ZN staining of the resected lung tissue (Figure 1D). We then successfully identified the Mycobacterium subtype after removal of the paraffin from a paraffin-embedded lung specimen, and detected the band indicating the MAC gene by tdPCR-NALFIA (Figure 1F). He was treated with CAM 800 mg/day, RIP 600 mg/day, and EB 750 mg/day for a year, with no tumor recurrence.
Case 2
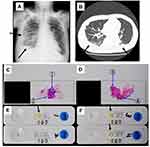
An 83- year-old male patient was suspected to have ileocecal cancer for which he underwent surgery, although no evidence of malignancy was identified in the resected specimen. He also had rheumatoid arthritis and was under treatment with immunosuppressive therapy, including prednisolone 5 mg/day and tocilizumab 8 mg/kg every month. Two months postoperatively, he developed chest abnormalities with pleural effusions (Figure 2A and B), and two skin tumors on his chest and waist, respectively, those were surgically resected. The tumors were initially suspected as metastasis of the ileocecal cancer, but malignant cells were not found in the skin section. However, granulomatous lesions were identified by H&E staining, suggesting the presence of mycobacterial infection (Figure 2C and D). Subsequent tdPCR-NALFIA using a tissue sample obtained by removal of the paraffin from the paraffin-embedded skin tissue section specimen led to identification of the Mycobacterium subtype and the MAC genes (Figure 2E and F). These results were matched with the MAC positive results of the culture from the skin tissue lesions at the 8 weeks after tumor removal surgery. He received CAM 800 mg/day, RIP 600 mg/day, and EB 750 mg/day for a year, with no further tumor recurrence.
Discussion
Identification of Mycobacterium and its subtype is essential for the diagnosis and treatment of mycobacterial infections, although its isolation from cultures is sometimes difficult.1 In addition, examination for mycobacteria, such as by ZN staining, PCR, or culture is overlooked in many cases that are initially suspected to be malignant tumors. It is important the knowledge of the Mycobacterium species involved in the disease because of the different treatments required dependent on each mycobacterium.1 For example, isoniazid are required for the treatment of M tuberculosis, but CAM are needed for the treatment for the MAC. Furthermore, differential diagnosis versus cancer is also required to prevent the wrong use of the antitumor agents and the recurrence of the Mycobacterium during the antitumor chemotherapy.5 In the present cases as well, we did not initially detect the Mycobacterium species and subtypes correctly, although we postoperatively identified the presence of mycobacteria and granulomas in the resected tissue.
Recently, a new test to identify bacterial nucleic acid in peripheral blood leukocytes (DiagnoSep) using cell direct PCR-NALFIA (cdPCR-NALFIA) has become clinically available in Japan (Fuso Pharmaceutical Industries, Osaka, Japan).6 The combination of cdPCR and NALFIA is a simple, rapid, and sensitive detection method for identifying seven representative bacterial species often isolated from blood cultures, including Staphylococcus aureus (S aureus).7 The use of cdPCR-NALFIA makes it possible to detect the DNA of bacteria phagocytized by leukocytes. Furthermore, the assay can be directly applied to blood samples without cultivation, and the results are obtained within 4–5 hours. In our cases, we next developed the new products and were able to detect mycobacterial genes by tdPCR-NALFIA and identified the subtypes as MAC, which enabled appropriate treatment with antimycobacterial agents, including CAM, RIP, and EB, rather than with antitumor agents although detection of mycobacterial genes from tissue sections were more difficult than the that of the other bacterium, such as S aureus from the blood.
tdPCR-NALFIA is similar to cdPCR-NALFIA for the detection of bacteria/fungus in the blood, and for the rapid detection of mycobacteria, including tuberculosis and MAC, in tissue sections. A basic scientific evaluation of this method was provided in our earlier paper.7 Briefly, the test procedure involves the following steps: removal of the paraffin from the tissue sections, preparing the tissue containing mycobacteria from the tissue section of a patient, coating the PCR tube with the tissues, increasing the membrane permeability of tissues and mycobacteria by enzyme/surfactant treatment, amplifying mycobacterial DNA in tissues through mycobacterial species-specific PCR which primers and systems were made by also Fuso Pharmaceutical Industries, Osaka, Japan, and evaluating the presence of PCR products using NALFIA.
In our cases, tdPCR-NALFIA methods that can specifically detect the two major mycobacterial species, M. tuberculosis and MAC, were applied. Next, the presence of PCR products was evaluated using a mycobacterial species-specific NALFIA for tissue samples containing granulomatous lesions. The results of the evaluation were the same as those of electrophoresis. If mycobacterial species-specific PCR products are present, red bands will appear within 20 minutes after chromatography is performed. The results can typically be obtained within 4–5 hours following tissue collection after removal of the paraffin from the tissue section, although if mycobacterial examination, such as ZN staining, PCR, or cultures are not performed before obtaining tissue sections, it typically takes 3–8 weeks to identify the pathogenic mycobacteria by conventional mycobacterial culture.
