Back to Journals » OncoTargets and Therapy » Volume 12
PRPF40A as a potential diagnostic and prognostic marker is upregulated in pancreatic cancer tissues and cell lines: an integrated bioinformatics data analysis
Authors Huo Z, Zhai S , Weng Y, Qian H, Tang X, Shi Y, Deng X, Wang Y, Shen B
Received 19 February 2019
Accepted for publication 31 May 2019
Published 28 June 2019 Volume 2019:12 Pages 5037—5051
DOI https://doi.org/10.2147/OTT.S206039
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Zhen Huo*, Shuyu Zhai*, Yuanchi Weng*, Hao Qian, Xiaomei Tang, Yusheng Shi, Xiaxing Deng, Yue Wang, Baiyong Shen
Department of General Surgery, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, People’s Republic of China
*These authors contributed equally to this work
Background: Pre-mRNA processing factor 40 homolog A (PRPF40A) is an important protein involved in pre-mRNA splicing and is expressed in a variety of cell types. However, the function of PRPF40A in pancreatic cancer remains unclear. Therefore, our study is to investigate the role of PRPF40A in the pathogenesis of pancreatic cancer.
Materials and methods: We extracted expression data and clinical information of PRPF40A from different online databases, including the Cancer Genome Atlas (TCGA), Oncomine and the Gene Expression Omnibus (GEO). Subsequently, samples were collected from patients to validate gene expression using qPCR, Western blotting and immunohistochemical (IHC) analyses. Receiver operating characteristic (ROC) and Kaplan-Meier curve were used to evaluate the diagnostic and prognostic potential. Colony formation assays and CCK-8 assays were performed to measure the proliferative capacity of pancreatic cancer. Finally, gene ontology (GO) and pathway enrichment analyses of co-expressed genes of PRPF40A were conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID).
Results: We found that PRPF40A was upregulated based on data from both the online databases and our samples. PRPF40A possessed a significant diagnostic value, and its overexpression was associated with poor prognosis. PRPF40A knockdown inhibited cell proliferation in pancreatic cancer. GO and pathway analysis showed that the co-expressed genes were mainly involved in viral processing, mRNA splicing and the AMPK signaling pathway.
Conclusion: The results suggest that PRPF40A is an oncogene and can serve as a diagnostic and prognostic biomarker for pancreatic cancer. However, the underlying mechanisms remain to be elucidated.
Keywords: PRPF40A, pancreatic cancer, diagnosis, prognosis, biomarker
Introduction
Pancreatic cancer is often not diagnosed until advanced stage due to its anatomical location and aggressive nature, which results in a high mortality rate. The prognosis of pancreatic cancer is typically worse than that of other digestive tumors, and the 5-year survival rate is less than 5%.1 Chemotherapy is not always effective and radical resection is currently the only curative treatment for pancreatic cancer.2,3 Therefore, more effective methods of making early diagnosis and indicating prognosis are required to improve pre-operative condition assessment and the personalized medical treatment for patients with pancreatic cancer.
At present, the use of targeted therapy for the treatment of pancreatic cancer has become a topic of increased interest.4 Previous studies have discovered several clinically useful markers of pancreatic cancer. For instance, CA19-9 and CA125 are well-established diagnostic and prognostic markers for pancreatic cancer.5,6 Moreover, MIC-1, PAM4, S100A6 and OPN can serve as diagnostic markers for pancreatic cancer, while SPARC, CISD2 and SMAD4 are identified as promising prognostic markers.7–13
PRPF40A is one of the two putative homologs of Pre-mRNA processing protein 40, which plays a vital role in the initiation of pre-mRNA splicing.14,15 It has been reported that these two homologs are associated with genetic diseases such as Rett syndrome and Huntington’s disease.16,17 Besides, PRPF40A may be one of the potential target genes of p53, which can cause malignant human cancer when it is mutated.18 Recent studies found that PRPF40A is associated with hypoxia response in lung cancer and is overexpressed in pancreatic ductal adenocarcinoma.19,20 However, the role of PRPF40A in pancreatic cancer and its underlying molecular mechanisms remain unclear.
