Back to Journals » International Journal of General Medicine » Volume 14
Prognostic Value and Significant Pathway Exploration Associated with TOP2A Involved in Papillary Thyroid Cancer
Authors Gong M, Chen W, Jin Z, Lyu J, Meng L, wu H, Chen F
Received 16 April 2021
Accepted for publication 27 June 2021
Published 15 July 2021 Volume 2021:14 Pages 3485—3496
DOI https://doi.org/10.2147/IJGM.S316145
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Mou-chun Gong,* Wei-qing Chen,* Zhao-qing Jin, Jia Lyu, Li-hao Meng, Hai-yan wu, Fei-hua Chen
Department of General Surgery, First People’s Hospital of Hangzhou Lin’an District, Hangzhou, Zhejiang, 311300, People’s Republic of China
*These authors contributed equally to this work.
Correspondence: Fei-hua Chen
Department of General Surgery, First People’s Hospital of Hangzhou Lin’an District, 548 Yijin Road, Street Jincheng, Hangzhou, Zhejiang, 311300, People’s Republic of China
Email [email protected]
Background: Topoisomerase 2-alpha (TOP2A) has been identified as a hub gene that played an important role in the initiation and progression of thyroid carcinoma (THCA). However, the exact function of TOP2A in papillary thyroid cancer (PTC) remained elusive. The current study aimed to evaluate the TOP2A expression, prognosis significance and key signaling pathways involved in PTC.
Methods: We firstly evaluated the expression of TOP2A in PTC via UALCAN, cBioportal, HPA and LinkdedOmics databases. Genetic alteration of TOP2A in PTC was then explored in cBioportal. Prognostic impacts of TOP2A expression on disease-free survival (DFS) of PTC patients were subsequently evaluated using Kaplan–Meier plotter and Gepia databases. Taking gender, age, cancer stage, T, N and M stages into consideration, we compared survival difference between TOP2A high and low expression groups. KEGG pathway analysis in WebGestalt and GSEA analysis were further performed to reveal the potential TOP2A-associated signaling pathways involved in PTC. Finally, the upstream microRNAs of TOP2A were assessed using DIANA, TargetScan, miRDB and miRWALK database, followed by mechanism exploration of upstream microRNAs.
Results: 1) The mRNA and protein of TOP2A were highly expressed in PTC tissue compared with normal thyroid tissue. TOP2A expression was associated with patient’s age, N stage and cancer stage (all P< 0.05). TOP2A protein was mainly localized to nucleoplasm. 2) Most of samples occurred the missense substitution, and mutation site was located at K1199E. Nucleotide mutations were mainly presented as G>A (35.29%). 3) TOP2A high expression significantly influenced the DFS of PTC patients (P=0.015). Restricted survival analysis showed that TOP2A high expression caused poorer DFS of female patients (P=0.003) and those with age < 60 years old (P=0.002), early clinical stage (P=0.012), N0 stage (P=0.002) or M0 stage (P=0.040). 4) Pathway analysis suggested that TOP2A positively participated in the cell cycle, oocyte meiosis and p53 signaling pathways (all P< 0.05) involved in thyroid cancer.
Conclusion: The expression of TOP2A was higher in PTC tissue, which resulted in a worse DFS of patients with PTC. TOP2A might act as an effective therapeutic target for PTC treatment.
Keywords: TOP2A, high expression, poor prognosis, pathway
Introduction
The morbidity of thyroid carcinoma (THCA) has been rising worldwide. Compared with other cancers, more THCA cases were diagnosed every year due to the increasing use of diagnostic equipment.1 Papillary thyroid cancer (PTC) was the most common type of thyroid cancer, representing up to 80% of all malignant thyroid tumors, and PTC was usually curable with 5-year survival of over 95%.2 Although the PTC patients usually had favorable overall survival (OS) after standard treatment, 20% of patients were still exposed to a risk of disease recurrence which resulted in aggressive diseases and lethal outcomes.3 Therefore, it was critical to identify the key biomarkers and effective therapeutic targets for prognosis improvement of PTC patients.
