Back to Journals » Infection and Drug Resistance » Volume 15
Prevalence of Multidrug-Resistant Pathogens Causing Bloodstream Infections in an Intensive Care Unit
Authors Golli AL , Cristea OM , Zlatian O, Glodeanu AD, Balasoiu AT, Ionescu M, Popa S
Received 30 July 2022
Accepted for publication 6 October 2022
Published 17 October 2022 Volume 2022:15 Pages 5981—5992
DOI https://doi.org/10.2147/IDR.S383285
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Andreea-Loredana Golli,1,* Oana Mariana Cristea,2,* Ovidiu Zlatian,2 Adina-Dorina Glodeanu,3,* Andrei Theodor Balasoiu,4 Mihaela Ionescu,5 Simona Popa6
1Department of Public Health and Management, University of Medicine and Pharmacy of Craiova, Craiova, Romania; 2Department of Microbiology, University of Medicine and Pharmacy of Craiova, Craiova, Romania; 3Department of Internal Medicine, University of Medicine and Pharmacy of Craiova, Craiova, Romania; 4Department of Ophthalmology, University of Medicine and Pharmacy of Craiova, Craiova, Romania; 5Department of Medical Informatics and Biostatistics, University of Medicine and Pharmacy of Craiova, Craiova, Romania; 6Department of Diabetes, Nutrition and Metabolic Diseases, University of Medicine and Pharmacy of Craiova, Craiova, Romania
*These authors contributed equally to this work
Correspondence: Andreea-Loredana Golli, Email [email protected]
Introduction: Bloodstream infections are the most severe infections that cause the highest mortality rate, especially in patients admitted to the intensive care unit (ICU). In this study, we aimed to analyze the distribution, resistance patterns and prevalence of MDR (multidrug-resistant) pathogens isolated in blood samples collected from patients with severe invasive infections hospitalized in the ICU.
Methods: A retrospective study of bacterial pathogens was performed on 490 patients admitted to the ICU between 2017 and 2020. The resistance patterns were analyzed using Vitek 2 Compact system.
Results: In total, 617 bacterial isolates were obtained. Four hundred and twenty-seven isolates (69.21%) were Gram positive and 190 isolates (30.79%) were Gram negative bacteria. The most frequently isolated micro-organisms identified in the blood samples for the entire period (2017– 2020) were Coagulase-negative staphylococci (CoNS) (318– 51.54%), followed by Klebsiella pneumoniae (70– 11.34%), Methicillin-Resistant Staphylococcus aureus (MRSA) (58– 9.40%), Acinetobacter baumannii (45– 7.29%) and Enterococcus faecalis (42– 6.80%). The number of Klebsiella pneumoniae strains significantly increased in 2020, compared to the previous year (p < 0.05). The Acinetobacter baumannii prevalence was significantly higher in the age group of 20– 64 years (10.89%) and over 65 years (3.53%) (p < 0.001). The difference between the prevalence of CoNS in the elderly (67.84%) and in adults (20– 64 years) (52.47%) was also statistically significant (p < 0.001). High rates of MDR were found for Acinetobacter baumannii (97.77%), Pseudomonas aeruginosa (65%), Klebsiella pneumoniae (50%), Enterococcus faecalis (47.61%) and MRSA (46.55%). More than 60% of the Klebsiella pneumoniae strains were found to be resistant to carbapenems.
Conclusion: The study revealed an alarming prevalence of MDR strains isolated in blood samples of the patients admitted to the ICU, indicating the necessity of consistent application of the measures to control.
