Back to Journals » Infection and Drug Resistance » Volume 12
Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) carriage and pattern of antibiotic resistance among sheep farmers from Southern Italy
Authors Mascaro V, Squillace L , Nobile CGA, Papadopoli R, Bosch T, Schouls LM , Casalinuovo F, Musarella R, Pavia M
Received 9 April 2019
Accepted for publication 14 June 2019
Published 20 August 2019 Volume 2019:12 Pages 2561—2571
DOI https://doi.org/10.2147/IDR.S211629
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Valentina Mascaro,1 Lorena Squillace,1 Carmelo GA Nobile,2 Rosa Papadopoli,1 Thijs Bosch,3 Leo M Schouls,3 Francesco Casalinuovo,4 Rosanna Musarella,4 Maria Pavia1
1Department of Health Sciences, University of Catanzaro “Magna Græcia”, Catanzaro, Italy; 2Department of Pharmacy, Health and Nutritional Sciences, University of Calabria, Cosenza, Italy; 3Centre for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, The Netherlands; 4Section of Catanzaro, Institute for Experimental Veterinary Medicine of Southern Italy, Catanzaro, Italy
Correspondence: Maria Pavia
Department of Health Sciences, University of Catanzaro “Magna Græcia”, Via Tommaso Campanella, Catanzaro 88100, Italy
Tel +39 96 171 2367
Fax +39 96 171 2382
Email [email protected]
Purpose: We conducted a cross-sectional study to measure the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) colonization, with a particular focus on livestock associated (LA)-MRSA in farmers working in contact with livestock (sheep) in one Italian region. Furthermore, we have assessed the antimicrobial resistance pattern of isolates and the association of carriage with specific characteristic of farms and working tasks.
Patients and methods: Demographic data, occupational history, and contact with animals information was collected. Nasal and oropharyngeal swabs were collected and all samples were tested for the isolation and identification of S. aureus. Isolates were examined for antimicrobial susceptibility and all MRSA strains underwent molecular analyses through multiple-locus variable number of tandem repeat analysis (MLVA).
Results: A total of 115 sheep farms and 275 sheep farmers were enrolled. MRSA colonized workers were found in three farms; S. aureus was isolated in 97 workers (35.5%), whereas MRSA was isolated in 3 (1.1%) workers. All MRSA isolates were classified as multidrug resistant. Two of the MRSA isolates were resistant to quinupristin/dalfopristin (QDA), mupirocin, erythromycin, and tetracycline. Among methicillin-susceptible S. aureus (MSSA), 32 (34%) were resistant to tetracycline, 31 (33%) to erythromycin, 26 (27.6%) to QDA, and 22 (23.4%) to linezolid and clindamycin. One MRSA belonged to MLVA complex (MC) 001, found to colonize both humans and animals.
Conclusion: The picture of MRSA transmission among sheep farmers does not seem to be critical, although there is the need to improve adequate control measures to prevent and minimize any biological risk in sheep farms for both animal and human health. Specific monitoring/surveillance programs would help in better understanding the epidemiology of resistant strains.