To study the expression pattern of PRPF40A and the biological mechanisms involved in tumorigenesis and the progression of pancreatic cancer, we extracted genetic expression data and clinical information from online databases (TCGA, Oncomine and GEO) and obtained co-expressed genes of PRPF40A from GEPIA. Subsequently, gene ontology and pathway analysis were performed using DAVID and a co-expression network was constructed using GeneMANIA and Cytoscape. Colony formation assays and CCK-8 assays were also performed. In addition, specimens and clinicopathological data were collected from patients treated at our hospital and analyzed to validate the results of the bioinformatic analysis.
Materials and methods
Bioinformatic analysis
Expression and survival data extraction
The results of the differential expression analysis performed on tumor and normal tissues from patients with cancer, including pancreatic cancer, were obtained and analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) server (http://gepia.cancer-pku.cn/index.html),21 which is based on the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) project and contains 171 normal and 179 cancer samples in total. Overall survival and disease-free survival curves were also generated based on this database. Differential expression analysis was performed using the Oncomine database (http://www.oncomine.com)22 with data from three datasets that were uploaded by Segara (6 normal and 11 cancer tissues),23 Pei (16 normal tissues and 36 cancer tissues)24 and Badea (39 tumor tissues and paired normal tissues).25 In addition, three datasets (GSE74629, GSE71729 and GSE62452) were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).26 GSE74629 contains 14 normal and 36 tumor samples, GSE71729 contains of 46 normal and 145 tumor samples, and GSE62452 is comprised of 69 tumor and paired normal samples. Six GEO datasets were used to determine the diagnostic power of PRPF40A, including GSE1542 (24 tumor vs 25 adjacent normal tissues), GSE15471 (39 tumor and paired adjacent normal tissues), GSE16515 (36 tumor vs 16 adjacent normal tissues), GSE28735 (45 tumor and paired adjacent normal tissues), GSE62452 (69 tumor vs 61 adjacent normal tissues) and GSE71729 (145 tumor vs 46 adjacent normal tissues).
Gene ontology (GO) and pathway enrichment analysis
The top 100 co-expressed genes targeted by PRPF40A in pancreatic cancer were screened using GEPIA and imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/)27 to perform the GO and pathway analysis. GO analysis is a bioinformatics method that is mainly used to annotate genes and categorize them according to biological process (BP), cellular component (CC) and molecular function (MF).28 Pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG), which is a database that contains various kinds of data from large molecular datasets generated using high-throughput experimental technologies.29 The results were visualized using GraphPad Prism 7.
Protein-protein interaction (PPI) network construction and analysis
The data of PRPF40A and its co-expressed genes was uploaded into GeneMANIA (http://genemania.org/), which is an online database to predict protein-protein interaction based on data collected from GEO, BioGRID, IRefIndex and I2D,30 and then imported into Cytoscape31 to generate a PPI network. GO analysis of these genes was performed and visualized using the BiNGO plugin in Cytoscape.32
Clinical samples
Forty-four surgical specimens from pancreatic cancers and adjacent normal tissues that were located at least 2 cm away from the tumor margin were collected from patients who were diagnosed based on clinical features and histopathological examination at Ruijin Hospital, which is affiliated with Shanghai Jiao Tong University. Twenty-nine males and 15 females, with an average age of 63 (ranged: 40~78) years old, were included in this study. The histological type was identified by two expert pathologists independently. The tumor samples used in this study were all derived from pancreatic ductal adenocarcinoma. The clinical-pathological data were gathered from medical records, and the tumor stages were classified according to the AJCC pancreatic cancer TNM staging system (2017).33 Written informed consents were obtained from all the participating patients and the experiments involving in human specimens were conducted in compliance with the Ethics Committee of Human Experimentation and with the Declaration of Helsinki as revised in 2013. The study protocol was approved by the Ethics Committee of Shanghai Ruijin Hospital (No.179 in 2017) for the scientific research of these clinical materials.
