Back to Journals » OncoTargets and Therapy » Volume 13
Progress in Understanding the IL-6/STAT3 Pathway in Colorectal Cancer
Authors Lin Y, He Z , Ye J, Liu Z, She X, Gao X, Liang R
Received 20 August 2020
Accepted for publication 1 December 2020
Published 21 December 2020 Volume 2020:13 Pages 13023—13032
DOI https://doi.org/10.2147/OTT.S278013
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Yan Lin,1 Ziqin He,1 Jiazhou Ye,2 Ziyu Liu,1 Xiaomin She,1 Xing Gao,1 Rong Liang1
1Department of Medical Oncology, Guangxi Medical University Cancer Hospital, Nanning, Guangxi 530021, People’s Republic of China; 2Department of Hepatobiliary Surgery, Guangxi Medical University Cancer Hospital, Nanning, Guangxi 530021, People’s Republic of China
Correspondence: Rong Liang
Department of Medical Oncology, Guangxi Medical University Cancer Hospital, Nanning, Guangxi 530021, People’s Republic of China
Tel +86 771 5333058
Email [email protected]
Abstract: As a pleiotropic cytokine, interleukin-6 (IL-6) not only regulates the cellular immune response, but it also promotes tumor development by activating multiple carcinogenic pathways. IL-6 expression is significantly elevated in colorectal cancer (CRC) and is closely related to CRC development and patient prognosis. In CRC, IL-6 activates signal transducers and activators of transduction-3 (STAT3) to promote tumor initiation and tumor growth. IL-6/STAT3 signalling has a profound effect on tumor-infiltrating immune cells in the tumor immune microenvironment in CRC. Additionally, IL-6/STAT3 pathway activates downstream target genes to protect tumor cells from apoptosis; drive tumor cell proliferation, cell cycle progression, invasion and metastasis; promote tumor angiogenesis; and stimulate drug resistance. Therefore, a thorough understanding of the many effects of the IL-6/STAT3 pathway in CRC is needed, which the present review examines.
Keywords: IL-6/STAT3 pathway, colorectal cancer, tumor development
Introduction
Colorectal cancer (CRC) is one of the 10 most frequent malignancies in the world. In 2018, there were more than 1.8 million new colorectal cancers worldwide, accounting for about 10% of all cancer cases, and the number of related deaths was 881,000, making it the second-deadliest cancer in the world.1 Incidence of CRC in China has been increasing due to changes in lifestyle and diet, with more than 521,000 new cases in 2018, when it was the second most common malignancy in the country.2 The increasing burden of CRC and related mortality highlight the need for early detection and prevention.3
Many factors are involved in the pathogenesis of CRC, including intestinal flora disorder, abnormal immune response, as well as genetic, environmental and lifestyle risk factors such as obesity, alcohol consumption, smoking, poor diet, and lack of exercise. Intestinal inflammation is one of the most important factors leading to CRC and is associated with dysregulation of numerous signaling pathways.4,5 Among them is the IL-6/STAT3 pathway, which promotes CRC cell proliferation and survival.6,7 CRC patients show significantly higher IL-6 levels than healthy individuals, and those levels correlate with tumor size, stage, metastasis, and survival rate.8–11 IL-6 appears to help drive CRC by activating the downstream signaling factor STAT3.12
The present review examines recent progress in understanding the many mechanisms through which the IL-6/STAT3 pathway contributes to CRC.
The IL-6/STAT3 Pathway
First cloned in 1986, IL-6 was found to be a pleiotropic cytokine playing roles in immune regulation, hematopoiesis, inflammation and tumorigenesis.13 IL-6 activates downstream signal pathways by forming complexes with its receptor, which consists of two subunits: a ligand-binding protein IL-6R (also called IL-6Ra or CD126), with a molecular weight of 80 kDa; and a signal-transducing glycoprotein-130 (gp130, IL-6Rb, CD130), with a molecular weight of 130 kDa.14
In the classical IL-6 signaling pathway, extracellular IL-6 and membrane-bound IL-6R (mIL-6R) combine to form a complex to which gp130 binds, yielding an iso-hexameric complex consisting of two IL-6, two IL-6R and two gp130 molecules. This complex activates the Janus kinase (JAK), which in turn causes STAT3 to dimerize and translocate to the nucleus to alter the expression of target genes.15 The main function of the classical pathway is to induce anti-inflammatory effects during the acute-phase response.16
Alternatively, trans IL-6 signaling can occur, which is the same as the classical pathway except that IL-6 binds to soluble IL-6R (sIL-6R) rather than mIL-6R.17 The soluble receptor sIL-6R is produced by limited proteolysis of mIL-6R or alternative splicing of the IL-6R mRNA.18,19 The main function of trans signaling is to promote an inflammatory response,16 so this pathway appears to contribute to cancers such as CRC (Figure 1).20,21
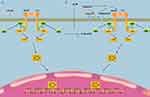 |
