Back to Journals » Journal of Pain Research » Volume 16
Proteomic Analysis of the Spinal Dorsal Horn in Mice with Neuropathic Pain After Exercise
Authors Bai J , Zhang J, Zhou L, Hua Y
Received 13 January 2023
Accepted for publication 9 March 2023
Published 18 March 2023 Volume 2023:16 Pages 973—984
DOI https://doi.org/10.2147/JPR.S403374
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Qi Fang
Jie Bai,1 Jingyu Zhang,1 Li Zhou,2 Yufang Hua1
1Department of Anesthesiology, Lanzhou University Second Hospital, Lanzhou, People’s Republic of China; 2Department of Pediatric Digestive, Gansu Provincial Maternity and Child-Care Hospital/Gansu Provincial Central Hospital, Lanzhou, People’s Republic of China
Correspondence: Yufang Hua, Department of Anesthesiology, Lanzhou University Second Hospital, Lanzhou, 730030, People’s Republic of China, Tel +86 139 1903 2553, Email [email protected]
Purpose: Neuropathic pain (NP) is a chronic pain state with a complex etiology that currently lacks effective treatment in clinical practice. Studies have found that exercise training can alleviate NP hyperalgesia, but the specific mechanism remains unclear. Here, we sought to identify proteins and signaling pathways critical for mediating the effects of treadmill training on NP in a mouse model of spared nerve injury (SNI).
Methods: We used Tandem Mass Tag (TMT) technology for proteins and signaling pathways identification. Functional enrichment analyses were conducted using DAVID and Metascape software. Ingenuity pathway analysis was used to conduct functional annotation and analyze alterations in canonical pathways and molecular networks. Reverse transcription quantitative PCR (RT-qPCR) was used to confirm the results of proteomics analysis.
Results: A total of 270 differentially expressed proteins were screened in the detrained and trained groups (P ≤ 0.05). Enrichment and ingenuity pathway analysis revealed the effects of treadmill training on autophagy, cAMP-mediated signaling, calcium signaling and NP signaling in dorsal horn nerves. Treadmill training reduced the expression of Akt3, Atf2, Gsk3b, Pik3c3, Ppp2ca, and Sqstm1, and increased the expression of Pik3cb in the autophagic pathway.
Conclusion: Our results suggest that treadmill training may alleviate nociceptive hyperalgesia in NP mice by modulating the autophagic pathway, providing unique mechanistic insights into the analgesic effects of exercise.
Keywords: proteomic changes, exercise, neuropathic pain, IPA, passway analysis
Introduction
Neuropathic pain (NP) is an incurable chronic pain caused by peripheral or central nervous system injury or dysfunction, with a prevalence of approximately 10% in the general population.1 NP treatment is difficult owing to the limited effectiveness and adverse effects of conventional analgesic drug therapy. Thus, there is an urgent need to find new directions and breakthroughs to study mechanisms underlying NP and its treatment.
Non-pharmacological treatments, including exercise, can significantly reduce pain. Clinical studies have shown that exercise is effective in improving arthritis, fibromyalgia, low back pain, intermittent claudication, neck pain, and knee osteoarthritis.2–6 Mechanistic studies have confirmed that exercise can relieve pain symptoms by increasing endogenous opioids, promoting the recovery of neurotrophic factors, activating serotonin (5-HT) receptors, modulating inflammatory factors, and enhancing pain line inhibition.7–10 Exploring the protective effect of exercise on NP and the specific molecular mechanisms involved is of great significance; however, limited studies have evaluated the role of treadmill training for alleviate NP.In recent years, tandem mass tagging (TMT) technology has been widely used in proteomics studies owing to its high precision, stability, and throughput. Hence, it can be used to determine the potential mechanisms by which exercise relieves pain. In this study, we used proteomic techniques to explore the impact of exercise on hyperalgesia in mice with NP and identify the specific mechanisms involved in this process.
Materials and Methods
Experimental Animals
Fifty-six male C57BL/6 mice, weighing 18–23 g, were provided by the Lanzhou Institute of Veterinary Medicine, Chinese Academy of Agricultural Sciences (SCXK (Gan) 2015–0001). Mice were housed in the Experimental Animal Center of Lanzhou University Second Hospital in a controlled room temperature of 23 ± 1°C, maintained on a 12-hour light cycle (lights on at 8 am.), and food/water was available ad libitum. Animal protocols were approved by the Committee on the Ethics of Animal Experiments of the Lanzhou University Second Hospital and conducted in conformity with the guidelines of the International Association for the Study of Pain.