Immunohistochemical analysis
The specimens were fixed in formalin, embedded in paraffin and cut into 4 μm sections. The sections were incubated overnight at 4 °C in the presence of primary antibody against PRPF40A (rabbit polyclonal; ab204371, 1:500; Abcam, USA). Negative controls were generated by incubating without the primary antibody. After the slides were stained with DAB and counterstained with hematoxylin, the PRPF40A staining was examined using Image-ProPlus (version 6.0, Media Cybernetics, Rockville, USA) as previously described.34 The specimens with negative or weak intensity staining (-/+) of PRPF40A were considered to have low expression of PRPF40A, while those with moderate or strong (++/+++) intensity staining were considered to have high expression.
Cell culture and Western blotting
Seven cell lines derived from pancreatic tumors (sw1990, Panc-1, patu8988, Bxpc3, Aspc1, CFPAC1, and Capan1) and human pancreatic ductal epithelial (HPDE) cell line were acquired from Cell bank of Chinese Academy of Sciences and were cultured in RPMI 1640, DMEM, IMDM supplemented with 10% fetal bovine serum (FBS) and antibiotics. Western blotting was used to analyze protein expression, the results of which were quantified using ImageJ. Briefly, each type of cell was collected in cold PBS and lysed using RIPA buffer (Sigma-Aldrich; R0278) supplemented with protease and phosphatase inhibitor cocktails (Sigma-Aldrich; P8340). The proteins were transferred onto nitrocellulose membranes following sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Antibodies against PRPF40A (rabbit polyclonal; ab204371, 1:500; Abcam, USA) and GAPDH (rabbit polyclonal; D16H11, 1:1000; CST, USA) were used to bind the corresponding proteins.
Cell proliferation assays
Colony formation assays were performed by plating pancreatic cancer cells (Capan1) treated with PRPF40A-siRNA or an empty vector in 6-well plates (1000 cells/well) with 5 mL culture medium. After 14 days of incubation, the colonies were stained with 0.1% crystal violet solution. The cell proliferation capacity was also measured using a cell counting kit8 assay (CCK8, Dojindo, Japan). Numbers of viable cells were quantified at each 24 h interval by measuring the OD450 using a microplate reader (Epoch; BioTek, Winooski, VT). The experiments were performed independently in triplicate.
Quantitative real-time PCR
Total RNA from 12 pairs of pancreatic cancer tissues were extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcription (RT) reactions were performed using random primers and an M-MLV Reverse Transcriptase kit (Invitrogen). Real-time PCR was performed using the standard protocol included with the SYBR Green PCR kit (Toyobo, Osaka, Japan) on a Rotor-Gene RG-3000A (Corbett Life Science, Sydney, NSW, Australia). GAPDH was used as a reference. The primer sequences used for the detection of PRPF40A and GAPDH are shown in Table 1. The ΔCt values were normalized based on the GADPH levels. Each sample was analyzed in triplicate.
 |
Table 1 Primer sequence used for mRNA detection |
Statistical analysis
SPSS version 20.0 (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis. The correlation between PRPF40A expression and the clinicopathological characteristics of the patients was analyzed using the Chi-squared test. ROC curve was plotted to assess the diagnostic value. Area under the ROC curve (AUC) was used to determine the diagnostic power. A graded AUC value of 0.5–0.7, 0.7–0.9 or 0.9–1.0 represented a poor, moderate or high diagnostic value. Overall survival was determined using Kaplan-Meier analysis with a log-rank test. Cox regression analysis was used to assess the hazard ratio (HR) and 95% confidence intervals (CI). A value of P<0.05 was considered to denote statistical significance.
Results
The expression of PRPF40A is upregulated in patients with pancreatic cancer
To measure the differential expression of PRPF40A in cancer, we extracted expression data from various types of cancers using Gene Expression Profiling Interactive Analysis (GEPIA), which showed that PRPF40A was overexpressed in many digestive cancers, such as gastric cancer, esophageal cancer, pancreatic cancer and colorectal cancer (Figure 1A). To further investigate the expression of PRPF40A in pancreatic cancer, differential expression analysis was performed in Oncomine using three datasets, all of which showed significant upregulation of PRPF40A (Figure 1B–D). GEPIA analysis demonstrated that PRPF40A was significantly overexpressed in tumor tissues compared with normal tissues (Figure 1E). Similarly, significant upregulation of PRPF40A was also detected in all three datasets obtained from the Gene Expression Omnibus (GEO) (Figure 1F).