Figure 1 The IL-6/STAT3 signaling pathway. (A) In the classical IL-6 signaling pathway, IL-6 binds to mIL-6 on the cell membrane to form a complex, which induces gp130 to form a heterohexamer, which then initiates the JAK/STAT3 pathway. (B) In the trans IL-6 signaling pathway, IL-6 complexes with sIL-6R, previously generated by variable splicing of the IL-6 mRNA or as a result of IL-6R cleavage by metalloproteinase (ADAM) 10/17 or meprin metalloproteinase α/β.18,19,25 The IL-6/sIL-6R complex then complexes with gp130 via intermolecular disulfide bonds26. The Box-1 and −2 domains in the cytoplasmic domain of gp130 bind and activate JAK,15 which phosphorylates tyrosine residues in the cytoplasmic region of gp130. The phosphorylated pTyr-X-X-Gln motif on gp130 (X = any amino acid) recruits the Src homology 2 (SH2) domain in STAT3.27,28 An adjacent JAK phosphorylates the conserved Tyr705 in STAT3, which then homodimerizes with another STAT3 via the SH2 domain. This dimer translocates to the nucleus, where it regulates expression of target genes.29 |
One or the other pathway may predominate in different cellular contexts. For example, mIL-6R is expressed mainly on neutrophils, monocytes, activated B cells, CD4+ cells, and hepatocytes, so these cell types participate primarily in the classical pathway.22 At the same time, the IL-6/sIL-6R complex in serum can potentially affect a broad range of cell types because gp130 is ubiquitously expressed in all cells.23,24
The Role of IL-6/STAT3 Signaling in the Immune Microenvironment of CRC
CRC formation is influenced by the intricate interactions between cancerous cells and the tumor microenvironment (TME). The immune components in the TME are called the tumor immune microenvironment (TIME), which can modulate tumour occurrence and development. The TIME is composed of various infiltrating immune cells [e.g. tumor-associated macrophages, T helper type 17 (Th17) cells, cancer-associated fibroblasts], tumor-associated endothelial cells as well as the extracellular matrix and complicated vasculature.30 Various signaling pathways, such as IL-6/STAT3 pathways, are activated in the TIME and influences the growth and progression of CRC.
During CRC progression from occurrence to development, IL-6 expression is significantly elevated and is involved in multiple processes of tumor development.8,9 Various cells have been identified as sources of IL-6 in the TIME. For instance, expression of IL-6 has been linked to macrophages, fibroblasts, dendritic cells, lymphocytes and CRC cells.31,32
Tumor-associated macrophages (TAMs) are major components of the TIME that are frequently associated with tumor metastasis in CRC. Studies have shown that TAMs promote migration and invasion by CRC cells.33 Emerging studies have suggested that tumor-derived IL-6 can enhance the phagocytic capacity and migration of macrophages in the TIME via STAT3 phosphorylation, but the exact mechanism requires further study.32 One study showed that TAM-derived IL-6 activates the JAK2/STAT3/miR-506-3p/FoxQ1 axis to modulate CRC cell migration and invasion.34 The available evidence suggests that IL-6/STAT3 signaling promotes interaction between macrophages and factors secreted by CRC cells into the TIME.
As a specialized subset of CD4+ cells, Th17 cells and their cytokines are involved in regulation of the immune system and cancer development.35 In the joint presence of IL-6 and TGF-β, Th17 cells differentiate from naive T cells are involved in intestinal inflammation.36 Those two and related cytokines also regulate the expansion of Th17 cells in CRC37. Down-regulation of IL-6 can reverse the Th17-driven carcinogenic process in murine colon cancer.38 IL-6 signaling drives Th17 cell differentiation in colitis-associated CRC by phosphorylating and activating STAT3, and Th17 cytokines overexpressed in CRC patients (IL-17A, IL-17F, IL-21, IL-22) can promote tumor angiogenesis and oncogenesis.30,39
Cancer-associated fibroblasts (CAFs) secrete factors that influence the TIME and CRC growth.40 Recently, CAFs have been demonstrated to be an important source of IL-6.31 IL-6-mediated STAT3 activation in CRC-CAFs promotes colorectal tumor development. STAT3-induced activation of vascular endothelial growth factor (VEGF) and proliferation-associated genes contribute to CRC initiation and growth.41 CRC cells further augment IL-6 secretion from CAFs, but specific mechanisms still need to be revealed.42 These studies indicate that CAFs produce abundant amounts of IL-6, and that CRC cells facilitate the process. IL-6/STAT3 signaling drives CAF activation in CRC, enhancing tumor progression.
So far, potential correlations between IL-6/STAT3 signaling and other immune cells in CRC, such as Treg cells, myeloid-derived stromal cells, and B lymphocytes, have not been reported. These are an important topic for future research.
The IL-6/STAT3 Signaling Pathway Promotes CRC Development
In CRC, the continuous activation of STAT3 by IL-6 signaling drives many malignant pathways in tumor cells, including cell cycle progression, proliferation, antiapoptosis, invasion and metastasis, the epithelial-mesenchymal transition (EMT), angiogenesis and drug resistance (Table 1).