Topoisomerase 2-alpha (TOP2A) was a key nuclear enzyme which implicated in several processes of DNA metabolism including replication, transcription and chromosome segregation during interphase and mitosis.4 The expression of TOP2A presented a significant cycle-specificity in proliferative cells, of which expression was low in G1 phase, increased in S phase and reached a peak after the G2-M phase.5 TOP2A was thus known to be a marker of cell proliferation in normal tissue, and over expression of TOP2A was obviously observed in cancer cells compared with non-malignant cells.6,7 Previous studies have reported that high expression of TOP2A was indicative of poor prognosis of patients in gastric cancer,8 breast cancer9 and prostate cancer.10 Recent studies suggested that TOP2A was a hub gene related with THCA,11 and recommended for the evaluation of thyroid tumor cell proliferation.12 Another study showed that TOP2A was a potential independent predictive gene for the recurrence of PTC.13 In addition, TOP2A overexpression might be a reliable marker for a rational application of targeted chemotherapeutic strategies in some subgroups of PTC patients,14 and especially the age group less than 40 years old might benefit from TOP2A chemotherapy.15 It follows that TOP2A is of great significance in PTC. However, the detailed information, correlation associated with clinical data, prognostic value and underlying mechanism of TOP2A in PTC lacks sufficient and systematic exploration.
Based on previous research, present study further evaluated expression of TOP2A in PTC and assessed its prognostic value. Furthermore, the TOP2A-associated potential regulatory pathway involved in PTC was explored by functional enrichment analysis. The study aimed to provide delightful insights to unearth prognostic biomarker candidates and new potential targets for PTC patients.
Methods
Expression Analysis of TOP2A
We first explored the mRNA expression of TOP2A across human cancers (tumor/normal samples) through UALCAN database (http://ualcan.path.uab.edu/index.html). The effect of subtypes of THCA on the TOP2A mRNA expression was also assessed. The effects of clinical characteristics on the TOP2A mRNA expression were explored through LinkedOmics database (http://www.linkedomics.org/). The clinical characteristics included patient’s age, radiation therapy status, T stage, N stage, M stage and clinical stage.
Furthermore, the protein expressions of TOP2A in PTC and normal clinical samples were verified through human protein atlas (HPA) database (https://www.proteinatlas.org/). HPA database maps all the human proteins in cells, tissues and organs using integration of various omics technologies. The subcellular distribution pattern of TOP2A proteins in human cells was also evaluated through HPA database. Learned from HPA database, in order to localize the whole human proteome on a subcellular level in one specific cell line, all proteins are stained in U-2 OS. Two additional cell lines are selected based on mRNA expression data. At present, originally three cell lines, U-2 OS, A-431 and U-251 MG, originating from different human tissues were chosen to be included in the analysis of protein subcellular location by indirect immunocytochemistry/immunofluorescence (ICC-IF). The human cells are stained in a standardized way where the antibody of interest is visualized in green, the microtubules red, the endoplasmic reticulum yellow, and nuclei counterstained in blue.
Genetic Alteration Analysis of TOP2A
A total of 388 patients with PTC was obtained from cBioPortal database to analyze the genetic alteration of TOP2A. The mutation frequency and sites of TOP2A in PTC can be found in the database. Detailed mutation types and nucleotide mutation of TOP2A in thyroid cancer was then explored from Catalogue Of Somatic Mutations In Cancer (COSMIC) database. (https://cancer.sanger.ac.uk/cosmic)
Prognosis Analysis of TOP2A in THCA
Gepia (http://gepia.cancer-pku.cn/) and Kaplan–Meier plotter (http://kmplot.com/) databases were applied to initially assess the influence of TOP2A expression on disease-free survival (DFS) of THCA patients. In survival analysis, the median of TOP2A expression was set as the group cutoff to define high expression (50%) and low expression (50%) groups. Furtherly, the association between TOP2A expression and clinical outcome of PTC patients was also evaluated based on gender (male/female), age (<55/≥55), clinical stage (I+II/III+IV), T stage (T1+T2/T3+T4), N stage (N0/N1) and M stage (M0/M1) by Kaplan–Meier and log rank analyses.