SNI Models
We modeled spared nerve injury (SNI) according to the method described by Decosterd.11 The mice were anesthetized with 40 mg/kg sodium pentobarbital by intraperitoneal injection before surgery. The femoris muscle was resected and separated to reveal the branches of the sciatic nerve. The tibial and common peroneal nerves were cut after ligation with 6–0 silk thread, and the stump of the nerve was cut at approximately 3 mm distal to the ligature. Stretching and stimulation of the peroneal nerve were avoided during this procedure. In the sham group (S), the sciatic nerve of the mice was exposed, but was neither ligated nor cut.
Treadmill Exercise
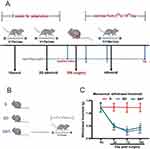
The mice were randomly assigned to three groups (n = 18): (1) S group, (2) SNI-detrained (SD) group, in which mice were not trained after surgery, and (3) SNI-trained (SNT) group: mice were trained after surgery. The treadmill training program (SA101; Jiangsu Sans Biotechnology Co. Ltd, Nanjing, China) was established based on the results of our previous studies and the related literature (Figure 1A and B).12,13 The mice in the SNT group were acclimatized for 2 weeks, 5 days per week, before surgery. Mice were run at 7 m/min for 10 min/day during week 1. In week 2, mice started with 20 minutes of running at 7 m/min per day, and the exercise time was increased by 10 min per day until 60 min. Mice in the SNT group were trained on a treadmill at a speed of 10 m/min for 60 min/day, starting on the third day after SNI. Stimuli, such as electric shocks from the treadmill, were avoided to not cause stress that could bias the results. The mice ran continuously on the treadmill by tapping their tails.
Behavioral Test
The mechanical hyperalgesia threshold (PWT) was described by Chaplan et al to assess mechanical hyperalgesia in mice.14 Before SNI surgery and 3, 7 and 14 days after SNI surgery, mice were first placed in a clear plastic box with a metal grid at the bottom for 30 min, and then researchers blindly stimulated the lateral edge of the left hind paw was blindly stimulated with different grams (0.6, 1.0, 1.4, 2.0 and 4.0 g; North Coast Medical, Morgan Hill, CA, USA), and then calculated according to the up-and-down rule to determine the PWT.
Proteomic Analysis
Three mice were randomly selected for proteomic analysis in the SD and SNT groups at the end of behavioural testing 14 days after SNI. Mice were anaesthetized with an intraperitoneal overdose of 1% sodium pentobarbital and then sacrificed. The L4–6 spinal cord segments were rapidly dissected and immediately stored at –80°C. SDT buffer was added to each tissue sample, which was homogenized with an MP homogenizer, and the supernatant was collected. Samples were subjected to repeated ultrafiltration using UA buffer. Then, 100 μL iodoacetamide was added to the samples and incubated in the dark for 30 min. After washing the filter twice with 100 μL UA buffer, and the filter was washed twice with 100 μL 40 mM NH4HCO3 buffer. The protein suspensions were then digested overnight at 37 °C in 40 μL TEAB buffer by adding 4 μg trypsin, and the filtrate was collected. The peptides were desalted, lyophilized mixed with 40 μ L of 0.1% formic acid solution, re-solubilized, and quantified (OD280). The peptides in each sample were labelled using TMT reagent (Thermo Fisher Scientific, Waltham, MA, USA). The TMT-labeled samples were fractionated by basic pH reversed-phase liquid chromatography using an Agilent 1260 Infinity II high-pressure liquid chromatography system. The samples were then loaded onto a trap column (1mL/min) and analytical column (Thermo Fisher Scientific, Waltham, MA, USA) before analysis. The samples were chromatographed and analyzed using a high-resolution Q-Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). Proteins were identified using the Proteome Discoverer 2.1 software. The proteome of Mus musculus was loaded from Uniprot, the maximum missed cleavage site was set to 2, the precursor mass tolerance was 10ppm, and the fragment mass tolerance was 0.05 Da, the quantitative method was TMT 6plex, and the FPR for peptide-spectrum match identification was 0.01. Differentially expressed proteins were identified using P < 0.05 as the screening criterion. We used R ggplot2 to plot volcano plots and heat maps to show the differentially expressed proteins in the SD and SNT groups.
Functional Enrichment Analyses of Differentially Expressed Proteins
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed on differentially expressed proteins using clusterProfiler and GGplot2 software packages. The most significantly enriched GO terms and KEGG pathways were screened based on P value.