 |
Table 1 Mechanisms Through Which the IL-6/STAT3 Signaling Pathway Promotes CRC Malignancy |
Cell Cycle Progression
The IL-6/STAT3 signaling pathway can drive progression through the cell cycle and thereby promote proliferation of CRC cells. Most CRC patients overexpress c-Myc, which up-regulates oncogenic proteins and non-coding RNAs that drive the cell cycle, differentiation, growth and metabolism,43,44 and higher c-Myc levels correlate with more severe disease and worse prognosis.45,46 Continuous STAT3 activation by the IL-6/IL-6R complex in CRC activates c-Myc and triggers metabolic disorder and tumor progression.47 STAT3 appears to up-regulate c-Myc by binding to the E2F site (98TTGGCGGGAAA106) in the c-Myc P2 promoter.48
The IL-6/STAT3 signaling pathway up-regulates cell cycle protein D1 (cyclinD1) in CRC. STAT3 binds to the so-called GAS site in the cyclinD1 promoter.49 CyclinD1 drives progression from G1 to S phase of the cell cycle, leading to cell proliferation.50,51 Cyclin D1 expression negatively correlates with overall and disease-free survival.52 A tea polysaccharide that inhibits IL-6/STAT3 signaling in CT26 mouse colon cancer cells concomitantly down-regulates cyclinD1.53
The IL-6/STAT3 signaling pathway promotes CRC cell growth also by up-regulating mitochondrial single-stranded DNA-binding protein (mtSBB). IL-6/STAT3 signaling induces the transcription factor FOXP1 to bind to the region between nt −800 and −700 in the mtSSB gene promoter, turning on the gene. The expressed mtSSB activates the ROS/Akt/mTOR pathway, which up-regulates telomerase reverse transcriptase (TERT), which stabilizes telomeres and thereby helps immortalize CRC cells.54
Inhibition of Apoptosis
STAT3 induces the expression of anti-apoptotic genes, which was demonstrated in studies of a mouse model of CRC treated with aspirin. The drug inhibited IL-6 as well as STAT3 phosphorylation, reversing STAT3-induced expression of the anti-apoptosis proteins Bcl-2 and Bcl-xl.55
The IL-6/STAT3 signaling pathway also up-regulates the anti-apoptotic protein Mcl-1, which protects CRC cells from apoptosis induced by tumor necrosis factor-associated apoptotic ligand (TRAIL).56 STAT3 binds to the so-called SIE element in the Mcl-1 gene promoter.57
IL-6/STAT3 signaling up-regulates the anti-apoptotic protein survivin,53 which inhibits apoptosis and promotes proliferation and angiogenesis. Survivin is up-regulated in various tumors,58 and its expression in CRC correlates positively with vascular infiltration and lymph node metastasis, and negatively with overall survival.59 STAT3 has been shown to bind the survivin promoter.60,61
Invasion, Metastasis and EMT
The IL-6/STAT3 signaling pathway can enhance invasiveness and metastasis in CRC by inducing the expression of related oncogenes such as the gene encoding Fos-related antigen-1 (Fra-1).62 After STAT3 is activated through acetylation of Lys685 and phosphorylation of Tyr705, it binds to the promoter of the Fra-1 gene approximately 600 bases upstream of the transcription initiation site and up-regulates its expression. The expressed Fra-1, in turn, up-regulates EMT-inducing transcription factors and matrix metalloproteinases (MMPs)-2 and −9.63
STAT3 in CRC down-regulates the microRNA-34a (miR-34a) by binding to its first intron. This miRNA normally inhibits the EMT and cancer development by down-regulating Snail 1. Thus, STAT3-mediated down-regulation of miR-34a up-regulates Snail 1. At the same time, it up-regulates IL-6R, because this mRNA is normally inhibited by miR-34a. The resulting up-regulation of IL-6R further enhances IL-6/STAT3 signaling.64,65 Third, the IL-6/STAT3 signaling pathway down-regulates E-cadherin and up-regulates vimentin and the transcription factor Twist.66 The net result of all these processes is promotion of the EMT and of CRC invasiveness and metastasis.
STAT3 can regulate FoxQ1 through miR-506-3p. As a transcription factor regulating the EMT, FoxQ1 promotes the EMT of CRC cells by inducing mesenchymal gene expression. Studies have shown that FoxQ1 is a direct target of miR-506-3p, and the latter can down-regulate FoxQ1 by directly binding its 3′UTR. STAT3, in contrast, inhibits miR-506-3p through its STAT3 binding site. In this way, IL-6 regulates the STAT3/miR-506-3p/FoxQ1 axis to induce the EMT and enhance CRC migration and invasion.34
Integrin β6 participates in IL-6-induced EMT and tumor cell invasion in CRC. IL-6-mediated activation of STAT3 can rapidly induce integrin β6 transcription, and the up-regulated integrin β6 inhibits E-cadherin and enhances vimentin expression to advance the EMT.42 Further investigations are needed to uncover how STAT3 induces integrin β6 expression.
STAT3 can up-regulate hypoxia-inducing factor-1α (HIF-1α),67,68 which binds to the positive regulatory element EP-1 (−153 to −148 bp) in the gene encoding carcinoembryonic antigen (CEA), inducing its expression.69 CEA helps drive migration, invasion and metastasis of CRC cells and is an independent prognostic factor.70,71
Tumor Angiogenesis
STAT3 binds to the promoter of the gene encoding vascular endothelial growth factor (VEGF), inducing its expression.72,73 VEGF stimulates the formation of tumor blood vessels, which provide nutrition and oxygen to sustain tumor growth and allow metastasis. Like levels of IL-6, levels of VEGF are elevated in CRC and correlate with disease progression.74,75
VEGF can bind to several receptors (VEGFRs), whose expression varies across tissue types. Which VEGFRs mediate the angiogenic effects of IL-6/STAT3 signaling in CRC remains to be established. A likely candidate is VEGFR2, which is up-regulated in intestinal epithelial cells by IL-6 and which mediates the angiogenic effects of VEGF in colitis-associated cancer.76
Tumor Resistance to Chemotherapy
Levels of STAT3 phosphorylated on Tyr705 positively correlate with resistance of CRC cells to chemoradiotherapy involving 5-fluorouracil, and inhibiting STAT3 renders CRC cells more sensitive to chemoradiotherapy.77 By up-regulating HIF-1α (see section 2.3), STAT3 down-regulates the downstream HIF-1α target miR-338-5P. This miRNA normally down-regulates IL-6, so the activation of STAT3 up-regulates IL-6, leading to a positive feedback loop that confers resistance to the drugs oxaliplatin and 5-fluorouracil, but details of the drug resistance mechanism are unclear.78 The caspase pathway may mediate the ability of exosomes to transfer STAT3 phosphorylated on Tyr705 and thereby promote resistance to 5-fluorouracil.79
High serum levels of IL-6 may reduce the therapeutic efficacy of the anti-VEGF antibody bevacizumab in metastatic CRC,80 and whether and how this involves STAT3 activity remains to be elucidated.