Keywords: antimicrobial resistance, blood cultures, bacterial strains, retrospective study
Introduction
Infections from resistant bacteria are a serious problem in health-care settings, causing life-threatening infections in the bloodstream, pneumonia, wound infections and others.1
Antimicrobial Resistance (AMR) is a considerable threat to public health both in the EU/EEA (European Union/European Economic Area) and worldwide. WHO has declared that AMR is one of the top 10 global public health threats facing humanity.2
The high levels of AMR for several important bacterial species–antimicrobial group combinations reported to the European Antimicrobial Resistance Surveillance Network (EARS-Net) for 2020 show that AMR remains a serious challenge in the EU/EEA. Recent estimates based on data from EARS-Net show that each year, more than 670,000 infections occur in the EU/EEA due to bacteria resistant to antibiotics and approximately 33,000 people die as a direct consequence of these infections.3
At the level of the intensive care unit, critically ill patients who require intensive antibiotic treatment and invasive procedures are hospitalized, procedures that are associated with a 5–10 times higher risk of occurrence of the hospital-acquired infections (HAIs).4
An important problem in intensive care units is the increasing percentage of multidrug-resistant pathogens, as a result of specific pathology and excessive use of antibiotics. According to the European Antimicrobial Resistance Surveillance Network (EARS-Net), Improving Patient Safety in Europe (IPSE) and European Centre for Disease Prevention and Control (ECDC) data, Romania is one of the South-Eastern European countries with one of the highest prevalence rates of MDR pathogens.5
MDR pathogens of greatest concern in the hospital environment include MDR gram-positive organisms (MDR-GPRs) such as Methicillin-Resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), and drug-resistant Streptococcus pneumoniae (DRSP), and the MDR gram-negative organisms (MDR-GNRs) including Pseudomonas, Acinetobacter, Klebsiella pneumoniae, Enterobacter, and other species.6
The aim of this study is to analyze the distribution, resistance patterns, and the prevalence of MDR pathogens isolated in blood samples collected from patients with severe invasive infections hospitalized in the ICU.
Materials and Methods
The research is a retrospective analysis of all the data on blood cultures collected from patients, admitted to the intensive care unit (ICU) of Emergency Clinical County Hospital of Craiova, Romania, a county hospital with 1518 beds (65 beds of ICU), which provides specialized healthcare to patients from Dolj county and South-West Region of Romania. In 2020, because of the COVID-19 pandemic, the ICU was restructured and 20 of the 65 beds were intended for patients with COVID-19.
Data were collected from January 2017 to December 2020 from the clinical pathology databases of the hospital. Blood culture specimens from all patients, including adults and children admitted to the ICU during the studied period, were sent to the Hospital’s Laboratory of Microbiology.
The protocol for this study was approved by the Institutional Review Board of the Emergency Clinical County Hospital of Craiova, Romania (No. 5200/01.02.2022). The informed consent was obtained from the study participants, because, at the time of admission, the patients/legal representatives have signed an informed consent by which they agreed to participate in the scientific research, and the signed consent form is attached to the medical record. The study is in accordance with the guidelines outlined in the Declaration of Helsinki.
Blood samples were collected in specialized bottles provided with the automated system Bact/Alert® 3D, harvesting for each patient a set of two culture bottles, including one bottle for aerobic bacteria and one for anaerobic bacteria.
The identification of the isolated strains and the analysis of the resistance patterns for the action of the appropriate antibiotics were performed using Vitek 2 Compact system.7 So were used VITEK 2 GN card for identification of fermentative and non-fermentative Gram-negative bacilli and VITEK 2 GP card to identify Gram-positive bacteria.7
The antimicrobial susceptibility test was carried out according to Clinical Laboratory Standard Institute (CLSI) guidelines.8
We also calculated the multiple antibiotic resistance index (MAR), as a ratio between the number of resistant antibiotics and the total number of antibiotics tested for a given strain. A MAR greater than 0.2 indicates that the high-risk source of infection is a region where drugs are widely used.9
Information about patients’ age, sex, hospital department, sample type, site of infection and antimicrobial resistance pattern have been stored in the WHONET System and in the Hospital`s Information System.
Data were entered and analyzed using Microsoft Excel. Continuous variables like age are expressed as mean±STDEV (standard deviation). The pattern of micro-organisms was analyzed and expressed as percentages. The χ2 test was used for counting data, and p < 0.05 meant the difference was statistically significant.
Multidrug-resistant (MDR) pathogen is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories. Extensively drug-resistant (XDR) is defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories.10
Pan-drug-resistant (PDR) is defined as non-susceptibility to all agents tested in the hospital.11
In our study, we have analyzed the percentage of multidrug-resistant (MDR) strains among the clinical isolates from ICU, by taking into consideration resistance to at least three different antibiotic groups: aminoglycosides, cephalosporins, carbapenems, tetracyclines and fluoroquinolones.10
Analysis of the type of microorganisms involved and their sensitivity to antibiotics includes CoNS, subject to the fact that a high percentage in the case of the presence of these microorganisms may be due to contamination of the samples, being necessary to repeat the harvest and to initiate the treatment according to the correlation with the clinical situation and with the result of other laboratory tests.