Keywords: methicillin-resistant Staphylococcus aureus, antimicrobial resistance, sheep farmers, colonization, epidemiology
Introduction
Antimicrobial resistance represents a major issue both in the hospital and in the community setting, and methicillin-resistant Staphylococcus aureus (MRSA) is one of the main pathogens affecting patients all over the world. Epidemiology of MRSA has slightly changed in recent years with the emergence of livestock-associated strain (LA-MRSA). Despite its low impact on the community, the introduction of LA-MRSA in the hospital setting could lead to the typical clinical pictures associated with the presence of S. aureus, including bacteremia cases and surgical site infections.1,2
Several animal species have been potential reservoirs of MRSA strains, and there is evidence supporting transmission to humans, with the major risk factor being the occupational exposure to livestock animals. The first report on LA-MRSA colonizing conventionally raised pigs3 was followed by several findings from European countries such as the Netherlands, Denmark, Germany, France, Italy,4 and later on from North America,5 Northern Africa,6 Asia,7 and Australia.8 Initial studies concerned only pigs, later veal calves,9 poultry,10 and dairy cattle.11 The emergence of LA-MRSA in livestock seems to correlate with farm size,12 and the spread of LA-MRSA among farms is often mediated by animal trading.13 The selection of multidrug-resistant (MDR) strains, as well as MRSA strains, is also related to the massive use of antimicrobials, both in human and in veterinarian medicine. In particular, it has been shown that the reduction of antibiotic use is associated with declining MRSA prevalence in pigs and LA-MRSA in humans.14
Only a few recent studies have assessed LA-MRSA carriage in sheep and sheep farmers, and they focused on carriage in samples of workers selected from MRSA positive sheep farms;15–17 therefore, knowledge on the prevalence of MRSA, and in particular of the LA-MRSA, among sheep farmers is still limited.
Hence, the main objective of the study was the assessment of the prevalence of MRSA colonization, with a particular focus on LA-MRSA, in farmers working in contact with livestock sheep from Calabria region farms. Furthermore, the antimicrobial resistance pattern of isolates was assessed and association of carriage with specific characteristic of farms and working tasks was evaluated.
Materials and methods
We conducted a cross-sectional study between March 2017 and February 2018 in a region of southern Italy (Calabria). Participants included in the study were workers in sheep farms (eg, sheep farmers and technical workers). Study participants were recruited through the help of the public veterinary service of the Local Health Units (LHUs). The LHUs provided lists of sheep farms subjected to periodic veterinary supervision; all farms involving sheep were considered eligible, and a sample of farms was randomly selected. Criterion for enrolment into the study was occupational exposure to sheep and the subjects were all well informed about the purpose and contents of the study. They were given an information sheet and asked to sign a consent form to document their voluntary participation. They were also informed that their participation was totally voluntary and that they could withdraw from the study at any time. This study was approved by the Institutional Ethical Committee (“Mater Domini” Hospital of Catanzaro, Italy) (2017/02/16).
Data collection and review instrument
Primarily, we gathered information about sheep farms, including number of sheep, origin of the sheep population, presence of other animals, and use of antibiotics in the farm. Then, we conducted an extensive face-to-face interview during working hours.
The questionnaire was developed by two researchers and based upon extensive review of the relevant literature and was pretested for clarity and consistency; refinements were made to improve flow and understandability. Confidentiality of responses was assured. The questionnaire included 55 questions divided into several sections. Each section elicited responses in a variety of formats: closed-ended questions with multiple answers possible, yes or no questions, and open option questions. The first section explored the sociodemographic characteristics of the workers. In the second section, the specific working activity was analyzed. In particular, the type of work, the number of years working in that setting, the mean weekly working hours, duties required under the specific task, and the use of personal protective equipment (PPE) were investigated. The third part of the questionnaire was designed to explore the contact with other animal species. The remaining sections were related to recent health history, access to health care facilities, previous MRSA infection, and eventual antibiotic treatment. The last section was related to educational prevention programs for infections in the farms. A copy of the questionnaire is available upon request from the corresponding author.