Clinical Investigations of the IL-6/STAT3 Signaling Pathway in CRC Therapy
Relevant literature has established that downstream target genes mediated by aberrant activation of the IL-6/STAT3 pathway are involved in the development and progression of CRC, and thus targeting the IL-6/STAT3 pathway is highly likely to be a viable and effective approach for the treatment of CRC. Currently, clinical trials are underway for a number of drugs targeting the CRC IL-6/STAT3 pathway, such as IL-6 inhibitors (siltuximab), JAK inhibitors (itacitinib), and STAT3 inhibitors (OPB-31,121, AZD9150, and TTI-101) (Table 2).
 |
Table 2 Clinical Trials Targeting the CRC IL-6/STAT3 Pathway |
Siltuximab, a monoclonal antibody against IL-6, is currently the only drug approved by the US Food and Drug Administration for multicentric Castleman disease (MCD).81 Siltuximab monotherapy, however, does not show efficacy against advanced solid tumors, including CRC. In that CRC trial, the Phase II primary efficacy endpoint was complete response, partial response, or stable disease > 6 weeks, and only 5 of the 84 patients achieved stable disease > 6 weeks. Although adverse events occurred in 98% of treated patients, they were driven primarily by underlying metastatic disease. The majority of drug-related adverse events were low-grade: 29 patients (35%) had grade 1–2 adverse events (fatigue, nausea, constipation, etc.), and 10% (8) had grade > 3 adverse events (neutropenia, leukopenia, lymphocytopenia). These results suggest that IL-6 inhibition alone offers limited clinical benefit to advanced CRC patients. Other parallel pathways, including the IL-6/STAT3 pathway, may similarly regulate the development of CRC, and therefore the development of combination therapies may provide more benefit for advanced CRC.82
Regorafenib is a multi-targeted kinase inhibitor that improves OS in patients with metastatic CRC and has been approved for the treatment of metastatic CRC (mCRC).83 Incyte Corporation performed a clinical trial to test the combination of ruxolitinib, a selective inhibitor of JAK1/JAK2, with regorafenib for the treatment of refractory mCRC.84 Although the combination did not lead to more adverse effects, it did not improve efficacy over regorafenib alone. The 396 patients included in the trial were randomized into two subgroups. In subgroup 1, the ruxolitinib group (n=87) showed median overall survival of 4.6 (95% CI, 3.5–5.4) months and median progression-free survival of 2.2 (95% CI, 1.9–3.0) months, while the corresponding survival times in the placebo group (n=88) were 5.3 (95% CI, 4.3–6.0) and 2.1 (95% CI, 1.8–2.7) months. In subgroup 2, the ruxolitinib group (n=110) showed corresponding survival times of 11.4 (95% CI, 9.0–13.2) and 3.5 (95% CI, 3.0–3.8) months, while the placebo group (n=111) showed times of 10.9 (95% CI, 7.2- not estimated) and 2.0 (95% CI, 1.9–3.1) months. The differences between overall and progression-free survival were not significant in either subgroup. A clinical trial of ruxolitinib in combination with trametinib for RAS-mutant CRC is currently underway (NCT04303403) and was expected to conclude in June 2020, with no results yet published.
Itacitinib, a novel oral JAK1 selective inhibitor, exerts anti-inflammatory effects by inhibiting IL-6-driven phosphorylation STAT3.85 A Phase I clinical trial of itacitinib in combination with pembrolizumab in patients with CRC has been conducted (NCT02646748) and was expected to end on September 31, 2020, but no results have been released yet.
The development of STAT3 inhibitors has been one of the main focuses of medical researchers. OPB-31,121 is a novel STAT3 inhibitor with high affinity for the SH2 domain of STAT3, and it has demonstrated significant anticancer activity in preclinical studies.86 The Otsuka Pharmaceutical-led Phase I study showed that OPB-31,121 is safe and well tolerated, with good antitumor activity in patients with advanced CRC. Most drug-related adverse events are grade 1–2 and a maximum tolerated dose of 800 mg/day has been established.87
AZD9150, a STAT3 antisense oligonucleotide, directly inhibits STAT3 by promoting the destruction of STAT3 mRNA or inhibiting its translation, and it is the only STAT3 antisense molecule to have entered clinical trials.88 The MD Anderson Cancer Center launched a Phase II clinical trial of AZD9150 in combination with durvalumab, an anti-PDL-1 antibody, in patients with CRC (NCT02983578), and the trial is expected to end in March 2021. TTI-101, an oral inhibitor of STAT3 developed by Tvardi Therapeutics, has been shown in preclinical studies to inhibit the growth of a variety of solid tumors in mice, including liver, breast, lung, and head and neck cancers. A Phase I trial of TTI-101 for the treatment of advanced solid tumors, including CRC, has been carried out (NCT03195699). The novel STAT3 inhibitor, Bruceantinol (BOL), strongly inhibits STAT3 DNA-binding ability and thus blocks IL-6-induced STAT3 activation in CRC. BOL showed potent anticancer activity in human CRC models in vivo and in vitro, but it has yet to be tested in clinical trials.89 To date, no inhibitors targeting the IL-6/STAT3 pathway have been approved for CRC treatment, and therefore drugs targeting the CRC IL-6/STAT3 pathway need to be further developed.