Functional Enrichment Analysis
Functional enrichment analysis can reveal the potential function of TOP2A in PTC. We first determined the co-expressed genes with TOP2A in PTC via cBioportal database, and a total of 20,033 co-expressed genes were found. These co-expressed genes with absolute Spearman correlation >0.5 and P-value <0.001 were selected for further enrichment analysis including GO and KEGG pathway analyses. GO analysis was performed using WebGestalt database (http://www.webgestalt.org/) to annotate the molecular function (MF), biological process (BP) and cellular component (CC) of co-expressed genes. Furthermore, KEGG analysis was also applied to dispose the potential biological pathways related to PTC, and the top 5 pathways were selected for presentation. Threshold value was set as P<0.05. GSEA analysis was subsequently performed to explore the significant pathways involved in thyroid cancer.
Potential Mechanism Exploration
According to the results of pathway analysis, we performed deeper investigation regarding pathways of cell cycle and microRNAs in cancer. For cell cycle pathway, we determined the exact influence of TOP2A on cell cycle through HPA database with U2-OS cancer cell. Staining of U2-OS FUCCI aimed to characterize the cell cycle dependency of the protein expression pattern. The FUCCI cells, Fluorescence Ubiquitination Cell Cycle Indicator cells, are cells tagged with different fluorescent proteins fused to two different cell cycle regulators Cdt1 (expressed in G1 phase) and Geminin (expressed in S and G2 phases) that allows cell cycle monitoring. When both proteins are present, the overlay of the images appeared in yellow marking the G1/S transition.
In terms of microRNAs in cancer, we predicted the target microRNAs of TOP2A from four databases including DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index), TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/) and miRWALK (http://mirwalk.umm.uni-heidelberg.de/). Then, the consistent microRNAs in four databases were identified by Venn analysis (http://bioinformatics.psb.ugent.be/webtools/Venn/). The influence of predicted upstream microRNAs on disease-free survival of patients was then analyzed through Kaplan–Meier plotter database. The potential regulatory pathway associated with upstream microRNAs was explored through DIANA database.
Statistical Analysis
Statistical analyses were performed using SPSS 19.0. Mann–Whitney U-test was performed for comparison between two groups. The Kruskal‑Wallis and Bonferroni’s post‑hoc tests were performed for multiple comparisons. P value less than 0.05 was considered as statistical significance.
Results
Expression Analysis of TOP2A
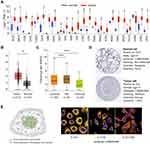
We first explored the mRNA expression of TOP2A in various cancers using UALCAN database. As shown in Figure 1A, TOP2A was obviously up-regulated in most of the cancers. Quantitative analysis confirmed TOP2A high expression in thyroid cancer compared with normal thyroid tissue (Figure 1B, P<0.05). Further analysis indicated that subtypes of thyroid cancer affected the mRNA expression of TOP2A (Figure 1C). In addition, TOP2A mRNA was significantly related to patient’s age, N stage and clinical cancer stage (Figure 2, all P<0.05).
 |
Figure 2 Correlation analysis between TOP2A mRNA expression and clinicopathological characteristics of patients. |
Subsequently, we assessed the protein expression of TOP2A in PTC by HPA database (Figure 1D). It was found that the protein staining of TOP2A was not detected in normal cell; however, medium staining and strong intensity were observed in PTC cell. The images in Figure 1E were the overlay containing 3 toggle channels of nucleus, microtubules and endoplasmic reticulum. The results indicated that the TOP2A protein was mainly localized to the nucleoplasm. In addition, it localized to the nucleoli.
The Genetic Alteration of TOP2A
A total of 388 samples with PTC were contained in genetic alteration analysis in cBioPortal database. The 6% of patients among 388 subjects appeared the TOP2A genetic alteration, and principal manifestation of alteration was mRNA high (Figure 3A). Most of samples occurred the missense substitution (Figure 3B), the nucleotide mutation was mainly presented as G>A (35.29%) and G>T (29.41%) (Figure 3C). The mutation site of TOP2A was located at K1199E (Figure 3D).