Validation of the PRPF40A expression level in pancreatic tissues and cell lines
To verify the expression of PRPF40A in online database, immunohistochemical analysis was performed to examine the 44 paired samples. The results are shown in Table 2, and indicate that pancreatic cancer tissues express more PRPF40A protein than adjacent tissue. Representative images are shown in Figure 2A. In addition, RNA expression was detected in 12 paired tissues using qRT-PCR, and we found that PRPF40A was significantly upregulated in tumor tissues (2.968±3.265 vs 6.323±4.885; P<0.05) (Figure 2B). Moreover, the expression level of PRPF40A was also studied in cell lines using qRT-PCR and Western blot, and the results showed that PRPF40A was upregulated in patu8988 and Capan1 cells (Figure 2C and D). All of the results mentioned above indicate that PRPF40A may be associated with carcinogenesis or the progression of pancreatic cancer.
 |
Table 2 Expression of PRPF40A in pancreatic cancer tissue and paired normal tissue by immunohistochemical analysis (n=44) |
The correlations between the expression of PRPF40A and clinicopathological parameters
To investigate the clinical relevance of PRPF40A expression in pancreatic cancer, baseline clinical characteristics of the 44 patients included in this study were collected and analyzed. As is shown in Table 3, the expression of PRPF40A was correlated with the level of tumor differentiation (P=0.0157) but not the TNM stage, tumor location, the degree of nerve and vascular invasion or distant metastasis. Moreover, we extracted and analyzed clinical and mRNA expression data for pancreatic cancer from the Cancer Genome Atlas (TCGA) database. The results in Table 4 suggest that no significant association exists between PRPF40A expression and the pathological stage, TNM stage or R stage.
 |
Table 3 The correlation between PRPF40A expression and clinicopathological features of patients with pancreatic cancer |
 |
Table 4 The association between PRPF40A expression and clinicopathological features of patients with pancreatic cancer in TCGA database |
The diagnostic and prognostic value of PRPF40A
To evaluate the diagnostic value of PRPF40A, ROC curves were plotted using data from six GEO datasets mentioned above. The values of AUC were 0.617, 0.811, 0.877, 0.732, 0.733 and 0.686, respectively (Figure 3A–F), suggesting that PRPF40A has a moderate diagnostic value for pancreatic cancer. To evaluate the prognostic potential of PRPF40A, survival data for patients with pancreatic cancer was analyzed and visualized using GEPIA. The results indicated that the high expression of PRPF40A was an unfavorable prognostic factor for overall survival (P=0.037) and disease-free survival (P=0.013) (Figure 4A and B). To further assess the prognostic value of PRPF40A, we collected follow-up information from 44 patients with pancreatic cancer that were treated at our hospital and divided them into two groups (high expression and low expression groups). The Kaplan-Meier analysis showed that patients with high expression of PRPF40A exhibited poor overall survival (Figure 4C). In addition, a Cox proportional hazard regression model was used to screen prognostic factors for patients in the TCGA database and those who were treated at our hospital. As is shown in Table 5, using univariate analysis, the N stage (P<0.001), R stage (P=0.02) and expression of PRPF40A (P=0.016) were significantly correlated with the survival of pancreatic cancer patients. Using multivariate analysis, the N stage (P=0.006) and expression of PRPF40A (P=0.041) were identified as independent prognostic factors for survival of pancreatic cancer patients. Similarly, as shown in Table 6, the results of the univariate analysis showed that tumor differentiation (P=0.04), distant metastasis (P=0.003) and the expression of PRPF40A (P=0.014) were correlated with patient survival. The multivariate analysis showed that distant metastasis (P=0.014) and the expression of PRPF40A (P=0.038) could independently predict survival status in pancreatic cancer patients. All of the above results suggest that PRPF40A could serve as a potential prognostic biomarker for pancreatic cancer.