Conclusion
In recent years, the high incidence and mortality rates of CRC have led to an increasing tumor burden. Investigating the mechanisms and treatments of CRC has become a major concern for researchers. As one of the key pathways in the development of CRC, the IL-6/SAT3 pathway not only directly regulates tumor immune cells and thus suppresses tumor immunity, but it also up-regulates the expression of numerous oncogenic proteins to help drive CRC. Thus, targeting components of the IL-6/STAT3 pathway can inhibit tumor cell progression and relieve immunosuppression in the TIME. Novel inhibitors of the IL-6/STAT3 pathway are being developed, and early phase clinical trials are also ongoing. However, many of the complex processes affected by IL-6/STAT3 signaling remain to be clarified, and such research may reveal new insights into CRC and how to combat it.
Abbreviations
IL-6, interleukin-6; CRC, colorectal cancer; STAT3, signal transducers and activators of transduction-3; IL-6R, IL-6 receptor; gp130, glycoprotein −130; mIL-6R, membrane-bound IL-6R; JAK, Janus kinase; sIL-6R, soluble IL-6R; TIME, tumor immune microenvironment; EMT, epithelial-mesenchymal transition; cyclinD1, cell cycle protein D1; mtSBB, mitochondrial single-strand DNA-binding protein; TERT, telomerase reverse transcriptase; TRAIL, tumor necrosis factor-associated apoptotic ligand; Fra-1, Fos-related antigen-1; MMPs, matrix metalloproteinases; miR-34a, microRNA-34a; HIF-1α, hypoxia-inducing factor-1α; VEGF, vascular endothelial growth factor; VEGFRs, vascular endothelial growth factor receptors.
Author Contributions
All authors made a significant contribution to the work reported. YL, RL and ZQH drafted and revised the manuscript. YL, RL, ZQH, ZYL and XG guided the writing of the manuscript, edited it and prepared it for submission. YL, JZY, RL and XMS critically revised the manuscript. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This research was supported by the National Natural Science Foundation of China (NO. 82060427,81660498 and 81803007), China Postdoctoral Science Foundation (NO.2019M663412), Guangxi Key Research and Development Plan (NO.GUIKEAB19245002), Guangxi Scholarship Fund of Guangxi Education Department, General Program of Guangxi Natural Science Foundation (NO. 2020GXNSFAA259080), Youth Talent Fund Project of Guangxi Natural Science Foundation (NO. 2018GXNSFBA281030, 2018GXNSFBA281091), Guangxi Medical and Health Appropriate Technology Development and Application Project (No. S2017101, S2018062), Guangxi Medical University Training Program for Distinguished Young Scholars, Science and Technology Plan Project of Qingxiu District, Nanning (NO.2020037, 2020038).
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 global cancer statistics. Cancer Commun (Lond). 2019;39(1):22. doi:10.1186/s40880-019-0368-6
3. Yin J, Bai Z, Zhang J, et al. Burden of colorectal cancer in China, 1990–2017: findings from the Global Burden of Disease Study 2017. Chin J Cancer Res. 2019;31(3):489–498. doi:10.21147/j.issn.1000-9604.2019.03.11
4. Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732.
5. Montalban-Arques A, Scharl M. Intestinal microbiota and colorectal carcinoma: implications for pathogenesis, diagnosis, and therapy. EBioMedicine. 2019;48:648–655. doi:10.1016/j.ebiom.2019.09.050
6. Francescone R, Hou V, Grivennikov SI. Cytokines, IBD, and colitis-associated cancer. Inflamm Bowel Dis. 2015;21(2):409–418. doi:10.1097/MIB.0000000000000236
7. Waldner MJ, Neurath MF. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin Immunol. 2014;26(1):75–79. doi:10.1016/j.smim.2013.12.003
8. Nikiteas NI, Tzanakis N, Gazouli M, et al. Serum IL-6, TNFalpha and CRP levels in Greek colorectal cancer patients: prognostic implications. World J Gastroenterol. 2005;11(11):1639–1643. doi:10.3748/wjg.v11.i11.1639
9. Zeng J, Tang ZH, Liu S, Guo SS. Clinicopathological significance of overexpression of interleukin-6 in colorectal cancer. World J Gastroenterol. 2017;23(10):1780–1786. doi:10.3748/wjg.v23.i10.1780
10. Chung YC, Chang YF. Serum interleukin-6 levels reflect the disease status of colorectal cancer. J Surg Oncol. 2003;83(4):222–226. doi:10.1002/jso.10269
11. Esfandi F, Mohammadzadeh Ghobadloo S, Basati G. Interleukin-6 level in patients with colorectal cancer. Cancer Lett. 2006;244(1):76–78. doi:10.1016/j.canlet.2005.12.003
12. Waldner MJ, Foersch S, Neurath MF. Interleukin-6–a key regulator of colorectal cancer development. Int J Biol Sci. 2012;8(9):1248–1253. doi:10.7150/ijbs.4614
13. Kishimoto T. IL-6: from its discovery to clinical applications. Int Immunol. 2010;22(5):347–352. doi:10.1093/intimm/dxq030
14. Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (review). Int J Oncol. 2014;44(4):1032–1040. doi:10.3892/ijo.2014.2259
15. Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248.