Biological sampling and microbiological identification
Swab samples were collected from both anterior nares and oropharynx of each participant, placed in a transport medium, transported to laboratory, and processed within 72 hrs of collection. The swabs were inoculated on Mannitol Salt Agar (MSA) plates and incubated at 37°C for 24–48 hrs. All typical S. aureus colonies were then subcultured into Nutrient Agar (NA) plates and incubated at 37°C for 24 hrs. Afterward, Gram stain, catalase, and coagulase tests (Pastorextm Staph-plus Bio-Rad, Marnes-la-Coquette, France) were performed. These agglutination tests use latex sensitized with fibrinogen and IgG, in order to detect the clumping factor and protein A. All suspect strains were identified to the species level using the API Staph identification system (bioMérieux, Marcy-l’Étoile, France). All S. aureus isolates were tested for their antibiotic susceptibility by Kirby–Bauer disk diffusion method (Oxoid Ltd, Basingstoke, United Kingdom); the antibiotics for which sensitivity was tested along with the disk content were: ciprofloxacin (5 µg), clindamycin (2 µg), erythromycin (15 µg), gentamicin (10 µg), linezolid (10 µg), mupirocin (200 µg), sulfamethoxazole/trimethoprim (25 µg), quinupristin/dalfopristin (QDA) (15 µg), rifampicin (5 µg), tetracycline (30 µg), cefoxitin (30 µg), and oxacillin (1 µg). Interpretation of results of the antibiotic susceptibility testing was performed according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines and clinical breakpoints (version 8.1),18 except for the oxacillin breakpoint, performed according to Clinical and Laboratory Standards Institute (CLSI).19 S. aureus strains were considered MRSA if resistant to cefoxitin and/or oxacillin, as indicated by the EUCAST expert rules on antimicrobial susceptibility testing.20
Multiple-locus variable number of tandem repeat analysis (MLVA)
MLVA was performed by the National Institute of Public Health and the Environment (RIVM, Bilthoven, the Netherlands) on the isolates found to be MRSA by antibiotic susceptibility testing, to have a confirmation of the presence of mecA or mecC genes. Ready-made PCR mixes (Eurogentec, Seraing, Belgium, art. no. CS-ALIQ-PROD-MLVA) were used. The MLVA also included the detection of the genes for mecA, mecC, and the lukF gene, indicative for Panton–Valentine leucocidin (PVL). Isolates belonging to MLVA complex 398 (MC398) were classified as LA-MRSA.
For comparative analysis, isolates from our study were compared with the MLVA profiles of LA-MRSA isolates from the Netherlands submitted in 2018 (n=1218).
Preparation of lysates
After inspection for purity of plates, two S. aureus colonies were suspended in 50 mL lysis mix in Tris-EDTA buffer (TE) (10 mM Tris. HCl, 1 mM EDTA, pH 8.0) supplemented with 100 mg/mL lysostaphin, incubated for 35 mins at 37°C and heated for 10 mins at 95°C.
After the inactivation step, 450 mL TE was added and the lysate was used either directly or stored at −20°C until use in PCR.
MLVA typing
Variable number of tandem repeats (VNTR) PCRs were performed in 25-mL volumes in Applied Biosystems 9700 PCR machines (Applied Biosystems, Foster City, USA).
VNTR loci were amplified as previously described.21 After PCR, samples underwent heat denaturation and fragments were separated on an ABI 3730 DNA sequencer. The resulting files were analyzed in the BioNumerics (version 7.6.3).
Each MLVA type consisted of an eight-string numeric code and received a unique number. Closely related MLVA types (MT) were clustered and classified into MLVA complexes (MC), which also correspond to a particular Multilocus sequence typing (MLST) clonal complex (CC) as described by Schouls et al.21 All typing data were imported into the Bionumerics software (Applied Maths), clustered using the appropriate settings and the relationships displayed using the graphing method called minimum spanning tree as described by Schouls et al.21
Data analysis
Data were stored and analyzed using an appropriate database.
Results were summarized using frequencies and percentages for categorical data and mean and standard deviations for continuous data.
Statistical analysis was performed using STATA software program, version 14.1 (Stata Corporation. College Station, Texas, USA).
Results
Characteristics of the farms
A total of 115 sheep farms located in Calabria region (Italy) were enrolled. The mean number of sheep per farm was 243.3 (SD±242.2). The vast majority of farms (98.3%) included only Italian sheep, whereas in the remaining two farms, foreign sheep from France were also present; 25.2% of the farms were exclusively dairy farms, whereas chickens and pigs were also present in 53.9% and 50.6% of the farms, respectively. Sheep of the selected farms were grazed during the day and were kept in barns for the night.