 |
Figure 3 The genetic alteration of TOP2A. (A) Alteration frequency in PTC. (B) Mutation types of TOP2A. (C) Mutation of nucleotide. (D) Mutation sites of TOP2A. |
Prognosis Analysis on TOP2A Expression in PTC
In view of the high expression and genetic alteration of TOP2A in THCA, it was necessary to evaluate the potential influence of TOP2A expression on the survival of THCA patients. As observed from Figure 4A, high expression of TOP2A significantly shortened the DFS time of THCA patients in Kaplan–Meier plotter database (P=0.0046). The association of TOP2A high expression with poor prognosis of THCA patients was also confirmed in Gepia database (Figure 4B, P=0.0023). Further analysis indicated that TOP2A high expression negatively correlated with disease-free survival of PTC patients (Figure 4C, P=0.015)
 |
Figure 4 Disease-free survival (DFS) analysis on TOP2A mRNA. (A) Kaplan–Meier Plotter analysis on THCA patients. (B) Gepia analysis on THCA patients. (C) Survival analysis on PTC patients. |
We further performed restricted survival analysis to evaluate the exact influence of TOP2A expression on the DFS of patients with papillary thyroid cancer based on clinical characteristics. The survival analysis showed that high expression of TOP2A was associated with shorter DFS time of patients with age <55 years old (Figure 5A, P=0.006). Female patients with TOP2A high expression showed poorer prognosis (Figure 5B, P=0.003). In addition, TOP2A high expression led to worse prognosis in patients with early clinical stage (Figure 5C, P=0.012). The expression of TOP2A did not influence the prognosis of patients under different T stage (Figure 5D). For N or M stage, high expression of TOP2A mainly shortened the survival time of patients with N0 (Figure 5E, P=0.002) or M0 stage (Figure 5F, P=0.040). It was noted that the sample size of patients with M1 stage was too small (N=9) to conduct the survival analysis; hence, the prognosis analysis was just performed in terms of M0 stage.
 |
Figure 5 Disease-free survival analysis (DFS) on PTC patients. (A) Age. (B) Gender. (C) Clinical stage. (D) T stage. (E) N stage. (F) M stage. |
Functional Enrichment Analysis
Given that the significance of TOP2A expression on the prognosis of cancer patients, we explored its potential role in cancer through enrichment analysis. The co-expressed genes with TOP2A in papillary thyroid cancer were firstly determined, and 168 genes with absolute Spearman correlation greater than 0.5 were selected for further enrichment analysis. GO annotation analysis was applied to reveal the function of these genes in PTC, and the significant terms were shown in Figure 6. For the biological process, the identified genes mainly participated in biological regulation, cellular component organization and metabolic process. For the cellular component, most of these genes were enriched in nucleus, membrane-enclosed lumen and cytosol. At the molecular function level, the co-expressed genes significantly correlated with binding function such as protein binding, ion binding and nucleotide binding.
 |
Figure 6 GO annotation analysis on co-expressed genes. |
KEGG analysis was also performed to reveal the possible regulating pathways. The top 5 pathways are shown in Table 1. The results indicated that those identified genes were significantly associated with pathways of cell cycle, oocyte meiosis, p53 signaling pathway, microRNAs in cancer and cellular senescence.
 |
Table 1 The KEGG Pathway Analysis on Co-Expressed Genes |
The GSEA analysis further screened out 3 consistent pathways involved in thyroid cancer with KEGG analysis. The result (Figure 7) further indicated that cell cycle, p53 signaling pathway and oocyte meiosis were positively associated with the TOP2A expression. The above survival analysis indicated that TOP2A high expression caused poor prognosis of patients, which might correlate with the activation of these pathways.
 |
Figure 7 The GSEA analysis on TOP2A involved in thyroid cancer. |
Potential Mechanism Exploration
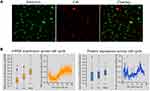
The pathway of cell cycle associated with TOP2A in thyroid cancer was predicted in our KEGG and GSEA analysis. We then explored the detailed changes about cell cycle associated with TOP2A expression. The custom assay in Figure 8A suggested that TOP2A was mostly expressed during S and/or G2, and during mitosis. Figure 8B showed that mRNA and protein expression of TOP2A increased with the time of cell cycle. It followed that the expression of TOP2A had cell cycle dependency.