16. Baran P, Hansen S, Waetzig GH, et al. The balance of interleukin (IL)-6, IL-6·soluble IL-6 receptor (sIL-6R), and IL-6·sIL-6R·sgp130 complexes allows simultaneous classic and trans-signaling. J Biol Chem. 2018;293(18):6762–6775. doi:10.1074/jbc.RA117.001163
17. Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457. doi:10.1038/ni.3153
18. Prenissl N, Lokau J, Rose-John S, Haybaeck J, Garbers C. Therapeutic blockade of the interleukin-6 receptor (IL-6R) allows sIL-6R generation by proteolytic cleavage. Cytokine. 2019;114:1–5. doi:10.1016/j.cyto.2018.11.023
19. Zunke F, Rose-John S. The shedding protease ADAM17: physiology and pathophysiology. Biochim Biophys Acta Mol Cell Res. 2017;1864(11 Pt B):2059–2070. doi:10.1016/j.bbamcr.2017.07.001
20. Taher MY, Davies DM, Maher J. The role of the interleukin (IL)-6/IL-6 receptor axis in cancer. Biochem Soc Trans. 2018;46(6):1449–1462. doi:10.1042/BST20180136
21. Becker C, Fantini MC, Wirtz S, et al. IL-6 signaling promotes tumor growth in colorectal cancer. Cell Cycle. 2005;4(2):217–220. doi:10.4161/cc.4.2.1413
22. Narazaki M, Kishimoto T. The two-faced cytokine IL-6 in host defense and diseases. Int J Mol Sci. 2018;19(11). doi:10.3390/ijms19113528
23. Mauer J, Denson JL, Brüning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36(2):92–101. doi:10.1016/j.it.2014.12.008
24. Aparicio-Siegmund S, Deseke M, Lickert A, Garbers C. Trans-signaling of interleukin-6 (IL-6) is mediated by the soluble IL-6 receptor, but not by soluble CD5. Biochem Biophys Res Commun. 2017;484(4):808–812. doi:10.1016/j.bbrc.2017.01.174
25. Arnold P, Boll I, Rothaug M, et al. Meprin metalloproteases generate biologically active soluble interleukin-6 receptor to induce trans-signaling. Sci Rep. 2017;7:44053. doi:10.1038/srep44053
26. Murakami M, Hibi M, Nakagawa N, et al. IL-6-induced homodimerization of gp130 and associated activation of a tyrosine kinase. Science. 1993;260(5115):1808–1810. doi:10.1126/science.8511589
27. Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin Sci (Lond). 2012;122(4):143–159. doi:10.1042/CS20110340
28. Tanaka T, Narazaki M, Kishimoto T. Interleukin (IL-6) immunotherapy. Cold Spring Harb Perspect Biol. 2018;10:8. doi:10.1101/cshperspect.a028456
29. Li B, Huang C. Regulation of EMT by STAT3 in gastrointestinal cancer (review). Int J Oncol. 2017;50(3):753–767. doi:10.3892/ijo.2017.3846
30. Zhang Y, Xu J, Zhang N, Chen M, Wang H, Zhu D. Targeting the tumour immune microenvironment for cancer therapy in human gastrointestinal malignancies. Cancer Lett. 2019;458:123–135. doi:10.1016/j.canlet.2019.05.017
31. Huynh PT, Beswick EJ, Coronado YA, et al. CD90(+) stromal cells are the major source of IL-6, which supports cancer stem-like cells and inflammation in colorectal cancer. Int J Cancer. 2016;138(8):1971–1981. doi:10.1002/ijc.29939
32. Yeh KY, Wu TH, Wu TL. Colorectal cancer cell-derived interleukin-6 enhances the phagocytic capacity and migration of THP-1 cells. Cytokine. 2016;79:82–89. doi:10.1016/j.cyto.2016.01.001
33. Zhong X, Chen B, Yang Z. The role of tumor-associated macrophages in colorectal carcinoma progression. Cell Physiol Biochem. 2018;45(1):356–365. doi:10.1159/000486816
34. Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. doi:10.1186/s12943-019-0976-4
35. Chang SH. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch Pharm Res. 2019;42(7):549–559. doi:10.1007/s12272-019-01146-9
36. Lee GR. The balance of Th17 versus treg cells in autoimmunity. Int J Mol Sci. 2018;19:3.
37. Wang J, Xu K, Wu J, et al. The changes of Th17 cells and the related cytokines in the progression of human colorectal cancers. BMC Cancer. 2012;12:418. doi:10.1186/1471-2407-12-418
38. Erdman SE, Poutahidis T. Roles for inflammation and regulatory T cells in colon cancer. Toxicol Pathol. 2010;38(1):76–87. doi:10.1177/0192623309354110
39. Velikova TV, Miteva L, Stanilov N, Spassova Z, Stanilova SA. Interleukin-6 compared to the other Th17/Treg related cytokines in inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2020;26(16):1912–1925. doi:10.3748/wjg.v26.i16.1912
40. Unterleuthner D, Neuhold P, Schwarz K, et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis. 2020;23(2):159–177. doi:10.1007/s10456-019-09688-8