The results were sorted according to P values, and the top 10 most significantly enriched GO entries and the top 20 most significantly enriched KEGG pathways were filtered. The STRING database was used to construct a protein interaction network (PPI) for differentially expressed proteins.
Ingenuity Pathway Analysis (IPA)
IPA software (Qiagen, Redwood City, CA, USA) was used to conduct functional annotation to analyze alterations in canonical pathways and molecular networks. We determined relative and absolute expression fold-change values for the identified peptides and performed core IPA analysis, biomarkers, and molecular and functional comparative analyses. IPA analysis was used to identify potential pathways and chart networks based on previously reported protein relationships.
RT- qPCR
After PWT was assessed at 14 days postoperatively in S, SD, and SNT groups, L4–6 spinal cord tissues were collected, weighed, and stored at −80 °C after rapid freezing in liquid nitrogen. Total RNA was fractionated using TRIzol reagent (Invitrogen, Carlsbad, USA) and qPCR was performed with SYBR Premix following the manufacturer’s instructions. Gene expression was normalized using the expression of gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2–ΔΔCT method. The primer sequences used for qPCR are listed in Table 1.
 |
Table 1 Primer Sequences Used for qRT-PCR |
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 8 and SPSS 26.0. Results are expressed as means ± standard deviation, and data were analyzed using one-way analysis of variance (ANOVA) or Student’s t-test, and statistical significance was set at P < 0.05.
Results
Treadmill Training Reduces Mechanical Nociceptive-Related Behaviors After SNI Surgery
There was no significant difference in the preoperative ipsilateral PWT of mice in each group (P > 0.05). As shown in Figure 1C and Table 2, compared with the S group, the SD and SNT groups had significantly decreased PWT from 3 days postoperatively until 14 days postoperatively (###P < 0.001), indicating that the SNI model of NP in mice was successfully constructed. The PWT was significantly higher in the SNT group than in the SD group at 7 and 14 days postoperatively (**P < 0.01), showing that treadmill training can greatly alleviate nociceptive hyperalgesia in mice with NP.
 |
Table 2 PWT Changes of Mice in Different Groups |
Identification of Differentially Expressed Proteins Following Treadmill Training
Differences in protein expression in the spinal cord tissue between the SD and SNT groups were quantified by proteomics. The results identified 47,716 unique peptides, corresponding to 6123 individual proteins. At a P value ≤ 0.05, 270 differentially expressed proteins were identified, of which 221 were downregulated and 49 were upregulated in the SNT group compared to the SD group (Supplemental Table 1). Figure 2A shows a cluster analysis of these results.
Functional Enrichment Analyses Differentially Expressed Proteins
GO enrichment analysis was conducted for differential expressed proteins, and the results were sorted based on P values in order of their significance (P < 0.05). “Protein localization to nucleus”, “vesicle-mediated transport in synapse”, “synaptic vesicle cycle”, “endosomal transport”, and “vesicle-mediated transport to the plasma membrane” were the biological functions most substantially activated by the differentially expressed proteins (Figure 2B).
The top 20 significantly enriched pathways are indicated in Figure 2C, among which the pathways of neurodegeneration, cAMP signaling pathway, nucleocytoplasmic transport, EGFR tyrosine kinase inhibitor resistance, dopaminergic synapses, autophagy, and proteasome potential were closely related to NP. PPI networks were used to identify differentially expressed proteins using STRING (Supplemental Figure 1).
Integrated IPA of Proteins
Table 3 shows the signaling pathways ranked based on P values, including the EIF2, synaptogenic, mTOR, ferritin, and cAMP-mediated signaling pathways as well as autophagy, and NP neuropathway in dorsal horn neurons. The direction of the functional changes was predicted using the z-score; a z-score ≥ 2 or ≤ −2 and P < 0.05 showed significantly activated or inhibited pathways, respectively, with the most significantly changed pathway being autophagy (Figure 2D). Supplemental Figures 2 and 3 show the highest-scoring autophagic pathway and another neural pathway for NP signaling in dorsal horn neurons, respectively, which may be of interest for future studies. There are eight differentially expressed proteins involved in the autophagic pathway, namely AKT3, ATF2, GNAI3, GSK3B, PIK3C3, PIK3CB, PPP2CA, and SQSTM1. However, while the classical pathway may provide a more detailed picture of the regulatory processes, new pathways and molecules could not be identified, mainly because the molecular mechanisms of neuropathic hyperalgesia are unknown; thus, additional signaling pathways that may be involved need to be identified. Figure 3A shows the expression trend of proteins with SQSTM1 as the core. Upstream and downstream molecules such as GSK3B, IKBKG, and HTT have been studied and confirmed to be closely related to autophagy, implying that autophagy may play an important part in exercise-induced hyperalgesia. Compared to the classical pathway shown in Supplement Figure 2, this network suggests more upstream and downstream molecules.