Results
In the analyzed interval (2017–2020), out of the 1339 blood culture specimens collected from all patients admitted to the ICU, 617 were positive (46.07%). The average age of the patients was 59±18.46 years. The gender distribution showed a higher share of men (279–56.82%). Almost half of the subjects were aged between 20 and 64 years. Depending on gender, almost 60% of female patients were over the age of 65, while in the case of men, almost 60% were between 20 and 64 years old (Figure 1).
 |
Figure 1 Distribution by gender and age of the patients admitted to the intensive care unit (ICU) of emergency clinical county hospital of craiova, Romania, 2017–2020 (Absolute numbers). |
In the first place among the identified microorganisms was MRSA - Methicillin-Resistant Staphylococcus aureus in 2017 and CoNS - Coagulase-negative staphylococci between 2018 and 2020. A total of 617 bacterial isolates were obtained, excluding cases where it was more than one isolate of the same pathogen from the same patient and the same site of infection. Among these, 427 isolates (69.21%) were Gram-positive and 190 isolates (30.79%) were Gram-negative bacteria (Table 1).
 |
Table 1 Distribution of the Microorganisms Isolated from Blood Samples from ICU Patients at Emergency Clinical County Hospital of Craiova, Romania, 2017–2020 |
The most frequently isolated of all micro-organisms identified in the blood samples for the entire period (2017–2020) were CoNS (318–51.54%), followed by Klebsiella pneumoniae (70–11.34%), MRSA (58–9.40%), Acinetobacter baumannii (45–7.29%) and Enterococcus faecalis (42–6.80%). The number of Klebsiella pneumoniae strains significantly increased in 2020, compared to the previous year (p < 0.05). An important increase, but without statistical significance, was also recorded in the case of E. coli and CoNS (Table 1).
The most common isolate of the Gram-negative pathogens was Klebsiella pneumoniae (70–36.65%), followed by Acinetobacter baumannii (45–23.56%), E.coli (27–14.13%), Pseudomonas aeruginosa (20–10.47%) nonfermenting Gram-negative bacilli, other than Pseudomonas and Acinetobacter (NFB) (11–5.76%) and Proteus (8–4.18%).
Among Gram-positive bacteria, on the first place were placed CoNS - Coagulase-negative staphylococci, representing over 50% of the total microorganisms isolated during the study period. The second predominant Gram-positive bacteria, accounting for 13.61% (58) of all isolates, was MRSA, followed by Enterococcus spp. (42–9.86%).
Forty percent of the total positive samples were identified in 2020, the year in which the highest number of positive samples with CoNS was registered (134–42.27% of the total CoNS identified) (Table 1).
Klebsiella pneumoniae ranked 2nd in frequency both in 2019 (8.33%), as well as in 2020 (15.26%), when the number of positive samples with this pathogen has doubled. MRSA ranked first among the isolated pathogens in 2017 (34.56%) and 3rd in 2018 (8.40%) and for the entire analyzed period (9.4%). The number of Acinetobacter baumannii strains identified in 2018–2020 was the same,12 but was in second place as the frequency in 2018 (11.76%), third in 2019 (8.33%) and fourth in 2020 (5.62%).
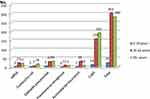
By gender, isolation rates indicate a higher value for male patients in the case of MRSA, Acinetobacter baumannii, CoNS, and more than double for Klebsiella pneumoniae, but without statistical significance (Figure 2).
In terms of germ distribution by age group, there was a significant difference in E.coli prevalence between adults 20–64 years of age and elderly people (over 65 years) (p < 0.01; 2.31% vs 6.36%). The Acinetobacter baumannii prevalence was significantly higher in the age group of 20–64 years (10.89%) and over 65 years (3.53%) (p < 0.001). The difference between the prevalence of CoNS in elderly people (67.84%) and in adults (20–64 years) (52.47%) was also statistically significant (p < 0.001) (Figure 3).
Approximately half of the MRSA (27–46.55%) and Enterococcus faecalis (20–47.61%) isolates were MDR. Almost all of the isolated Acinetobacter baumannii strains were MDR (44–97.77%), 8 strains were sensitive to a single antibiotic from all those tested, while one strain was resistant to all antibiotics used for testing. Only 2 of the E.coli strains were MDR, while 5 showed resistance to cephalosporins and fluoroquinolones.
Sixty-five percent13 of the Pseudomonas aeruginosa strains were MDR, 12 of them were sensitive only to one antibiotic (Colistin) of those used for testing, while one of the strains was found resistant to all 23 antibiotics used for testing. Half of the Klebsiella pneumoniae strains34 were MDR.