41. Heichler C, Scheibe K, Schmied A, et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut. 2020;69(7):1269–1282.
42. Sun Q, Shang Y, Sun F, Dong X, Niu J, Li F. Interleukin-6 promotes epithelial-mesenchymal transition and cell invasion through integrin β6 upregulation in colorectal cancer. Oxid Med Cell Longev. 2020;2020:8032187. doi:10.1155/2020/8032187
43. Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi:10.1016/j.cell.2012.03.003
44. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi:10.1038/nature11252
45. Rochlitz CF, Herrmann R, de Kant E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology. 1996;53(6):448–454. doi:10.1159/000227619
46. Lee KS, Kwak Y, Nam KH, et al. c-MYC copy-number gain is an independent prognostic factor in patients with colorectal cancer. PLoS One. 2015;10(10):e0139727. doi:10.1371/journal.pone.0139727
47. Zhang S, Li J, Xie P, et al. STAT3/c-Myc axis-mediated metabolism alternations of inflammation-related glycolysis involve with colorectal carcinogenesis. Rejuvenation Res. 2019;22(2):138–145. doi:10.1089/rej.2018.2089
48. Kiuchi N, Nakajima K, Ichiba M, et al. STAT3 is required for the gp130-mediated full activation of the c-myc gene. J Exp Med. 1999;189(1):63–73. doi:10.1084/jem.189.1.63
49. Leslie K, Lang C, Devgan G, et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 2006;66(5):2544–2552. doi:10.1158/0008-5472.CAN-05-2203
50. Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology. 2004;145(12):5439–5447. doi:10.1210/en.2004-0959
51. Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14(24):3102–3114. doi:10.1101/gad.854900
52. Li Y, Wei J, Xu C, Zhao Z, You T, Katoh M. Prognostic significance of cyclin D1 expression in colorectal cancer: a meta-analysis of observational studies. PLoS One. 2014;9(4):e94508. doi:10.1371/journal.pone.0094508
53. Liu LQ, Nie SP, Shen MY, et al. Tea polysaccharides inhibit colitis-associated colorectal cancer via interleukin-6/STAT3 pathway. J Agric Food Chem. 2018;66(17):4384–4393. doi:10.1021/acs.jafc.8b00710
54. Wang G, Wang Q, Huang Q, et al. Upregulation of mtSSB by interleukin-6 promotes cell growth through mitochondrial biogenesis-mediated telomerase activation in colorectal cancer. Int J Cancer. 2019;144(10):2516–2528. doi:10.1002/ijc.31978
55. Tian Y, Ye Y, Gao W, et al. Aspirin promotes apoptosis in a murine model of colorectal cancer by mechanisms involving downregulation of IL-6-STAT3 signaling pathway. Int J Colorectal Dis. 2011;26(1):13–22. doi:10.1007/s00384-010-1060-0
56. Lee DH, Sung KS, Bartlett DL, Kwon YT, Lee YJ. HSP90 inhibitor NVP-AUY922 enhances TRAIL-induced apoptosis by suppressing the JAK2-STAT3-Mcl-1 signal transduction pathway in colorectal cancer cells. Cell Signal. 2015;27(2):293–305. doi:10.1016/j.cellsig.2014.11.013
57. Liu H, Ma Y, Cole SM, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102(1):344–352.
58. Ryan BM, O’Donovan N, Duffy MJ. Survivin: a new target for anti-cancer therapy. Cancer Treat Rev. 2009;35(7):553–562. doi:10.1016/j.ctrv.2009.05.003
59. Krieg A, Werner TA, Verde PE, Stoecklein NH, Knoefel WT, Srinivasula SM. Prognostic and clinicopathological significance of survivin in colorectal cancer: a meta-analysis. PLoS One. 2013;8(6):e65338. doi:10.1371/journal.pone.0065338
60. Boidot R, Végran F, Lizard-Nacol S. Transcriptional regulation of the survivin gene. Mol Biol Rep. 2014;41(1):233–240. doi:10.1007/s11033-013-2856-0
61. Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132:7. doi:10.1242/jcs.223826
62. Iskit S, Schlicker A, Wessels L, Peeper DS. Fra-1 is a key driver of colon cancer metastasis and a Fra-1 classifier predicts disease-free survival. Oncotarget. 2015;6(41):43146–43161. doi:10.18632/oncotarget.6454
63. Liu H, Ren G, Wang T, et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial-mesenchymal transition. Carcinogenesis. 2015;36(4):459–468. doi:10.1093/carcin/bgv017
64. Rokavec M, Öner MG, Li H, et al. Corrigendum. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2015;125(3):1362. doi:10.1172/JCI81340
65. Li H, Rokavec M, Hermeking H. Soluble IL6R represents a miR-34a target: potential implications for the recently identified IL-6R/STAT3/miR-34a feed-back loop. Oncotarget. 2015;6(16):14026–14032. doi:10.18632/oncotarget.4334
66. Han C, Sun B, Zhao X, et al. Phosphorylation of STAT3 promotes vasculogenic mimicry by inducing epithelial-to-mesenchymal transition in colorectal cancer. Technol Cancer Res Treat. 2017;16(6):1209–1219. doi:10.1177/1533034617742312