 |
Table 3 Ingenuity Canonical Pathways of IPA |
RT-qPCR Confirmed the Results of Proteomics Analysis
Differentially expressed proteins were analyzed by GO, KEGG, and IPA, and the results suggested that autophagy is a key cue for exercise-induced hyperalgesia in mice withNP and that several proteins are closely related to the autophagic pathway. To determine the fitness of the TMT proteomic screening results, we examined the expression of genes encoding Akt3, Atf2, Gnai3, Gsk3b, Pik3c3, Pik3cb, Ppp2ca, and Sqstm1 in the S, SD, and SNT groups using RT-qPCR (Figure 3B). Compared to that in the S group, the expression of Akt3, Atf2, Gnai3, Gsk3b, Pik3c3, Pik3cb, Ppp2ca, and Sqstm1 encoding genes was elevated in the ipsilateral L4–L6 spinal cord of mice in the SD group. The expression of genes encoding Akt3, Atf2, Gsk3b, Pik3c3, Ppp2ca, and Sqstm1 in the spinal cord of mice in the SNT group decreased after treadmill training, expression of gene encoding Pik3cb increased, and that of gene encoding Gnai3 remained unchanged. These results indicate that treadmill training downregulates the expression of Akt3, Atf2, Gsk3b, Pik3c3, Ppp2ca, and Sqstm1 encoding genes and upregulates the expression of Pik3cb in the spinal cord of SNI mice, which is consistent with the results of the TMT proteomic screening. These proteins may play important roles in exercise-induced hyperalgesia in mice with NP.
Discussion
Exercise training can alleviate NP. However, the specific molecular mechanisms underlying this effect remain unclear. In this study, we used TMT-based proteomics techniques to quantify differentially expressed proteins and investigated the underlying pathway whereby treadmill training relieves pain in mice experiencing NP.
In this study, 270 differentially expressed proteins were identified between the SD and SNT groups. Of these, 49 were up-regulated and 221 were down-regulated. GO and KEGG analyses showed that differentially expressed proteins were mainly enriched in pain-related biological processes and signaling pathways such as cAMP signaling pathway, EGFR tyrosine kinase inhibitor resistance, dopaminergic synapses, and autophagy. Analysis of the classical pathways involving differentially expressed proteins using the IPA bioinformatic analysis revealed that among the significantly enriched classical pathways, the EIF2, synaptogenesis, mTOR, and cAMP-mediated signaling pathways as well as autophagy and NP signaling in dorsal horn neurons were associated with the generation and persistence of NP, further demonstrating the accuracy of the proteomic results.
The significant enrichment of EIF2 signaling suggests an enhancement of ElF2 signaling in the spinal dorsal horn of SNI mice after treadmill training; this may be possibly related to the endoplasmic reticulum stress involved in the occurrence and continuation of NP.15 In addition, spinal mTOR signaling plays an essential role in regulating spinal sensitization in NP.16 Targeting mTOR may provide a new strategy for pain treatment. Synaptic plasticity underlies spinal cord sensitization in NP, and synaptic transmission can be enhanced by synaptogenic signaling pathways, which are closely related to NP onset.17 CAMP-mediated signaling in the spinal cord may be critical for the development of painful neuropathy,18 and the autophagic pathway is a potential therapeutic target for NP.19 These pathways mainly involve differentially expressed proteins, such as AGO2, AKT3, ATF2, and CAMK2A.
Autophagy is involved in NP development. In a rat model of spinal nerve ligation (SNL), the expression of both microtubule-associated protein light chain 3 (LC3) and P62 was upregulated at 7 days postoperatively, suggesting that NP onset is associated with impaired lysosomal degradation and dysregulated autophagic processes.20 Intrathecal injection of the autophagy inducer rapamycin three days after surgery in the SNL model induced autophagy in spinal microglia and inhibited inflammatory factor IL-1β, which relieved pain, suggesting that autophagy induction can relieve NP.21 Autophagy dysfunction is involved in the onset and development of NP, and appropriate induction of autophagy may be an effective measure for NP treatment. Recent studies have found that exercise can inhibit nociceptive hyperalgesic behavior and effectively alleviate pain in various animal models of NP.22,23 Whether exercise can improve pain and its specific mechanisms remain unclear and require further exploration. Increased exercise intensity early in the peripheral nerve chronic constriction injury (CCI) model rats relieved nociceptive hyperalgesia in the sciatic nerve region while increasing neurotrophic factor levels.24 Kami et al reported that mice with a partial sciatic nerve ligation PSL model had significantly reduced pain after exercise.12 Although these studies explored the biological role of exercise in NP, but the specific molecular mechanisms by which exercise reduces pain remain unclear. Autophagy was the most significantly enriched classical signaling pathway in IPA analysis in this study, indicating that autophagy may play an essential role in exercise-induced hypoalgesia. The differential proteins involved in this pathway include ATF2, AKT3, GNAI3, GSK3B, PIK3C3, PPP2CA, and SQSTM1.