In 20 (17.4%) farms, the owners declared they had used antibiotics in their sheep herd for mastitis prophylactic and therapeutic purposes in the previous year, and the most used antibiotics were tetracyclines (68.8%). Other used antibiotics were penicillins (43.8%), streptomycin (6.3%), cephalosporins (6.3%), and colistin (6.3%); in the remaining cases, the used antibiotics were not specified. The most frequent microorganisms isolated from sheep mastitis in our area in the period 2016–2018 were S. aureus (41.8%), Enterobacteria (E.coli, Enterobacter spp., etc.) (20%), Streptococcus (agalactiae, ovis, etc.) (14.5%), Enterococcus spp. (9%), and Coagulase-negative Staphylococci (9%) (Regional Veterinary Public Health Laboratory, data not published). Moreover, in 4.4% of the sheep farms, education prevention programs had been carried out.
MRSA colonized workers were found in three farms; in two of these farms, antibiotics were used.
Characteristics of workers
Overall, 275 workers were invited to participate in the study. Of these, one refused to complete the questionnaire but accepted to undergo nasal and oropharyngeal swab, whereas two workers completed the questionnaire but refused to undergo swabs. Therefore, 99.6% of the workers completed the questionnaire and 99.3% accepted to undergo nasal and/or oropharyngeal swabs.
Characteristics of the included workers are depicted in Table 1. The mean age of workers was 46±15.3 years (range 18–80 years); 70.6% of them were sheep farmers and 29.4% were technical workers, ie, employees who mainly take care of sheepfold cleaning. The mean time of employment in the sheep sector was 23.8 years (SD±14.6) and the mean working hours per week were 53.4 (SD±26.5).
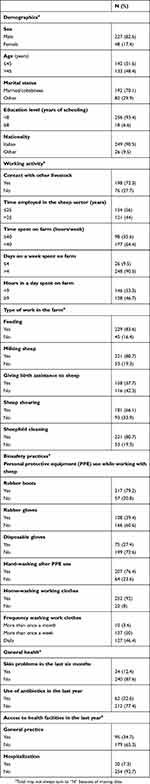 |
Table 1 Characteristics of workers (N=275) |
During working activities with livestock, 20.8% of the workers declared they did not use rubber boots, 60.6% rubber gloves, and 72.6% disposable gloves. Furthermore, 23.6% of the workers stated they did not wash their hands after PPEs usage.
S. aureus was isolated in 95 workers (34.8%), whereas MRSA in 3 (1.1%) workers. Two of the three MRSA colonized workers reported skin problems in the previous 6 months, and one of these had had a hospitalization in the previous year; none of them reported having used antibiotics in the previous year. S. aureus colonization was significantly more likely (χ2=5.95 p=0.015) among those who did not use rubber gloves (40.6%) compared to those who did (26.2%) and in those who did not wash their hands after PPE use (55.6%) compared to those who did (28.6%) (χ2=15.37 p<0.001).
Antimicrobial susceptibility testing
Table 2 shows the prevalence of antimicrobial resistance among the recovered isolates. The antimicrobial resistance profiles were similar among the isolated MRSA strains. All of them were resistant to both cefoxitin and oxacillin and were classified as MDR S. aureus (MDRSA), because of their resistance to ≥3 classes of antibiotics. Two of the isolates were resistant to QDA, mupirocin, erythromycin, and tetracycline (Figure 1).
 |
Table 2 Prevalence of antibiotic resistance among the 95 Staphylococcus aureus isolates |
Concerning methicillin-susceptible S. aureus (MSSA), 30.4% was classified as MDRSA. Among them, 31 (33.7%) were resistant to tetracycline and erythromycin, 25 (27.2%) to QDA, and 21 (22.8%) to linezolid and clindamycin. Twenty-six (27.4%) S. aureus isolates were susceptible to all antimicrobials tested.