In the present study, tdPCR-NALFIA was used to identify mycobacterial genes in patients with suspected MAC infection after surgery, although we initially diagnosed them as having cancers. Culture tests remain the gold standard for identifying the causative organism in infectious diseases, however, mycobacterial cultures are sometimes neglected when cancers are strongly suspected, and we frequently found negative culture results.1,5
Shimada et al previously compared the results of using a PCR and in situ hybridization kit (PCR-ISH, named HybriSep; Fuso Pharmaceutical Industries, Osaka, Japan), and the PCR-based assay was reported to provide a significantly higher bacterial detection rate than blood cultures within 12 hours in sepsis patients,8 although the bacterial detection rate was higher with blood cultures in our previous similar study using DiagnoSep (Fuso Pharmaceutical Industries, Osaka, Japan).7 In our cases, granulomatous lesions and mycobacteria were found and MAC genes were identified by tdPCR-NALFIA using formalin-fixed paraffin-embedded tissue sections, following which the patients were successfully treated with antimycobacterial agents. Concurrence between the results of cultures and tdPCR-NALFIA in the patient 2 and the resultant clinical improvement following appropriate treatment in the two patients in the present study are suggestive of the diagnostic accuracy of this method. Additionally, the more rapid results of tdPCR-NALFIA rather than the standard culture methods might make this method important as an auxiliary diagnostic test for identifying infecting mycobacterium. In our cases, we could immediately start the anti-mycobacterial therapy without waiting for positive culture results at 3–8 weeks after surgeries.
Our report has certain limitations. First, this was a single center study and the sample size was small. In addition, we did not confirm the accuracy of the method for the detection of M. tuberculosis. Furthermore, we did not show the no negative and positive results of these clinical case series, and need to more detailed basic experiments. Moving forward, studies with a larger number of patients are needed to confirm the utility of tdPCR-NALFIA for the diagnosis of MAC infections.
Conclusions
In these two cases, although we did not initially perform conventional Mycobacterium examinations, identification of granulomatous lesions and mycobacterial structures in the resected specimen led us to perform tdPCR-NALFIA for the detection of MAC genes after removal of paraffin from the paraffin-embedded tissue sections. Clinical improvement with antimycobacterial agents confirmed the diagnosis. In the future, given its accuracy and speed of diagnosis, tdPCR-NALFIA has the potential to be a clinically useful diagnostic tool for identifying and predicting infecting organisms after surgery. When performed simultaneously with tissue pathological analysis, tdPCR-NALFIA might have diagnostic value as an auxiliary diagnostic test for identifying infecting organisms, in particular for the detection of DNA of mycobacterium, especially in cases where the previous culture examinations are insufficient.
Acknowledgments
The authors are grateful to all doctors, clinical technicians, and students in the microbiological laboratory, whose sacrifices, efforts, devotion to patients, and passion have made this report possible. The authors performed this study as a collaborative research with Fuso Pharmaceutical Industry (Osaka, Japan), and received 1,500,000 yen for scientific expenses, although no other funding was received from this company.
Disclosure
The authors report no competing interests in this work.
References
1. Daley CL, Iaccarino JM, Lange C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline: executive summary. Clin Infect Dis. 2020;71:e1–e36.
2. Griffith DE. Treatment of Mycobacterium avium Complex (MAC). Semin Respir Crit Care Med. 2018;39:351–361. doi:10.1055/s-0038-1660472
3. Wallace RJ
4. Koyama K, Ohshima N, Kawashima M, et al. Characteristics of pulmonary Mycobacterium avium complex disease diagnosed later in follow-up after negative mycobacterial study including bronchoscopy. Respir Med. 2015;109:1347–1353. doi:10.1016/j.rmed.2015.08.016
5. Charoenratanakul S, Dejsomritrutai W, Chaiprasert A. Diagnostic role of fiberoptic bronchoscopy in suspected smear negative pulmonary tuberculosis. Respir Med. 1995;89:621–623. doi:10.1016/0954-6111(95)90231-7
6. Sugimoto N, Kubo S, Uehara H, Kaku M. Development and validation of a combined cell-direct polymerase chain reaction-based nucleic acid lateral flow assay for the rapid detection of bacterial pathogens associated with sepsis. J Japanese Assoc Infect Dis. 2017;91:752–758.
7. Imai H, Watanabe Y, Shimada D, et al. Utility of a cell-direct polymerase chain reaction-based nucleic acid lateral flow immunoassay for detection of bacteria in peripheral blood leukocytes of suspected sepsis cases. Infect Drug Resist. 2021;4:5137–5144. doi:10.2147/IDR.S345361
8. Shimada J, Hayahsi I, Inamatsu T, et al. Clinical trial of in-situ hybridization method for the rapid diagnosis of sepsis. J Infect Chemother. 1999;5:21–31. doi:10.1007/s101560050004
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.