The transcription factor ATF2 is a downstream target of p38 mitogen-activated protein kinase (p38-MAPK) in dorsal horn neurons of the spinal cord, mainly found in dorsal root ganglia and spinal cord tissue. Transcriptionally activated ATF2 regulates various genes that regulate cellular responses associated with pain and inflammatory responses.25 Activation of p38-MAPK and ATF2 is important in central NP sensitization and spinal nerve ligation in chronic pain models has been shown to induce mechanical nociceptive hyperalgesia and thermal nociceptive hypersensitivity in rats, leading to increased expression of ATF2 and ATF3 in the ipsilateral dorsal root ganglia and spinal cord. Intrathecal injection of ATF siRNA reverses nociceptive hypersensitivity.26,27 Studies suggest that ATF2 is involved in the maintenance of NP in animal models of nerve injury.
Glycogen synthase kinase 3 beta (GSK3B) is one of the rate-limiting enzymes of glycogen synthesis and acts on several intracellular signaling pathways to regulate glial cell activation and production of pro-inflammatory factors in the central nervous system.28 Zhang et al reported that GSK3B may participate in NP maintenance by regulating the relationship between proinflammatory and anti-inflammatory factors.29 Conversely, the administration of GSK3B inhibitors reduces the response to injurious stimuli and tolerance to morphine in mice with pathological pain.30,31 GSK3B is associated with the onset and development of NP through various mechanisms, including promotion of peripheral sensitization, central sensitization, and glial cell activation. Thus, it is a potential new pharmacological target for NP treatment. In the present study, exercise training suppressed ATF2 and GSK3B expression, which is consistent with published reports.
The phosphatidylinositol 3-kinase (PI3K) pathway is involved in the regulation of central and peripheral nerve sensitization, and its inhibitors can reduce nociceptive sensitization induced by nerve injury.32 The PI3K pathway has recently received increasing attention as a potential therapeutic target for pain management. PIK3CB, a member of the PI3K family, is associated with insulin resistance, sensory neuron inflammation, neuropathy, and NP.33–35 By establishing a model of NP caused by brachial plexus nerve injury, Wang et al found that the neuropathic PIK3C3 is an autophagy-related isoform of the PI3K family and plays a very vital role in activating autophagy.36,37 PIK3C3 may regulate autophagy through STAT3 and is associated with NP maintenance.38,39 In the present study, exercise training-induced PIK3CB expression and decreased PIK3C3 expression, which is consistent with published reports. Thus, PIK3CB and PIK3C3 may be novel NP biomarkers.
Protein phosphatase 2A (PP2A) is an critical protein phosphatases in the central nervous system. Earlier, a model of central sensitization to pain was established by subcutaneous injection of capsaicin in the hind paw of rats to induce mechanical and thermal nociceptive sensitization; this resulted in a notable upregulation of PP2A expression in the rat spinal cord, which regulates the phosphorylation status of basic proteins involved in central sensitization.40 In the present research, exercise training inhibited the expression of GNAI3 and PPP2CA, which is consistent with previous reports.
SQSTM1/P62 is a critical protein in the autophagic pathway and a potential indicator of the extent of autophagy.41 Autophagy dysfunction is closely associated with the onset and development of NP, with rats exhibiting reduced mechanical and thermal pain thresholds and hyperalgesia seven days after the establishment of an SNL model in animals with NP. In contrast, the expression of the spinal autophagy markers LC3II and SQSTM1 was significantly increased and co-expressed in microglia of the spinal dorsal horn. After intrathecal injection of an autophagy inducer, the pain threshold started to increase, LC3II expression increased, SQSTM1 expression decreased, and the number of autophagosomes in microglia in the dorsal horn of the spinal cord increased significantly. The opposite result was observed with intrathecal injection of autophagy inhibitors.21 In NP models, dysregulation of autophagy in the spinal dorsal horn is a fundamental cause of chronic pain progression.20,42 Increasing the autophagic activity of microglia in the spinal cord horn plays a vital role in the alleviation of NP development. Figure 2D shows that SQSTM1 is located more centrally and interacts directly with GSK3B, CPLV2, CHCHD2, HTT, MTX2, DNAJA2, USP7, and IKBKG. These upstream and downstream regulatory relationships need further experimental verification.