Among the MDR Gram-positive pathogens, between 2017 and 2020, around 50% of the MRSA strains were MDR (Figure 4) and the number of isolated MDR strains was three times lower in 2020, compared to 2017. This can be explained by the intensification of infection control measures at the hospital level. The percentage of MDR strains of Enteroccocus faecalis isolated dropped by half in the analyzed period (from 75% to 37.5%), but the absolute number of MDR strains was three times higher. The number of isolated CoNS strains was seven times higher in 2020 compared to 2017, the percentage of MDR strains being almost 50% in 2018–2019 and decreasing to 30% in 2020. The resistance was found in at least three different antibiotic groups: aminoglycosides, tetracyclines and fluoroquinolones.
In the case of the most frequently isolated gram-negative bacteria, our research revealed the fact that all Acinetobacter species isolated in 2017 (3), 2019 (14) and 2020 (14) were MDR, and only one was not MDR in 2018. Also, a PDR Acinetobacter strain was isolated. In the first two years of the analyzed period, MDR Pseudomonas spp was isolated in a single blood sample, but in 2019–2020 the number of identified strains was increasing, and the percentage of MDR strains was around 60%. The percentage of MDR strains of Klebsiella pneumoniae decreased in 2018 (37.5%), but in the following years, half of the strains were MDR, also increasing the total number of isolated strains. The resistance was found to at least three different antibiotic groups: cephalosporins, carbapenems and fluoroquinolones.
Multiple antibiotic resistance (MAR) index was determined for each isolate. The average MAR index resulting from the study was for Klebsiella pneumoniae strains of 0.80, for E. coli of 0.26, for the Acinetobacter baumannii strains of 0.79, for the Pseudomonas aeruginosa strains of 0.67 and for Enterococcus faecalis strains of 0.64. Klebsiella pneumoniae strains showed high resistance to cephalosporins, over 80% of the strains isolated in our study being resistant to third-generation cephalosporins, and over 70% to fourth-generation cephalosporins. Over 70% of the Klebsiella pneumoniae strains were resistant to meropenem and ertapenem, and almost 60% to imipenem. A very high percentage was registered also for fluoroquinolones (up to 80%) and amoxicillin/ clavulanic acid (almost 80%). In terms of resistance to monobactams (Aztreonam), it was also around 80% (Table 2).
 |
Table 2 Antimicrobial Resistance Pattern of Enterobacteriaceae GNB (Number and Percentage) from Blood Samples from ICU Patients at Emergency Clinical County Hospital Craiova, Romania, 2017–2020 |
One-third of E. coli isolates were resistant to third-generation cephalosporins and fluoroquinolones, while less than 4% were resistant to carbapenems. The research highlighted that there was statistical difference between the drug resistance rate of Klebsiella pneumonia and E. coli strains to ceftazidime and ceftriaxone (p < 0.001).
In the case of MRSA isolates, all were resistant to oxacillin and penicillin, and also a higher drug resistance rate was found to be against erythromycin (87.93%), fluoroquinolones (around 70%) and clindamycin (almost 80%). Around 15% of the tested MRSA strains were resistant to vancomycin and around 20% to teicoplanin (Table 3).
 |
Table 3 Antimicrobial Resistance Pattern of Gram Positive Coci (Number and Percentage) from Blood Samples from ICU Patients at Emergency Clinical County Hospital Craiova, Romania, 2017–2020 |
Almost 80% of the isolated strains of CoNS were resistant to penicillin, 60% to clindamycin and ciprofloxacin, and over 70% to erythromycin, oxacillin and tetracycline (Table 3). But the results must be interpreted with caution because we have to take into consideration the possibility of contamination of the samples. Repeated blood cultures should have been harvested to highlight the same germ to support the diagnosis, and that did not happen in our study.
In the case of Pseudomonas aeruginosa strains, it was identified a high resistance (75%) to third-generation cephalosporins and also to fourth-generation (over 70% from the tested antibiotics), fluoroquinolones (over 70%), aminoglycosides and carbapenems (65–75%). Only 12.5% of the tested strains were found resistant to Colistin, which is very often used in this hospital for the treatment of patients with severe infections with MDR Gram-negative pathogens.
The Enterococci isolates were found resistant in high proportion to ciprofloxacin (83.33%) and erythromycin (88.09%). Almost 12% of the isolates were resistant to linezolid.