 |
Table 5 Univariate and multivariate analysis of clinicopathological parameters of patients with pancreatic cancer in TCGA database by cox-regression |
 |
Table 6 Univariate and multivariate analysis of clinicopathological parameters of patients with pancreatic cancer by cox-regression |
Downregulation of PRPF40A inhibits pancreatic cell proliferation
To further investigate the role of PRPF40A in the progression of pancreatic cancer, cell lines in which PRPF40A was stably downregulated were established. Colony formation assays and CCK-8 assays were performed. As is shown in Figure 5A, after culturing for 14 days, Capan1 cell lines with downregulated PRPF40A expression formed significantly fewer colonies compared to untreated control cell lines. Similarly, the CCK-8 assays also indicated that the downregulation of PRPF40A inhibited pancreatic cell proliferation (Figure 5B). Overall, these results suggested that PRPF40A could enhance the proliferative capacity of pancreatic cancer.
Gene ontology and pathway enrichment analysis
To reveal the molecular mechanisms operating downstream of PRPF40A in pancreatic cancer, top 100 co-expressed genes targeted by PRPF40A were determined using GEPIA and then imported into the Database for Annotation, Visualization and Integrated Discovery (DAVID) to perform gene ontology and pathway analysis. The results are presented in Figure 6. It was found that, in the biological process (BP) category, the co-expressed genes were mainly involved in regulation of serine/threonine kinase activity, mRNA splicing, gene expression, regulation of cell differentiation and cell adhesion. In the cellular component (CC) category, they were mainly located in the cytosol, membrane and nucleoplasm. In regard to the molecular function (MF) category, the co-expressed genes were mainly associated with RNA and protein binding. Pathway analysis using KEGG database indicated that the enriched genes were mainly involved in the AMPK signaling pathway, endocytosis and the mRNA surveillance pathway.
Co- expression network construction and analysis
To investigate the protein-protein interactions mediated by the co-expressed genes, a protein-protein interaction (PPI) network was constructed (Figure 7A). Similarly, a PRPF40A associated PPI network was constructed to identify the potential molecular targets (Figure 7B). In addition, a network based on the gene ontology analysis was constructed using the BiNGO plugin (Figure 8).
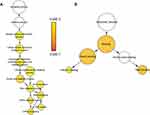 |
Figure 8 Gene ontology analysis network of co-expressed genes. Notes: (A) Biological process and (B) molecular function. The color and size of the node indicates the significance of the interaction. |
Discussion
In the present study, we used bioinformatic methods to extract PRPF40A expression data from public databases, including the Cancer Genome Atlas (TCGA), Oncomine and the Gene Expression Omnibus (GEO), and revealed that PRPF40A was significantly overexpressed in pancreatic cancer tissues. Furthermore, we collected 44 paired tissue samples from pancreatic cancer patients for immunohistochemical analysis, and 12 for qRT-PCR study. The results indicated that PRPF40A was upregulated in pancreatic cancer. In addition, it is also upregulated in patu8988 and Capan1 cell lines. All of the above results are consistent with previous evidence which suggests that PRPF40A was upregulated in pancreatic cancer and may be involved in carcinogenesis and/or the progression of pancreatic cancer.20,35
There are several molecules that are significantly upregulated in pancreatic cancer and well established to be diagnostic markers for pancreatic cancer, such as CA19-9, VEGFR2 and IGF-I.36 Our study has identified PRPF40A as a potential diagnostic marker for pancreatic cancer using data from six GEO datasets. The values of AUC were 0.617, 0.811, 0.877, 0.732, 0.733 and 0.686 respectively, indicating that PRPF40A is, to some extent, capable of distinguishing pancreatic cancer patients from healthy individuals, which may contribute to preoperative diagnosis and assessment. Combined with serum level of CA19-9, the sensitivity and specificity of diagnosis by PRPF40A could be even higher, which requires further investigation to validate with large patient cohort.