The pathway of microRNAs in cancer was also identified from KEGG. Therefore, we subsequently predicted the target microRNAs of TOP2A in 4 databases including miRDB, miRWALK, TargetScan and DIANA. We predicted 98, 65, 3314 and 353 target microRNAs from DIANA, miRDB, miRWALK and TargetScan, respectively. Finally, two consistent microRNAs, including hsa-miR-96-5p and hsa-miR-635, were identified by Venn analysis (Figure 9A). Kaplan–Meier plotter analysis (Figure 9B) showed that hsa-miR-96 high expression decreased the survival time of patients (P=0.021), but hsa-miR-635 high expression caused a favorable prognosis of patients (P<0.001). The results suggested that hsa-miR-96-5p and hsa-miR-635 might play a vital role in cancer progression via different regulation.
 |
Figure 9 The upstream microRNAs analysis of TOP2A. (A) Venn analysis for screening consistent upstream microRNAs. (B) Survival analysis in thyroid cancer. |
We then used DIANA database to predict the significant KEGG pathways associated with hsa-miR-96-5p and hsa-miR-635, respectively. The 5 consistent pathways were observed between two groups. Table 2 shows that both hsa-miR-96-5p and hsa-miR-635 participated in the ECM-receptor interaction, p53 signaling pathway, Oocyte meiosis, cell cycle and PI3K-Akt signaling pathway.
 |
Table 2 The KEGG Pathway Analysis on hsa-miR-96-5p and hsa-miR-635 |
As observed from Table 1, Figure 7 and Table 2, p53 signaling pathway, oocyte meiosis and cell cycle became the consistent pathways associated with TOP2A and its upstream microRNAs (Figure 10). It can be speculated that TOP2A possibly influenced the progression of PTC patients via regulating p53 signaling pathway, oocyte meiosis and cell cycle.
 |
Figure 10 The 3 consistent pathways associated with hsa-miR-96-5p, hsa-miR-635 and TOP2A. |
Discussion
Papillary thyroid cancer (PTC) has relatively good clinical outcome; however, the partial patients ultimately developed recurrence or disease-related death.16 It was necessary to find appropriate prognostic factors and biomarkers for treatment of PTC. Accumulating evidences demonstrated the participation of multiple genes and cellular pathways in the occurrence and development of cancer.17,18 TOP2A has been identified as a key gene that played an important role in the initiation and progression of THCA.11,19 But the exact role of TOP2A in PTC was not reported until now. In present study, we initially evaluated the expression of TOP2A in human cancers. The results indicated the high expression of TOP2A in most cancers. Our study further found that the expression of TOP2A was higher in thyroid cancer tissues including PTC, and TOP2A expression did not correlate with the subtypes of thyroid cancer.
We then assessed the prognostic significance of TOP2A expression on THCA patients. It was found that high expression of TOP2A was indicative the shorter DFS of THCA patients (HR=2.6), but did not influence the OS. Previous study revealed that pancreatic ductal adenocarcinoma patients with high expression of TOP2A had a significantly shorter OS time (HR=1.5) and DFS time (HR=2.1).20 Patients with bladder urothelial carcinoma with high TOP2A expression also showed significantly poorer cancer-specific survival, progression-free survival and recurrence-free survival.21 These researches indicated the significance of TOP2A expression on clinical outcome of cancer patients. Our study further showed that high expression of TOP2A mainly influenced the DFS of patients with PTC. For the PTC patients, TOP2A high expression penalized the DFS of female patients and those with age <55 years old, early clinical stage, N0 stage or M0 stage. The results reminded us that more attention should be placed to those subjects for improving their clinical outcome. TOP2A over expression has been demonstrated to be an independent factor for worse distant metastasis-free survival in luminal breast cancer with early stage.22 These findings revealed that TOP2A could be identified as a significant prognostic factor for several cancers.
Subsequently, we conducted enrichment analysis on the co-expressed genes with TOP2A to uncover the potential pathways involved in PTC. Our results revealed that these genes significantly participated in proliferation-associated pathways such as cell cycle and oocyte meiosis. Knockdown of TOP2A in pancreatic cancer cell lines can inhibit cell proliferation and migration.23 Proliferation and invasion of colon cancer cells were also suppressed by knockdown of TOP2A.24 Further, we explored the cell cycle regulation of TOP2A through HPA database analysis. The result suggested that TOP2A was mostly expressed during S and/or G2, and during mitosis. The expression of TOP2A had cell cycle dependency. TOP2A transcript accumulation was also observed during the S- and G2/M- phase of cell cycle, which indicating the active requirement of TOP2A during these stages of the cell cycle.25 In addition, several key cancer-related pathways, including p53 signaling pathway, microRNAs in cancer and cellular senescence, were found in our study. For the pathway of microRNAs in cancer, the present study finally predicted two consistent target microRNAs of TOP2A including miR-96-5p and miR-635. Previous study has found that miR-96-5p was remarkably up-regulated in PTC tumor tissues and cells, and miR-96-5p promoted the proliferation, invasion and metastasis of PTC through down-regulating CCDC67.26 However, low expression of miR-96-5p seemed to be associated with poor clinical outcome in colorectal cancer.27 The miR-635 has been identified as a target of prostate androgen-regulated transcript 1 (PART1), PART1 could sponge miR-635 and promoted progression of non-small cell lung cancer cells via JAK-STAT signaling pathway.28 The potential effect of microRNAs in cancer progression has attracted more and more attention of scholars.