A very high level of resistance was found for the Acinetobacter strains to the third (97.77%- Ceftazidime, 88.88% - Ceftriaxone) and fourth-generation cephalosporins (97.77% - Cefepime). High resistance was found also to the carbapenems (84–95%), piperacillin-tazobactam (93.33%), fluoroquinolones (80–95%) and aminoglycosides (70–97%) (Table 4).
 |
Table 4 Antimicrobial Resistance Pattern of Non-Fermenting GNB (Number and Percentage) from Blood Samples from ICU Patients at Emergency Clinical County Hospital Craiova, Romania, 2017–2020 |
Discussion
The research we carried out aimed at detecting the main pathogens involved in bloodstream infections (BSI) in patients hospitalized in the ICU, over a 4 years period, as well as the analysis of antibiotic resistance profile in the case of these microorganisms and the identification of the MDR strains.
Our study revealed that almost half of the blood culture specimens harvested from the patients admitted to the ICU were positive. The most common isolated strains identified as being involved in bloodstream infections belonged to CoNS (51.354%), followed by Klebsiella pneumoniae (11.34%), and MRSA (9.40%). CoNS ranked first among isolated microorganisms in other researchers’ studies.13
In other studies conducted with the aim of highlighting the MDR strains which caused other types of infections besides BSI, Klebsiella ranks first among isolated microorganisms,12,14,15 with the same findings in other researchers’ studies.16
In other investigations were highlighted as the main pathogens causing BSIs, E. coli and Staphylococcus aureus17–20 or Acinetobacter baumannii.21,22 In the opinion of other authors, the most common bacterial pathogen was Escherichia coli, followed by Pseudomonas aeruginosa and Klebsiella species.23
Although antibiotics are considered to be the most effective method of fighting against infections, their empirical, indiscriminate, prolonged, or incorrect usage contributes significantly to the selection of MDR strains.24,25
Our research revealed that half of the strains of Klebsiella pneumoniae are MDR, in the conditions in which, according to the European Antimicrobial Resistance Surveillance Network (EARS-Net), in 2019, more than one-third (34.5%) of the Klebsiella pneumoniae isolates were resistant to at least one of the antimicrobial groups under surveillance (fluoroquinolones, third-generation cephalosporins, aminoglycosides and carbapenems), and combined resistance to several antimicrobial groups was frequent.3 According to ECDC Annual Epidemiological Report for 2019,3 the percentage of invasive isolates resistant to third-generation cephalosporins (cefotaxime or/and ceftriaxone or/and ceftazidime) was over 50% in Romania, which also highlights our research, identifying the resistance of over 80% to third-generation cephalosporins, and over 70% to fourth-generation cephalosporins, because of the circulation of ESBL strains in the hospital. The results were consistent with those of other researches.9,21,26,27
The high percentage of Klebsiella MDR strains, which is the most common of the isolated Gram-negative microorganisms highlighted in our research, has a negative impact on the health of the population, as it is associated with increased mortality due to these infections.
More than 60% of the Klebsiella pneumoniae strains were found to be resistant to carbapenems, higher than the one specified for Romania in the data from ECDC Surveillance Atlas (48.3%),28 but consistent with the highest percentages of carbapenem resistance observed in south and south-eastern Europe, reflected in other European surveillance initiatives European Centre for Disease Prevention and Control29,30 and with other findings.31 This high resistance to the carbapenems is due to the circulation of the plasmids encoding carbapenemase-producing genes.
Twenty-one percent of the Klebsiella strains tested were resistant to colistin, which can represent a substantial public health risk because it represents an option for the treatment of the patients with infections caused by multidrug-resistant gram-negative bacteria.
A small percentage of E. coli strains have been carbapenem-resistant and 7.4% of strains were MDR, consistent with other studies18,19 and with the results published in the ECDC Annual Epidemiological Report 2019,3 according to which in Romania, the percentage of invasive isolates resistant to carbapenems was under 5%. In other studies, a high percentage of MDR strains was found.32 E coli is also a major cause of bloodstream infections in Europe, with no significant EU/EEA trend for third-generation cephalosporin resistance and a small, but statistically significant, decreasing EU/EEA trend for fluoroquinolone resistance.3
Approximately half of the MRSA strains have been MDR, less than the percentage identified in other studies performed in Romania,5 but consistent with other findings,4,31 while Staphylococcus aureus is one of the most common causes of bloodstream infections, exhibiting a high burden in terms of morbidity and mortality.3 Among the most important risk factors for MRSA colonization and infection are considered to be ICU admission, intravascular devices, prior or prolonged antibiotic therapy.33
In our research, both Coagulase-negative Staphylococci (CoNS) and MRSA showed high resistance to penicillin and fluoroquinolones, which is similar to other studies.12,14,15,34
Sixty five percent of the Pseudomonas strains were MDR, and 60%.10 were sensitive to only one antibiotic (Colistin), outcomes consistent with other findings.4,23,35,36, According to the European Antimicrobial Resistance Surveillance Network (EARS-Net), the percentage of invasive isolates with resistance to carbapenems was over 50% in Romania, Pseudomonas remaining one of the major causes of healthcare-associated infection in Europe.