Through the investigation of cross antimicrobial resistance among the recovered isolates (Table 3), we found that most of the S. aureus isolates showing resistance to linezolid were also resistant to clindamycin (57.1%) and erythromycin (57.1%), whereas 48.2% S. aureus showing resistance to QDA were also resistant to tetracycline and erythromycin.
 |
Table 3 Cross-resistance of Staphylococcus aureus isolates |
Molecular analyses
Results of MLVA are shown in Figure 2.
In total, 11 isolates underwent MLVA, including isolates showing borderline profiles of identification and of resistance at the phenotypical analyses. Of them, three isolates were confirmed to be MRSA, as they harbored mecA gene. Furthermore, the three isolates harbored different MTs and belonged to different MCs. None of the MRSA isolates was defined as LA-MRSA. One isolate had MT0461 and MC0022, a CC corresponding to the MLST CC22, that is associated to hospital-associated MRSA (HA-MRSA) strains. However, it was not isolated from the worker who had been hospitalized in the previous year. The two remaining MRSA isolates had MT0149 with MC0001 (corresponding to CC1), associated both with community-acquired MRSA (CA-MRSA) and with cases of mastitis in bovine and small ruminants,17,22 and MT0388 with MC0030 (corresponding to CC30), associated with CA-MRSA. All strains were PVL-negative.
Among isolates classified as MSSA, two pairs, originating from the same farms, had the same MLVA-type. In particular, isolates 2 and 3 shared MT0319 belonging to MC0008 and isolates 11 and 14 shared MT0491 belonging to MC0022.
Discussion
This study is one of the few evaluations of the prevalence of MRSA colonization among sheep farmers. So far, LA-MRSA carriage in this context has been assessed only in small samples of workers selected from MRSA positive sheep farms,15–17 whereas this study was designed to assess the circulation of MRSA, as well as LA-MRSA, among sheep farmers.
Among isolated MRSA isolated, we found one strain carrying mecA gene and belonging to MLVA MC001, corresponding to MLST CC1. In Italy, the CC1 lineage has been increasingly detected in dairy cattle mastitis23,24 and colonized small ruminants.25 Although the isolate belongs to a possible LA lineage, it lacks fluoroquinolone resistance, which is a typical feature of porcine MDRSA CC1 isolates from Italy.26 A pathway of human-to-cattle exchange may be direct contact between farmworkers and animals or indirect exposure through farm environment could be however possible and the epidemiology of such resistant strain requires to be monitored at the farm level.
Previous studies have assessed the prevalence of LA-MRSA among different livestock farmers and in most European countries. CC 398 remains the most commonly identified type of LA-MRSA,2 but among sheep farms, different sequence types (STs) and CCs of LA-MRSA by human or animal isolates have been found, in particular, ST(CC)1,17 ST(CC)130,15 and ST(CC)153.27 Furthermore, LA strains are mainly associated with swine, calves, and poultry exposure.28–30 In particular, these kinds of livestock animals are more subjected to the use of antimicrobials,31 which may result in a low prevalence of MRSA among sheep.17,27 According to this evidence, in our study, antibiotics were reported to be used only in 17.4% of the examined sheep farms.
Moreover, MRSA strains were detected in 1.1% of the farmers; this figure is higher than that found in the general population studied by Zanelli et al in Italy (0.12%),32 but in line with the European prevalence, ranging between 0% in Sweden and 2.1% in Belgium.33
Our results also reflect the low prevalence of MRSA and LA-MRSA found in small ruminant bulk tank milk (BTM) in several Italian,16,25 Spanish,34 and Greek studies,35 which reported an MRSA prevalence ranging from 0% to 2%, as well as the detection of MRSA isolates from dairy products.36,37
Apart from the use in the farms, antimicrobials are often administered to animals to prevent or to treat infections due to stressful situations that can weaken their immune systems and make them more susceptible to different pathogens. Among these situations, the long-distance transport is included, especially for those livestock animals originating from abroad. Almost all farms included in our study contained only Italian sheep which have not been subjected to such kind of conditions associated with a greater use of antimicrobials.