The 5-year survival of patients with pancreatic cancer has remained almost unchanged for many decades.37 Previous studies have identified several prognostic biomarkers in pancreatic cancer, such as TRPM8, CD74 and LKB1.38–40 In our study, we investigated the association between clinicopathological features and PRPF40A expression in pancreatic cancer using samples from our patients and the TCGA database. We confirmed that expression of PRPF40A was associated with tumor differentiation, and that the overexpression of PRPF40A was significantly correlated with poor overall survival and disease-free survival in pancreatic cancer patients. In addition, multivariate Cox regression analysis revealed that the expression of PRPF40A could serve as an independent prognostic factor of pancreatic cancer. The results above suggest that PRPF40A is a promising prognostic biomarker to provide individualized treatment for pancreatic cancer patients, as well as a therapeutic target to improve patients’ long-term survival. To sum up, our study for the first time revealed the diagnostic and prognostic value of PRPF40A in pancreatic cancer. However, a study in a larger sample of patients is required for further validation.
Malignant tumors are characterized by unsupervised cell proliferation that is caused by the mis-regulation of multiple proteins. Previous research has demonstrated that PRPF40A is involved in cell migration in untreated acute wounds and chronic wounds treated via debridement.41 In this study, we performed colony formation assays and CCK-8 assays using the Capan1 cell line. The results showed that inhibition of PRPF40A reduced the proliferation of pancreatic cancer, indicating that PRPF40A may be positively correlated with the proliferative capacity and serve as an oncogene in pancreatic cancer. However, the underlying mechanisms remain to be elucidated.
To uncover the probable molecular mechanisms underlying the involvement of PRPF40A in the initiation and development of pancreatic cancer, we determined the top 100 co-expressed genes of PRPF40A in the Gene Expression Profiling Interactive Analysis (GEPIA) and imported them into the Database for Annotation, Visualization and Integrated Discovery (DAVID) to perform gene ontology and pathway analysis. In the BP category of gene ontology, the results were enriched in genes involved in regulation of serine/threonine kinase activity, mRNA splicing, gene expression, regulation of cell differentiation and cell adhesion. Each of these processes could result in the pathogenesis or progression of pancreatic cancer. Pathway analysis indicated that the enriched genes were mainly involved in the AMPK signaling pathway, which is known as a vital mediator in maintaining cellular energy homeostasis and has been recently found associated with invasion and metastasis of pancreatic cancer.42,43 As is shown in Figure 7B, we used GeneMANIA to predict genes that have physical interaction with PRPF40A. Among the top 5 related genes, FMNL3 has been well-established as an oncogene in many cancers that regulates cytoskeletal mediation, invasion and metastasis of cancer cells.44 Therefore, we are interested in the relationship between PRPF40A and FMNL3. This axis may be subject to crosstalk with the AMPK signaling pathway during the regulation of pancreatic cancer cell proliferation or other biological processes. Therefore, we will focus on this axis to further reveal the underlying molecular mechanisms. Interestingly, according to previously published study, PRPF40A is significantly associated with hypoxia markers (HIF-1α, CYGB and VEGFa) in non-small cell lung cancer,19 while in our study we found two of the top 100 co-expressed genes of PRPF40A, PGM2 (Spearman’s rho=0.632, P<10−4) and RPE (Spearman’s rho=0.751, P<10−4), were correlated with pentose phosphate pathway (not significant enough to be listed in Figure 6), which initiates glycolysis that responses to hypoxia stress. This may account for hypoxia-induced upregulation of PRPF40A in non-small cell lung cancer. However, the underlying specific mechanisms remain to be discovered.