The present study demonstrated the expression of TOP2A and assessed its clinical significance in PTC. However, several shortcomings must be acknowledged. The sample size for restricted survival analysis was insufficient, which might influence the power of test. Hence, the real effect of TOP2A on the patient survival was uncertain. In addition, TOP2A protein expression and subcellular location lacked the verification using clinical PTC samples. Although several significant related pathways involved in PTC have been determined, the details of the involvement of TOP2A in these pathways need further study.
Conclusion
This study aimed to evaluate the expression, prognostic impacts as well as potential regulatory pathway of TOP2A involved in PTC. Our results indicated that TOP2A was highly expressed in PTC, which was unfavorable for disease-free survival of PTC patients, especially for the female patients and those with age <55 years old, early clinical stage, N0 stage and M0 stage. The pathway analysis indicated that TOP2A might play an important role in the cancer progression by positively regulating cell cycle, p53 signaling pathway and oocyte meiosis pathways.
Data Sharing Statement
Data were analyzed from cBioPortal database (https://www.cbioportal.org/); UALCAN database (http://ualcan.path.uab.edu/index.html); HPA database (https://www.proteinatlas.org/); Gepia database (http://gepia.cancer-pku.cn/); WebGestalt database (http://www.webgestalt.org/); DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index); TargetScan (http://www.targetscan.org/); miRDB (http://mirdb.org/) and miRWALK (http://mirwalk.umm.uni-heidelberg.de/). We declare that the data and materials in this study will be provided free of charge to scientists for noncommercial purposes.
Funding
The authors received no funding for this work.
Disclosure
Wei-qing Chen and Mou-chun Gong should be considered as co-first authors. The authors declare there are no competing interests.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Li WB, Zhou J, Xu L, et al. Identification of genes associated with Papillary Thyroid Carcinoma (PTC) for diagnosis by integrated analysis. Horm Metab Res. 2016;48(04):226–231. doi:10.1055/s-0035-1569289
3. Toniato A, Boschin I, Casara D, et al. Papillary thyroid carcinoma: factors influencing recurrence and survival. Ann Surg Oncol. 2008;15(5):1518–1522. doi:10.1245/s10434-008-9859-4
4. Jain M, Zhang L, He M, et al. TOP2A is overexpressed and is a therapeutic target for adrenocortical carcinoma. Endocr Relat Cancer. 2013;20(3):361–370. doi:10.1530/ERC-12-0403
5. Wang J, Yang H, Si Y, et al. Iodine promotes tumorigenesis of thyroid cancer by suppressing Mir-422a and up-regulating MAPK1. Cell Physiol Biochem. 2017;43(4):1325–1336. doi:10.1159/000481844
6. Lynch BJ, Guinee DG
7. Turley H, Comley M, Houlbrook S, et al. The distribution and expression of the two isoforms of DNA topoisomerase II in normal and neoplastic human tissues. Br J Cancer. 1997;75(9):1340–1346. doi:10.1038/bjc.1997.227
8. Cao Y, Zhang G, Wang P, et al. Clinical significance of UGT1A1 polymorphism and expression of ERCC1, BRCA1, TYMS, RRM1, TUBB3, STMN1 and TOP2A in gastric cancer. BMC Gastroenterol. 2017;17(1):2. doi:10.1186/s12876-016-0561-x
9. Zheng H, Li X, Chen C, et al. Quantum dot-based immunofluorescent imaging and quantitative detection of TOP2A and prognostic value in triple-negative breast cancer. Int J Nanomedicine. 2016;11:5519–5529. doi:10.2147/IJN.S111594
10. de Resende MF, Vieira S, Chinen LT, et al. Prognostication of prostate cancer based on TOP2A protein and gene assessment: TOP2A in prostate cancer. J Transl Med. 2013;11(1):36. doi:10.1186/1479-5876-11-36
11. Gao X, Wang J, Zhang S. Integrated bioinformatics analysis of hub genes and pathways in anaplastic thyroid carcinomas. Int J Endocrinol. 2019;2019:9651380. doi:10.1155/2019/9651380
12. Ludvíková M, Holubec L, Ryska A, et al. Proliferative markers in diagnosis of thyroid tumors: a comparative study of MIB-1 and topoisomerase II-a immunostaining. Anticancer Res. 2005;25:1835–1840.