Concerning MLVA results, a previous study conducted by Yan et al38 analyzing S. aureus isolates randomly selected from the European survey database already found a low prevalence of MC0030 (0.9%), corresponding to MLST CC30, but, differently from our results, no MC0001 (0%), corresponding to CC1, among MRSA were found. On the contrary, MC0022, corresponding to CC22, resulted in a common MRSA clone (17.2%), and it seems to be a typical HA-MRSA clone. All MRSA isolates were PVL negative, while in the literature it is well known that the presence of PVL is widespread among CA-MRSA, but not among HA-MRSA.39 However, the line between “community” and “hospital” strains is now becoming blurred, since strains responsible for CA-MRSA infections have already entered the healthcare setting.40
Of note, a substantial number of workers declared they did not use PPEs during their working activities in contact with livestock. In particular, only a small percentage used rubber gloves or disposable gloves during their job. Concerning the use of PPE during working activities in animal farms, a previous study showed contradictory data,41 although it is possible that hygiene practices help to reduce the prevalence of MRSA among farmers.42 Indeed, our findings that showed a lower S. aureus colonization among PPE users seem to confirm a protective role of PPE in sheep farmers.
Resistance found for some tested antibiotics is relevant, since it attained to molecules that play a potential role as therapeutic alternatives to treat MRSA and MSSA infections. In our study, the highest resistance among S. aureus was found to tetracycline (34.7%). This figure is quite different from the prevalence of resistance found in the European general population, where resistance to tetracycline ranges from 1.8% in Spain to 7.2% in Croatia43 and in Italy (2.8%),32 whereas it is lower than the prevalence found among pig farmers (52%)44 and sheep milk samples (58.1%).45 The resistance to linezolid and QDA is of concern since, although not first-line options, they are considered useful in the treatment of MRSA infections.46 Consistent with a previous study,47 we found 100% of susceptibility to linezolid among MRSA strains, but 22.8% among MSSA isolates, that was not detected in a recent study conducted in Europe.43 Regarding QDA, in our study 28.4% of S. aureus showed resistance, in particular, we found that two out of three MRSA showed resistance, contrary to a previous study.47
Resistance to clindamycin among S. aureus isolates (22.1%) is higher than that reported in healthy subjects in Italy (7.6%)32 and in Europe (14.6%).43 Clindamycin is an alternative drug for the treatment of skin and soft-tissue infections (SSTIs) caused by both MSSA and MRSA, particularly in Europe.48 SSTIs are intended as clinical entities of variable presentation, etiology, and severity that involve microbial invasion of the layers of the skin and underlying soft tissues which could range from mild infections, such as pyoderma, to serious life-threatening infections, such as necrotizing fasciitis.49 In addition, this antibiotic is an alternative drug to be used in penicillin-allergic patients.46 Although resistance to methicillin was detected in a small proportion of S. aureus isolates, the resistance patterns that we found are relevant. In fact, the choice of an antibiotic therapy also in MSSA infections should be appropriate, since there is no compelling evidence that MRSA is more virulent than MSSA.50
It is also of concern the presence of two MRSA resistant to mupirocin, given that the administration of nasal mupirocin is the common standard of care for peri-interventional prophylaxis of MRSA carriers.51
As far as we know, previous studies have assessed the prevalence of MRSA or LA-MRSA among sheep farmers only in the farms where MRSA was previously detected. Instead, our study was conducted independently of the status of sheep farms, representing a strength of the study, since it allows a more realistic picture of the prevalence of LA-MRSA among sheep farmers. Other host species were similarly investigated.28,30,52
There are several limitations in the present study. Samples were not collected from animals on the farms, so the presence of LA-MRSA strains and transmission of antibiotic resistance and strains between animals and farmers could not be directly addressed. Moreover, we could not consider the prevalence of clinical or subclinical mastitis, because there is no standardized monitoring system in our area. Although previous studies have investigated hand skin samples,17 other possible sites of colonization than the nares and the oropharynx were not cultured, so we may have underestimated MRSA detection. Different from a previous study,25 the persistence of MRSA carriage was not determined in our survey, since the study was conducted at one time point; therefore, it is unclear whether colonization with MRSA is transient or permanent. Finally, MLVA was not performed on all MSSA isolated but only on phenotypic MRSA isolates, to either confirm or exclude the presence of mecA or mecC genes.