A cross-sectional survey that also included Romania among other 24 countries, conducted by Infectious Diseases International Research Initiative (ID-IRI) between the 1st and 30th November 2019, showed meropenem susceptibility rates for K. pneumoniae and Pseudomonas aeruginosa, as the prominent Gram-negative bacilli isolated in ICU, of 42.1%, respectively 36.8%.10
More than half (53.4%) of the Acinetobacter species isolates reported by EU/EEA countries to EARS-Net for 2019 were resistant to at least one of the antimicrobial groups under surveillance (fluoroquinolones, aminoglycosides and carbapenems). In our study, almost all of the isolates were MDR and it was found a very high level of resistance of Acinetobacter strains to the third and fourth-generation cephalosporins, carbapenems, piperacillin-tazobactam and fluoroquinolones, which is consistent with the results published by other authors.4,37,38
An important issue is related to the response of patients to antibiotic treatment, which depends on the presence of functional insufficiency of the organs, like chronic heart failure, abdominal aorta aneurysm or renal dysfunction, being patients who need intensive care services. Patients with abdominal aorta aneurysm present an increased risk of infections, during the diagnostic invasive procedures.39 Renal dysfunction is also a common comorbidity in diabetes, heart failure, and is a strong independent predictor of an increased risk of anaemia and even death.40 A worrying phenomenon in Romania is also the existence of the MDR-TB and XDR-TB cases in socio-economic conditions (malnutrition, agglomeration, stress), including the cases of extrapulmonary tuberculosis which originates from the hematogenous metastatic affects developed during the prime TB infection period.41
The results of our study draw attention to the need for rapid implementation of public health measures, including monitoring the prescribing of state-of-the-art antibiotics and the application of measures to prevent and control hospital-acquired infections (HAIs).
Although the samples collected came from patients hospitalized in one of the largest hospitals in Romania, our study has several limitations. The limits are related to the fact that the results are not confirmed by highlighting the same pathogen in repeated blood sample (2–3 samples/24 hours). The analysis also included CoNS, stating that the possibility of contamination cannot be ruled out.
Conclusion
The study revealed an alarming prevalence of MDR strains isolated in blood samples of the patients admitted to the ICU, in South-West region of Romania, exceeding the national average, which limits the therapeutic possibilities, leading to increased mortality and to the risk as a part of these resistant strains to spread outside the hospital causing infections in the community. It is necessary to intensify the measures for preventing hospital acquired bloodstream infections and the constant surveillance of antibiotic consumption, in order to reduce the proliferation of MDR strains.
Abbreviations
ICU, intensive care unit; MDR, multidrug-resistant; CoNS, Coagulase-negative staphylococci; MRSA, Methicillin-Resistant Staphylococcus aureus; AMR, Antimicrobial Resistance; HAIs, hospital-acquired infections; EARS-Net, European Antimicrobial Resistance Surveillance Network; ECDC, European Centre for Disease Prevention and Control; MDR-GPRs, MDR gram-positive organisms; VRE, vancomycin-resistant enterococci; DRSP, drug-resistant Streptococcus pneumoniae; MDR-GNRs, MDR gram-negative organisms; MAR, multiple antibiotic resistance index; XDR, Extensively drug-resistant; PDR, Pan-drug-resistant.
Funding
The article Processing Charges were funded by the University of Medicine and Pharmacy of Craiova, Romania.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Global action plan on antimicrobial resistance 2016. Available from: https://ahpsr.who.int/publications/i/item/global-action-plan-on-antimicrobial-resistance.
2. Antimicrobial resistance. Available from: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
3. Antimicrobial resistance in the EU/EEA (EARS-Net) – annual epidemiological report for 2019; 2020. Available from: http://www.ecdc.europa.eu.