There are some limitations in our study. First, we only collected 44 tumor and adjacent tissue samples from patients with pancreatic cancer for immunohistochemical analysis, and only 12 of these were used for qRT-PCR. Due to an inadequate number of samples, we cannot exclude the possibility that sampling error may have led to the conclusion that PRPF40A is upregulated in pancreatic cancer. Second, the 44 samples were all harvested from patients with pancreatic ductal adenocarcinoma, which accounts for almost 90 percent of pancreatic carcinomas; most previous research has focused on this type of pancreatic cancer.20 Therefore, the expression of PRPF40A in other types of pancreatic cancer, such as intraductal papillary mucinous neoplasms or neuroendocrine neoplasms, requires further investigation. Third, according to clinical correlation analysis, the expression of PRPF40A is correlated with the pathological grade (Table 3) only when the data from TCGA (Table 4) was excluded, which yielded contradictory results. Hence, a larger sample would be required to reveal the correlation of PRPF40A with clinical attributes. Fourth, to measure the effects of PRPF40A on cell proliferation, we performed the colony formation assays and CCK-8 assays in only one pancreatic cell line (Capan1), which is not sufficient to draw a solid conclusion. Thus, we will need to perform further functional testing to validate the results.
Conclusion
In conclusion, using bioinformatic approaches followed by biological validation, this study clearly confirmed that PRPF40A was significantly upregulated in pancreatic cancer, exhibited diagnostic value and was associated with poor prognosis. Downregulation of PRPF40A inhibited cell proliferation in pancreatic cancer. Overall, the results suggest that PRPF40A is an oncogene and can serve as a diagnostic and independent prognostic biomarker in pancreatic cancer. However, the underlying molecular mechanisms require further elucidation.
Acknowledgments
This study was supported by grants from the Shanghai Anti-cancer Association (EYAS PROJECT; no. SACA-CY1C19).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. doi:10.1038/nrclinonc.2015.53
3. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
4. Chiorean EG, Coveler AL. Pancreatic cancer: optimizing treatment options, new, and emerging targeted therapies. Drug Des Devel Ther. 2015;9:3529–3545. doi:10.2147/DDDT.S60328
5. Cwik G, Wallner G, Skoczylas T. Cancer antigens 19-9 and 125 in the differential diagnosis of pancreatic mass lesions. Arch Surg. 2006;141(10):968–973. doi:10.1001/archsurg.141.10.968
6. Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. J Gastrointest Oncol. 2012;3(2):105–119. doi:10.3978/j.issn.2078-6891.2012.005
7. Chen YZ, Liu D, Zhao YX. Diagnostic performance of serum macrophage inhibitory cytokine-1 in pancreatic cancer: a meta-analysis and meta-regression analysis. DNA Cell Biol. 2014;33(6):370–377. doi:10.1089/dna.2013.2237
8. Gold DV, Gaedcke J, Ghadimi BM, et al. PAM4 enzyme immunoassay alone and in combination with CA 19-9 for the detection of pancreatic adenocarcinoma. Cancer. 2013;119(3):522–528. doi:10.1002/cncr.27762
9. Zihao G, Jie Z, Yan L, et al. Analyzing S100A6 expression in endoscopic ultrasonography-guided fine-needle aspiration specimens: a promising diagnostic method of pancreatic cancer. J Clin Gastroenterol. 2013;47(1):69–75. doi:10.1097/MCG.0b013e3182601752
10. Koopmann J, Fedarko NS, Jain A, et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2004;13(3):487–491.
11. Han W, Cao F, Chen M-B. Prognostic value of SPARC in patients with pancreatic cancer: a systematic review and meta-analysis. PLoS One. 2016;11:e0145803. doi:10.1371/journal.pone.0145803
12. Yang Y, Bai YS, Wang Q. CDGSH iron sulfur domain 2 activates proliferation and EMT of pancreatic cancer cells via Wnt/β-catenin pathway and has prognostic value in human pancreatic cancer. Oncol Res. 2017;25(4):605–615. doi:10.3727/096504016X14767450526417
13. Xia X, Wu W, Huang C, et al. SMAD4 and its role in pancreatic cancer. Tumour Biol. 2015;36(1):111–119. doi:10.1007/s13277-014-2883-z
14. Kao HY, Siliciano PG. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16(3):960–967. doi:10.1128/mcb.16.3.960
15. Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89(3):403–412.
16. Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7(9):1463–1474. doi:10.1093/hmg/7.9.1463
17. Buschdorf JP, Strätling WH. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J Mol Med (Berl). 2004;82(2):135–143. doi:10.1007/s00109-003-0497-9
18. Garritano S, Inga A, Gemignani F, Landi S. More targets, more pathways and more clues for mutant p53. Oncogenesis. 2013;2:e54. doi:10.1038/oncsis.2013.15
19. Oleksiewicz U, Liloglou T, Tasopoulou KM, et al. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143(7):1133–1141. doi:10.1007/s00432-017-2381-y
20. Wang J, Dumartin L, Mafficini A, et al. Splice variants as novel targets in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7(1):2980. doi:10.1038/s41598-017-03354-z
21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
22. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180.
23. Segara D, Biankin AV, Kench JG, et al. Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin Cancer Res. 2005;11(9):3587–3596. doi:10.1158/1078-0432.CCR-04-1813
24. Pei H, Li L, Fridley BL, et al. FKBP51 affects cancer cell response to chemotherapy by nagetively regulating Akt. Cancer Cell. 2009;16(3):259–266. doi:10.1016/j.ccr.2009.07.016
25. Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55(88):2016–2027.
26. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi:10.1093/nar/30.1.207
27. Huang D, Sherman B, Tan Q, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi:10.1186/gb-2007-8-5-r81
28. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi:10.1038/75556
29. Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101; discussion 101–103, 119–128, 244–152.
30. Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Tumour Biol. 2014;35(10):9725–9733. doi:10.1007/s13277-014-2206-4
31. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi:10.1093/bioinformatics/btq675
32. Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi:10.1093/bioinformatics/bti551
33. Allen PJ, Kuk D, Castillo CF, et al. Multi-institutional validation study of the American Joint Commission on Cancer (8th edition) changes for T and N staging in patients with pancreatic adenocarcinoma. Ann Surg. 2017;265(1):185–191.
34. Fan XJ, Wan XB, Yang ZL, et al. Snail promotes lymph node metastasis and twist enhances tumor deposit formation through epithelial-mesenchymal transition in colorectal cancer. Hum Pathol. 2013;44(2):173–180. doi:10.1016/j.humpath.2012.03.029
35. Thakur A, Bollig A, Wu J, Liao DJ. Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol Cancer. 2008;7:11. doi:10.1186/1476-4598-7-11
36. Chu LC, Goggins MG, Fishman EK. Diagnosis and detection of pancreatic cancer. Cancer J. 2017;23(6):333–342. doi:10.1097/PPO.0000000000000290
37. Kleeff J, Korc M, Apte M, et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022.
38. Du JD, Zheng X, Chen YL, et al. Elevated Transient Receptor Potential Melastatin 8 (TRPM8) expression is correlated with poor prognosis in panreatic cancer. Med Sci Monit. 2018;24:3720–3725. doi:10.12659/MSM.909968
39. Zhang JF, Hua R, Liu DJ, Liu W, Huo YM, Sun YW. Effect of CD74 on the prognosis of patients with resectable pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2014;13(1):81–86.
40. Yang JY, Jiang SH, Liu DJ, et al. Decreased LKB1 predicts poor prognosis in pancreatic ductal adenocarcinoma. Sci Rep. 2015;5:10575. doi:10.1038/srep10575
41. Ashrafi M, Sebastian A, Shih B, et al. Whole genome microarray data of chronic wound debridement prior to application of dermal skin substitutes. Wound Repair Regen. 2016;24(5):870–875. doi:10.1111/wrr.12460
42. Li W, Saud SM, Young MR, Chen G, Hua B. Targeting AMPK for cancer prevention and treatment. Oncotarget. 2015;6(10):7365–7378. doi:10.18632/oncotarget.3629
43. Chen K, Qian W, Li J, et al. Loss of AMPK activation promotes the invasion and metastasis of pancreatic cancer through an HSF1-dependent pathway. Mol Oncol. 2017;11(10):1475–1492. doi:10.1002/1878-0261.12116
44. Wu Y, Shen Z, Wang K, et al. High FMNL3 expression promotes nasopharyngeal carcinoma cell metastasis: role in TGF-β1-induced epithelia-to-mesenchymal transition. Sci Rep. 2017;7:42507. doi:10.1038/srep42507
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.