Conclusion
In conclusion, the picture of LA-MRSA transmission among sheep farmers does not seem to be critical in our area, although there is the need to further investigate sheep and related sheep farmers colonization.
Moreover, although a low prevalence of MRSA, and in particular of LA-MRSA was detected, poor use of biosafety measures was found. The implementation of PPE use and good hygiene practices could be useful to prevent S. aureus and other microorganisms spread at the farm level and to minimize exposure in the community. Further studies are needed to better understand MRSA and other S. aureus resistance traits in ruminant herds, including the risk factors involved in animal colonization and infection and in within-herd transmission. Indeed, the demonstration that mecC-MRSA is present in livestock in Europe16,17,53 highlights the need for monitoring the presence and evolution of mecC-MRSA in animal and environmental reservoirs.
Abbreviations
BTM, bulk tank milk; CA-MRSA, community-associated methicillin-resistant S. aureus; CC, Clonal complex; CLSI, Clinical and Laboratory Standards Institute; EUCAST, European committee on antimicrobial susceptibility testing; HA-MRSA, health care-associated methicillin-resistant S. aureus; LA-MRSA, livestock-associated methicillin-resistant S. aureus; LHUs, local health units; MC, MLVA complex; MDR, multidrug resistant; MDRSA, multidrug-resistant S. aureus; MLST, multilocus sequence typing; MLVA, multiple-locus variable number of tandem repeat analysis; MRSA, methicillin-resistant S. aureus; MSA, Mannitol Salt Agar; MSSA, methicillin-susceptible S. aureus; MT, MLVA type; NA, Nutrient Agar; PPE, personal protective equipment; PVL, Panton–Valentine leucocidin; QDA, quinupristin/dalfopristin; SSTI, skin and soft-tissue infection; ST, sequence type; TE, tris-EDTA; VNTR, variable number of tandem repeats.
Ethics approval
The Ethics Committee of “Mater Domini” Hospital of Catanzaro (Italy) approved the protocol of the study on February 16, 2017.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
We extend our sincere thanks to the Collaborative Working Group, who are as follows: Carmelo Salviati, Giovanni Iaquinta, Rosario Fruci, Pietro Ruffa, Francesco Massara, for their substantial contribution to identification and sampling of sheep farms, and to data collection. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Layer F, Cuny C, Strommenger B, Werner G, Witte W. Aktuelle daten und trends zu methicillin-resistenten [Staphylococcus aureus (MRSA)]. Bundesgesundheitsbl. 2012;55(11–12):1377–1386. doi:10.1007/s00103-012-1560-x
2. Cuny C, Wieler LH, Witte W. Livestock-associated MRSA: the impact on humans. Antibiotics (Basel). 2015;4(4):521–543. doi:10.3390/antibiotics4040521
3. Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11(12):1965–1966. doi:10.3201/eid1112.050428
4. European Food Safety Authority (EFSA). Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs in the EU, Part A: MRSA prevalence estimates. Eur Food Saf Auth. 2009;7(11):1376.