67. Holmer R, Wätzig GH, Tiwari S, Rose-John S, Kalthoff H. Interleukin-6 trans-signaling increases the expression of carcinoembryonic antigen-related cell adhesion molecules 5 and 6 in colorectal cancer cells. BMC Cancer. 2015;15:975. doi:10.1186/s12885-015-1950-1
68. Middleton K, Jones J, Lwin Z, Coward JI. Interleukin-6: an angiogenic target in solid tumours. Crit Rev Oncol Hematol. 2014;89(1):129–139. doi:10.1016/j.critrevonc.2013.08.004
69. Kokkonen N, Ulibarri IF, Kauppila A, et al. Hypoxia upregulates carcinoembryonic antigen expression in cancer cells. Int J Cancer. 2007;121(11):2443–2450. doi:10.1002/ijc.22965
70. Blumenthal RD, Hansen HJ, Goldenberg DM. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (carcinoembryonic antigen). Cancer Res. 2005;65(19):8809–8817. doi:10.1158/0008-5472.CAN-05-0420
71. Thirunavukarasu P, Sukumar S, Sathaiah M, et al. C-stage in colon cancer: implications of carcinoembryonic antigen biomarker in staging, prognosis, and management. J Natl Cancer Inst. 2011;103(8):689–697. doi:10.1093/jnci/djr078
72. Xiong H, Zhang ZG, Tian XQ, et al. Inhibition of JAK1, 2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10(3):287–297. doi:10.1593/neo.07971
73. Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21(13):2000–2008. doi:10.1038/sj.onc.1205260
74. Martins SF, Reis RM, Rodrigues AM, Baltazar F, Filho AL. Role of endoglin and VEGF family expression in colorectal cancer prognosis and anti-angiogenic therapies. World J Clin Oncol. 2011;2(6):272–280. doi:10.5306/wjco.v2.i6.272
75. Eldesoky A, Shouma A, Mosaad Y, Elhawary A. Clinical relevance of serum vascular endothelial growth factor and interleukin-6 in patients with colorectal cancer. Saudi J Gastroenterol. 2011;17(3):170–173. doi:10.4103/1319-3767.80378
76. Waldner MJ, Wirtz S, Jefremow A, et al. VEGF receptor signaling links inflammation and tumorigenesis in colitis-associated cancer. J Exp Med. 2010;207(13):2855–2868. doi:10.1084/jem.20100438
77. Spitzner M, Roesler B, Bielfeld C, et al. STAT3 inhibition sensitizes colorectal cancer to chemoradiotherapy in vitro and in vivo. Int J Cancer. 2014;134(4):997–1007.
78. Xu K, Zhan Y, Yuan Z, et al. Hypoxia Induces drug resistance in colorectal cancer through the HIF-1α/miR-338-5p/IL-6 Feedback Loop. Mol Ther. 2019;27(10):1810–1824. doi:10.1016/j.ymthe.2019.05.017
79. Zhang Q, Liu RX, Chan KW, et al. Exosomal transfer of p-STAT3 promotes acquired 5-FU resistance in colorectal cancer cells. J Exp Clin Cancer Res. 2019;38(1):320. doi:10.1186/s13046-019-1314-9
80. Hara M, Nagasaki T, Shiga K, Takahashi H, Takeyama H. High serum levels of interleukin-6 in patients with advanced or metastatic colorectal cancer: the effect on the outcome and the response to chemotherapy plus bevacizumab. Surg Today. 2017;47(4):483–489. doi:10.1007/s00595-016-1404-7
81. Sarosiek S, Shah R, Munshi NC. Review of siltuximab in the treatment of multicentric castleman’s disease. Ther Adv Hematol. 2016;7(6):360–366. doi:10.1177/2040620716653745
82. Angevin E, Tabernero J, Elez E, et al. A phase I/II, multiple-dose, dose-escalation study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with advanced solid tumors. Clin Cancer Res. 2014;20(8):2192–2204. doi:10.1158/1078-0432.CCR-13-2200
83. Grothey A, Van Cutsem E, Sobrero A, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, Phase 3 trial. Lancet. 2013;381(9863):303–312. doi:10.1016/S0140-6736(12)61900-X
84. Fogelman D, Cubillo A, García-Alfonso P, et al. Randomized, double-blind, phase two study of ruxolitinib plus regorafenib in patients with relapsed/refractory metastatic colorectal cancer. Cancer Med. 2018;7(11):5382–5393. doi:10.1002/cam4.1703
85. Covington M, He X, Scuron M, et al. Preclinical characterization of itacitinib (INCB039110), a novel selective inhibitor of JAK1, for the treatment of inflammatory diseases. Eur J Pharmacol. 2020;885:173505. doi:10.1016/j.ejphar.2020.173505
86. Brambilla L, Genini D, Laurini E, et al. Hitting the right spot: mechanism of action of OPB-31121, a novel and potent inhibitor of the Signal Transducer and Activator of Transcription 3 (STAT3). Mol Oncol. 2015;9(6):1194–1206. doi:10.1016/j.molonc.2015.02.012
87. Oh DY, Lee SH, Han SW, et al. Phase I Study of OPB-31121, an oral STAT3 inhibitor, in patients with advanced solid tumors. Cancer Res Treat. 2015;47(4):607–615. doi:10.4143/crt.2014.249
88. Lau YK, Ramaiyer M, Johnson DE, Grandis JR. Targeting STAT3 in cancer with nucleotide therapeutics. Cancers (Basel). 2019;11(11):1681.
89. Wei N, Li J, Fang C, et al. Targeting colon cancer with the novel STAT3 inhibitor bruceantinol. Oncogene. 2019;38(10):1676–1687. doi:10.1038/s41388-018-0547-y
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