4. Moolchandani K, Sastry AS, Deepashree R, Sistla S, Harish BN, Mandal J. Antimicrobial resistance surveillance among intensive care units of a Tertiary Care Hospital in Southern India. J Clin Diagn Res. 2017;11(2):DC01–DC07. doi:10.7860/JCDR/2017/23717.9247
5. Licker M, Moldovan R, Hogea E, et al. Multidrug-resistant pathogens in a Romanian intensive care unit – trends and affordable costs. Farmacia. 2017;65(6):929–933.
6. Fraimow HS, Tsigrelis C. Antimicrobial resistance in the intensive care unit: mechanisms, epidemiology, and management of specific resistant pathogens. Crit Care Clin. 2011;27:163–205. doi:10.1016/j.ccc.2010.11.002
7. Zlatian O, Balasoiu AT, Balasoiu M, et al. Antimicrobial resistance in bacterial pathogens among hospitalised patients with severe invasive infections. Exp Ther Med. 2018;16:4499–4510. doi:10.3892/etm.2018.6737
8. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th Informational Supplement, M100-S127. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI); 2017.
9. Ghenea AE, Cioboată R, Drocaș AI, et al. Prevalence and antimicrobial resistance of Klebsiella strains isolated from a County Hospital in Romania. Antibiotics. 2021;10:868. doi:10.3390/antibiotics10070868
10. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
11. INSP-CRSPB. Sentinel surveillance methodology for healthcare associated infections and antimicrobial resistance; 2018. Available from: https://www.cnscbt.ro.
12. Golli AL, Nitu FM, Balasoiu M, et al. Antibiotic resistance pattern of bacterial pathogens in elderly patients admitted in the intensive care unit. Rev Chim. 2018;69(12):3433–3438. doi:10.37358/RC.18.12.6764
13. Japoni A, Vazin A, Hamad M, Davarpanah MA, Alborzi A, Rafaatpour N. Multidrug-resistant bacteria isolated from intensive-care-unit patient samples. Braz J Infect Dis. 2009;13(1):82–86. doi:10.1590/S1413-86702009000200009
14. Golli AL, Nitu FM, Balasoiu M, et al. The characterization of antibiotic resistance of bacterial isolates from intensive care unit patient samples in a university affiliated hospital in Romania. Rev Chim. 2019;70(5):1778–1783. doi:10.37358/RC.19.5.7214
15. Golli AL, Nitu FM, Balasoiu M, et al. Bacterial isolates from endotracheal aspirates and their antimicrobial resistance pattern in patients from intensive care unit. Rev Chim. 2019;70(9):3299–3304. doi:10.37358/RC.19.9.7538
16. Licker M, Anghel A, Moldovan R, et al. Microarray technology for the detection of antimicrobial resistance genes present in hospital and community acquired methicillin resistant S.aureus strains. Rev Chim. 2017;68(11):2546–2550. doi:10.37358/RC.17.11.5925
17. Holmbom M, Möller V, Nilsson LE, et al. Low incidence of antibiotic-resistant bacteria in south-east Sweden: an epidemiologic study on 9268 cases of bloodstream infection. PLoS One. 2020;15(3):e0230501. doi:10.1371/journal.pone.0230501
18. Tian L, Zhang Z, Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: a 20-year surveillance study (1998–2017). Antimicrob Resist Infect Control. 2019;8:86. doi:10.1186/s13756-019-0545-z
19. Sommerstein R, Damonti L, Marschall J, et al. Distribution of pathogens and antimicrobial resistance in ICU-bloodstream infections during hospitalization: a nationwide surveillance study. Sci Rep. 2021;11(1):16876. doi:10.1038/s41598-021-95873-z
20. Buetti N, Marschall J, Timsit JF, et al; Swiss Centre for Antibiotic Resistance (ANRESIS). Distribution of pathogens and antimicrobial resistance in bacteraemia according to hospitalization duration: a nationwide surveillance study in Switzerland. Clin Microbiol Infect. 2021;27(12):1820–1825. doi:10.1016/j.cmi.2021.04.025
21. Lachhab Z, Frikh M, Maleb A, et al. Bacteraemia in intensive care unit: clinical, bacteriological and prognostic prospective study. Can J Infect Dis Med Microbiol. 2017;2017:4082938. doi:10.1155/2017/4082938
22. Dimopoulos G, Koulenti D, Tabah A, et al. Bloodstream infections in ICU with increased resistance: epidemiology and outcomes. Minerva Anestesiol. 2015;81(4):405–418.