13. Jingni H, Zhong T, Xu Y, et al. A novel RNA sequencing-based risk score model to predict papillary thyroid carcinoma recurrence. Clin Exp Metastasis. 2020;37(2):257–267. doi:10.1007/s10585-019-10011-4
14. Manaios L, Tsiambas E, Alevizaki M, et al. Comparative topoisomerase IIa and ki 67 protein expression in papillary thyroid carcinoma based on tissue microarrays and image analysis. J BUON. 2008;13:537–541.
15. Karayan-Tapon L, Menet E, Guilhot J, et al. Topoisomerase II alpha and telomerase expression in papillary thyroid carcinomas. Eur J Surg Oncol. 2004;30(1):73–99. doi:10.1016/j.ejso.2003.10.002
16. Lin JD, Liou MJ, Hsu HL, et al. Circulating epithelial cell characterization and correlation with remission and survival in patients with thyroid cancer. Thyroid. 2018;28(11):1479–1489. doi:10.1089/thy.2017.0639
17. Hossain MA, Asa TA, Rahman MM, et al. Network-based genetic profiling reveals cellular pathway differences between follicular thyroid carcinoma and follicular thyroid adenoma. Int J Environ Res Public Health. 2020;17.
18. Pan Z, Li L, Fang Q, et al. Integrated bioinformatics analysis of master regulators in anaplastic thyroid carcinoma. Biomed Res Int. 2019;2019:9734576. doi:10.1155/2019/9734576
19. Hu S, Liao Y, Chen L. Identification of key pathways and genes in anaplastic thyroid carcinoma via integrated bioinformatics analysis. Med Sci Monit. 2018;24:6438–6448. doi:10.12659/MSM.910088
20. Zhou Z, Liu S, Zhang M, et al. Overexpression of topoisomerase 2-alpha confers a poor prognosis in pancreatic adenocarcinoma identified by co-expression analysis. Dig Dis Sci. 2017;62(10):2790–2800. doi:10.1007/s10620-017-4718-4
21. Zeng S, Liu A, Dai L, et al. Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer. 2019;19(1):604. doi:10.1186/s12885-019-5814-y
22. An X, Xu F, Luo R, et al. The prognostic significance of topoisomerase II alpha protein in early stage luminal breast cancer. BMC Cancer. 2018;18(1):331. doi:10.1186/s12885-018-4170-7
23. Pei YF, Yin XM, Liu XQ. TOP2A induces malignant character of pancreatic cancer through activating beta-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(1):197–207. doi:10.1016/j.bbadis.2017.10.019
24. Zhang R, Xu J, Zhao J, et al. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J Cell Biochem. 2018;119(9):7256–7263. doi:10.1002/jcb.26916
25. Sapetto-Rebow B, McLoughlin SC, O’Shea LC, et al. Maternal topoisomerase II alpha, not topoisomerase II beta, enables embryonic development of zebrafish top2a-/- mutants. BMC Dev Biol. 2011;11(1):71. doi:10.1186/1471-213X-11-71
26. Liu ZM, Wu ZY, Li WH, et al. MiR-96-5p promotes the proliferation, invasion and metastasis of papillary thyroid carcinoma through down-regulating CCDC67. Eur Rev Med Pharmacol Sci. 2019;23:3421–3430.
27. Ress AL, Stiegelbauer V, Winter E, et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol Carcinog. 2015;54(11):1442–1450. doi:10.1002/mc.22218
28. Zhu D, Yu Y, Wang W, et al. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 2019;8(13):6064–6081. doi:10.1002/cam4.2494
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.