AKT3 belongs to the Akt kinase family, which is closely associated with NP, and is a potential target for treatment. Akt inhibition attenuates nociceptive sensitization and downregulates pro-inflammatory cytokine expression in vivo.43 The differentially expressed proteins PIK3C3, PI3CB, GNAI3, and SQSTM1 in this study were shown to be associated with Akt signaling, further suggesting that the AKT pathway may play an essential role in the induction of exercise-induced hypoalgesia via autophagy.
To further explore the potential molecular changes and specific bioinformatic functions involved, we used high-throughput proteomic tools to identify proteins differentially expressed in the spinal cord after treadmill training. Some molecular relationships among the predicted networks have been confirmed in the literature. The molecular mechanisms of exercise-induced hypoalgesia remain unknown and may not be necessarily regulated by known classical pathways; therefore, further studies are needed. Additional signaling pathways that may be involved in this process need to be identified.
This study has some limitations. First, the proteomic sample size was too small to draw definite conclusions. Second, the clinical significance of the genes determined to play a role in exercise-induced hyperalgesia remains unknown, and thus, further functional studies are warranted. Finally, the present study presents data only at the gene level, and further validation at the protein level is required.
Conclusion
This is the first proteomic study using a murine NP model to identify a role for the autophagy pathway in treadmill training to modulate NP nociceptive hypersensitivity. Our study offers a basis for the application of exercise therapy to improve chronic pain and provides clues for finding new therapeutic targets.
Data Sharing Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.
Ethics Approval
All experimental protocols were approved by Animal Use and Ethics Committee of the Second Hospital of Lanzhou University.
Acknowledgments
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the Natural Science Foundation of Gansu Province of China (20JR10RA726, 21JR11RA178); and Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (CY2022-QN-B01).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Raja SN, Carr DB, Cohen M, et al. The revised international association for the study of pain definition of pain: concepts, challenges, and compromises. Pain. 2020;161(9):1976–1982. doi:10.1097/j.pain.0000000000001939
2. Geneen LJ, Moore RA, Clarke C, Martin D, Colvin LA, Smith BH. Physical activity and exercise for chronic pain in adults: an overview of Cochrane reviews. Cochrane Database Syst Rev. 2017;1(1):Cd011279. doi:10.1002/14651858.CD011279.pub2
3. Cooper MA, Kluding PM, Wright DE. Emerging relationships between exercise, sensory nerves, and neuropathic pain. Front Neurosci. 2016;10:372. doi:10.3389/fnins.2016.00372
4. Sluka KA, Frey-Law L, Hoeger Bement M. Exercise-induced pain and analgesia? Underlying mechanisms and clinical translation. Pain. 2018;159(Suppl1):S91–S97. doi:10.1097/j.pain.0000000000001235
5. Song JS, Yamada Y, Kataoka R, et al. Training-induced hypoalgesia and its potential underlying mechanisms. Neurosci Biobehav Rev. 2022;141:104858. doi:10.1016/j.neubiorev.2022.104858
6. Cheng X, Yu Z, Hu W, et al. Voluntary exercise ameliorates neuropathic pain by suppressing calcitonin gene-related peptide and ionized calcium-binding adapter molecule 1 overexpression in the lumbar dorsal horns in response to injury to the cervical spinal cord. Exp Neurol. 2022;354:114105. doi:10.1016/j.expneurol.2022.114105
7. Chuganji S, Nakano J, Sekino Y, Hamaue Y, Sakamoto J, Okita M. Hyperalgesia in an immobilized rat hindlimb: effect of treadmill exercise using non-immobilized limbs. Neurosci Lett. 2015;584:66–70. doi:10.1016/j.neulet.2014.09.054
8. Lopez-Alvarez VM, Puigdomenech M, Navarro X, Cobianchi S. Monoaminergic descending pathways contribute to modulation of neuropathic pain by increasing-intensity treadmill exercise after peripheral nerve injury. Exp Neurol. 2018;299(Pt A):42–55. doi:10.1016/j.expneurol.2017.10.007
9. Ma XQ, Qin J, Li HY, Yan XL, Zhao Y, Zhang LJ. Role of exercise activity in alleviating neuropathic pain in diabetes via inhibition of the pro-inflammatory signal pathway. Biol Res Nurs. 2019;21(1):14–21. doi:10.1177/1099800418803175
10. Tian J, Yu T, Xu Y, et al. Swimming training reduces neuroma pain by regulating neurotrophins. Med Sci Sports Exerc. 2018;50(1):54–61. doi:10.1249/MSS.0000000000001411
11. Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87(2):149–158. doi:10.1016/S0304-3959(00)00276-1
12. Kami K, Taguchi S, Tajima F, Senba E. Improvements in impaired GABA and GAD65/67 production in the spinal dorsal horn contribute to exercise-induced hypoalgesia in a mouse model of neuropathic pain. Mol Pain. 2016;12:
13. Bai J, Geng B, Wang X, et al. Exercise facilitates the M1-to-M2 polarization of microglia by enhancing autophagy via the BDNF/AKT/mTOR pathway in neuropathic pain. Pain Physician. 2022;25(7):E1137–E1151.
14. Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi:10.1016/0165-0270(94)90144-9
15. Zhang E, Yi MH, Shin N, et al. Endoplasmic reticulum stress impairment in the spinal dorsal horn of a neuropathic pain model. Sci Rep. 2015;5:11555. doi:10.1038/srep11555
16. Ma X, Du W, Wang W, et al. Persistent Rheb-induced mTORC1 activation in spinal cord neurons induces hypersensitivity in neuropathic pain. Cell Death Dis. 2020;11(9):747. doi:10.1038/s41419-020-02966-0
17. Wu Y, Fu Q, Huang X, et al. NWD1 facilitates synaptic transmission and contributes to neuropathic pain. Neuropharmacology. 2022;205:108919. doi:10.1016/j.neuropharm.2021.108919
18. Feng H, Lu G, Li Q, Liu Z. Inhibition of adenylyl cyclase in the spinal cord alleviates painful diabetic neuropathy in Zucker diabetic fatty rats. Can J Diabetes. 2017;41(2):177–183. doi:10.1016/j.jcjd.2016.09.006
19. Liu X, Zhu M, Ju Y, Li A, Sun X. Autophagy dysfunction in neuropathic pain. Neuropeptides. 2019;75:41–48. doi:10.1016/j.npep.2019.03.005
20. Berliocchi L, Maiarù M, Varano GP, et al. Spinal autophagy is differently modulated in distinct mouse models of neuropathic pain. Mol Pain. 2015;11:3. doi:10.1186/1744-8069-11-3
21. Feng T, Yin Q, Weng ZL, et al. Rapamycin ameliorates neuropathic pain by activating autophagy and inhibiting interleukin-1β in the rat spinal cord. J Huazhong Univ Sci Technolog Med Sci. 2014;34(6):830–837. doi:10.1007/s11596-014-1361-6
22. Hutchinson KJ, Gómez-Pinilla F, Crowe MJ, Ying Z, Basso DM. Three exercise paradigms differentially improve sensory recovery after spinal cord contusion in rats. Brain. 2004;127(Pt 6):1403–1414. doi:10.1093/brain/awh160
23. Stagg NJ, Mata HP, Ibrahim MM, et al. Regular exercise reverses sensory hypersensitivity in a rat neuropathic pain model: role of endogenous opioids. Anesthesiology. 2011;114(4):940–948. doi:10.1097/ALN.0b013e318210f880
24. López-álvarez VM, Modol L, Navarro X, Cobianchi S. Early increasing-intensity treadmill exercise reduces neuropathic pain by preventing nociceptor collateral sprouting and disruption of chloride cotransporters homeostasis after peripheral nerve injury. Pain. 2015;156(9):1812–1825. doi:10.1097/j.pain.0000000000000268
25. Aoki Y, Nishizawa D, Yoshida K, et al. Association between the rs7583431 single nucleotide polymorphism close to the activating transcription factor 2 gene and the analgesic effect of fentanyl in the cold pain test. Neuropsychopharmacol Rep. 2018;38(2):86–91. doi:10.1002/npr2.12012
26. Salinas-Abarca AB, Velazquez-Lagunas I, Franco-Enzástiga Ú, Torres-López JE, Rocha-González HI, Granados-Soto V. ATF2, but not ATF3, participates in the maintenance of nerve injury-induced tactile allodynia and thermal hyperalgesia. Mol Pain. 2018;14:1744806918787427. doi:10.1177/1744806918787427
27. Zhuang ZY, Kawasaki Y, Tan PH, Wen YR, Huang J, Ji RR. Role of the CX3CR1/p38 MAPK pathway in spinal microglia for the development of neuropathic pain following nerve injury-induced cleavage of fractalkine. Brain Behav Immun. 2007;21(5):642–651. doi:10.1016/j.bbi.2006.11.003
28. Peng Z, Zha L, Yang M, Li Y, Guo X, Feng Z. Effects of ghrelin on pGSK-3β and β-catenin expression when protects against neuropathic pain behavior in rats challenged with chronic constriction injury. Sci Rep. 2019;9(1):14664. doi:10.1038/s41598-019-51140-w