23. Sligl WI, Dragan T, Smith SW. Nosocomial gram-negative bacteremia in intensive care: epidemiology, antimicrobial susceptibilities, and outcomes. Int J Infect Dis. 2015;37:129–134. doi:10.1016/j.ijid.2015.06.024
24. Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance - the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–1098. doi:10.1016/S1473-3099(13)70318-9
25. Llor C, Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther Adv Drug Saf. 2014;5(6):229–241. doi:10.1177/2042098614554919
26. Radji M, Fauziah S, Aribinuk N. Antibiotic sensitivity pattern of bacterial pathogens in the intensive care unit of Fatmawati Hospital, Indonesia. Asian Pac J Trop Biomed. 2011;1(1):39–42. doi:10.1016/S2221-1691(11)60065-8
27. Balkhy HH, El-Saed A, Alshamrani MM, et al. High burden of resistant gram negative pathogens causing device-associated healthcare infections in a tertiary care setting in Saudi Arabia, 2008–2016. J Glob Antimicrob Resist. 2020;23:26–32. doi:10.1016/j.jgar.2020.07.013
28. Data from the ECDC surveillance atlas - antimicrobial resistance. Available from: www.ecdc.europa.eu/en/antimicrobial-resistance/surveillance-and-disease-data/data-ecdc.
29. ECDC. Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in European Acute Care Hospitals, 2016–2017. Stockholm: ECDC; 2021.
30. Brolund A, Lagerqvist N, Byfors S, et al; European Antimicrobial Resistance Genes Surveillance Network EURGen-Net Capacity Survey Group. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill. 2019;24(9):1900123. doi:10.2807/1560-7917.ES.2019.24.9.1900123
31. Akter T, Murshed M, Begum T, Nahar K, Duza SS, Shahnaz S. Antimicrobial resistance pattern of bacterial isolates from intensive care unit of a tertiary care hospital in Bangladesh. Bangladesh J Med Microbiol. 2014;8(1):7–11. doi:10.3329/bjmm.v8i1.31052
32. Mutair AA, Alhumaid S, Alawi ZA, et al. Five-year resistance trends in pathogens causing healthcare-associated infections at a multi-hospital healthcare system in Saudi Arabia, 2015–2019. J Glob Antimicrob Resist. 2021;25:142–150. doi:10.1016/j.jgar.2021.03.009
33. Brusselaers N, Vogelaers D, Blot S. The rising problem of antimicrobial resistance in the intensive care unit. Ann Intensive Care. 2011;23(1):47. doi:10.1186/2110-5820-1-47
34. Bhatia A, Kalra J, Kohli S, Kakati B, Kaushik R. Antibiotic resistance pattern in intensive care unit of a tertiary care teaching hospital. Int J Basic Clin Pharmacol. 2018;7(5):906–911. doi:10.18203/2319-2003.ijbcp20181633
35. Gill JS, Arora S, Khanna SP, Kumar KH. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J Glob Infect Dis. 2016;8(4):155–159. doi:10.4103/0974-777X.192962
36. Axente C, Muntean D, Baditoiu L, et al. Beta-lactam resistance mechanisms in pathogens isolated from patients admitted to intensive care unit. Rev Chim. 2017;68(6):1223–1226. doi:10.37358/RC.17.6.5646
37. Stratchounskil LS, Kozlov RS, Rechedko GK, Stetsioukl OU, Chavrikova EI. Antimicrobial resistance patterns among aerobic gram-negative bacilli isolated from patients in intensive care units: results of a multicenter study in Russia. Clin Microbiol Infect. 1998;4:497–507. doi:10.1111/j.1469-0691.1998.tb00404.x
38. Al Bshabshe A, Joseph MRP, Al Hussein A, Haimour W, Hamid ME. Multidrug resistance Acinetobacter species at the intensive care unit, Aseer Central Hospital, Saudi Arabia: a one year analysis. Asian Pac J Trop Med. 2016;9(9):903–908. doi:10.1016/j.apjtm.2016.07.016
39. Carstea D, Streba LA, Glodeanu AD, Carstea AP, Vancu M, Ninulescu AM. The accuracy of combined physical examination and ultrasonography for the detection of abdominal aorta aneurysm. Rom J Morphol Embryol. 2008;49(4):569–572.
40. Zaharie M, Carstea D, Streba CT, et al. Renal dysfunction – a possible marker of severity of heart failure. Rev Chim. 2018;69(6):1435–1440. doi:10.37358/RC.18.6.6341
41. Olteanu M, Niţu M, Golli A. Tuberculosis mesenteric adenopathy and polyserositis. Rom J Morphol Embryol. 2012;53(3 Suppl):835–840.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.