29. Zhang LY, Liu ZH, Zhu Q, et al. Resolvin D2 relieving radicular pain is associated with regulation of inflammatory mediators, Akt/GSK-3β signal pathway and GPR18. Neurochem Res. 2018;43(12):2384–2392. doi:10.1007/s11064-018-2666-9
30. Martins DF, Rosa AO, Gadotti VM, et al. The antinociceptive effects of AR-A014418, a selective inhibitor of glycogen synthase kinase-3 beta, in mice. J Pain. 2011;12(3):315–322. doi:10.1016/j.jpain.2010.06.007
31. Parkitna JR, Obara I, Wawrzczak-Bargiela A, Makuch W, Przewlocka B, Przewlocki R. Effects of glycogen synthase kinase 3beta and cyclin-dependent kinase 5 inhibitors on morphine-induced analgesia and tolerance in rats. J Pharmacol Exp Ther. 2006;319(2):832–839. doi:10.1124/jpet.106.107581
32. Guan XH, Lu XF, Zhang HX, et al. Phosphatidylinositol 3-kinase mediates pain behaviors induced by activation of peripheral ephrinBs/EphBs signaling in mice. Pharmacol Biochem Behav. 2010;95(3):315–324. doi:10.1016/j.pbb.2010.02.007
33. Le Stunff C, Dechartres A, Mariot V, et al. Association analysis indicates that a variant GATA-binding site in the PIK3CB promoter is a Cis-acting expression quantitative trait locus for this gene and attenuates insulin resistance in obese children. Diabetes. 2008;57(2):494–502. doi:10.2337/db07-1273
34. Zhuang ZY, Xu H, Clapham DE, Ji RR. Phosphatidylinositol 3-kinase activates ERK in primary sensory neurons and mediates inflammatory heat hyperalgesia through TRPV1 sensitization. J Neurosci. 2004;24(38):8300–8309. doi:10.1523/JNEUROSCI.2893-04.2004
35. Yang RH, Lin J, Hou XH, et al. Effect of docosahexaenoic acid on hippocampal neurons in high-glucose condition: involvement of PI3K/AKT/nuclear factor-κB-mediated inflammatory pathways. Neuroscience. 2014;274:218–228. doi:10.1016/j.neuroscience.2014.05.042
36. Jaber N, Dou Z, Chen JS, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109(6):2003–2008. doi:10.1073/pnas.1112848109
37. Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi:10.1038/nri3532
38. Tang J, Li ZH, Ge SN, et al. The inhibition of spinal astrocytic JAK2-STAT3 pathway activation correlates with the analgesic effects of triptolide in the rat neuropathic pain model. Evid Based Complement Alternat Med. 2012;2012:185167. doi:10.1155/2012/185167
39. Wang ZF, Li Q, Liu SB, et al. Aspirin-triggered Lipoxin A4 attenuates mechanical allodynia in association with inhibiting spinal JAK2/STAT3 signaling in neuropathic pain in rats. Neuroscience. 2014;273:65–78. doi:10.1016/j.neuroscience.2014.04.052
40. Zhang X, Wu J, Lei Y, Fang L, Willis WD. Protein phosphatase 2A regulates central sensitization in the spinal cord of rats following intradermal injection of capsaicin. Mol Pain. 2006;2:9. doi:10.1186/1744-8069-2-9
41. Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18(9):1865. doi:10.3390/ijms18091865
42. Berliocchi L, Russo R, Maiarù M, Levato A, Bagetta G, Corasaniti MT. Autophagy impairment in a mouse model of neuropathic pain. Mol Pain. 2011;7:
43. Shi J, Jiang K, Li Z. MiR-145 ameliorates neuropathic pain via inhibiting inflammatory responses and mTOR signaling pathway by targeting Akt3 in a rat model. Neurosci Res. 2018;134:10–17. doi:10.1016/j.neures.2017.11.006
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.