5. Molla B, Byrne M, Abley M, et al. Epidemiology and genotypic characteristics of methicillin-resistant Staphylococcus aureus strains of porcine origin. J Clin Microbiol. 2012;50(11):3687–3693. doi:10.1128/JCM.01263-11
6. Chairat S, Gharsa H, Lozano C, et al. Characterization of Staphylococcus aureus from raw meat samples in Tunisia: detection of clonal lineage ST398 from the African continent. Foodborne Pathog Dis. 2015;12(8):86–92. doi:10.1089/fpd.2015.1958
7. Chuang YY, Huang YC. Livestock-associated methicillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int J Antimicrob Agents. 2015;45(4):334–340. doi:10.1016/j.ijantimicag.2014.08.014
8. Groves MD, O’Sullivan MV, Brouwers HJ, et al. Staphylococcus aureus ST398 detected in pigs in Australia. J Antimicrob Chemother. 2014;69(5):1426–1428. doi:10.1093/jac/dkt526
9. Graveland H, van Duijkeren E, van Nes A, et al. Evaluation of isolation procedures and chromogenic agar media for the detection of MRSA in nasal swabs from pigs and veal calves. Vet Microbiol. 2009;139(1–2):121–125. doi:10.1016/j.vetmic.2009.05.019
10. Nemati M, Hermans K, Lipinska U, et al. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob Agents Chemother. 2008;52(10):3817–3819. doi:10.1128/AAC.00613-08
11. Vanderhaeghen W, Cerpentier T, Adriaensen C, Vicca J, Hermans K, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet Microbiol. 2010;144(1–2):166–171. doi:10.1016/j.vetmic.2009.12.044
12. Alt K, Fetsch A, Schroeter A, et al. Factors associated with the occurrence of MRSA CC398 in herds of fattening pigs in Germany. BMC Vet Res. 2011;7:69. doi:10.1186/1746-6148-7-69
13. Broens EM. MRSA CC398 in the pig production chain. Prev Vet Med. 2011;98(2–3):182–189. doi:10.1016/j.prevetmed.2010.10.010
14. Dorado-García A, Dohmen W, Bos ME, et al. Dose-response relationship between antimicrobial drugs and livestock-associated MRSA in pig farming. Emerg Infect Dis. 2015;21(6):950–959. doi:10.3201/eid2101.140256
15. Harrison EM, Paterson GK, Holden MTG, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. 2013;5(4):509–515. doi:10.1002/emmm.201201876
16. Caruso M, Latorre L, Santagada G, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in sheep and goat bulk tank milk from Southern Italy. Small Rumin Res. 2015;135:26e31.
17. Carfora V, Giacinti G, Sagrafoli D, et al. Methicillin-resistant and methicillin-susceptible Staphylococcus aureus in dairy sheep and in-contact humans: an intra-farm study. J Dairy Sci. 2016;99(6):4251–4258. doi:10.3168/jds.2016-10912
18. EUCAST breakpoint tables for interpretation of MICs and zone diameters version 8.1. 2018. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf.
19. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: informational supplement. CLSI Document M100-S20. 2011.
20. Leclercq R, Cantón R, Brown DF, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013;19(2):141–160. doi:10.1111/j.1469-0691.2011.03703.x
21. Schouls LM, Spalburg EC, van Luit M, et al. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One. 2009;4(4):e5082. doi:10.1371/journal.pone.0005082
22. Petersen A, Stegger M, Heltberg O, et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19(1):E16–E22. doi:10.1111/1469-0691.12036
23. Pilla R, Castiglioni V, Gelain ME, et al. Long-term study of MRSA ST1, t127 mastitis in a dairy cow. Veterinary Record. 2012;170:312a. doi:10.1136/vr.100510
24. Luini M, Cremonesi P, Magro G, et al. Methicillin-resistant Staphylococcus aureus (MRSA) is associated with low within-herd prevalence of intra-mammary infections in dairy cows: genotyping of isolates. Vet Microbiol. 2015;178(3–4):270–274. doi:10.1016/j.vetmic.2015.05.010
25. Cortimiglia C, Bianchini V, Franco A, et al. Short communication: prevalence of Staphylococcus aureus and methicillin-resistant S. aureus in bulk tank milk from dairy goat farms in northern Italy. J Dairy Sci. 2015;98(4):2307–2311. doi:10.3168/jds.2014-8923
26. Franco A, Hasman H, Iurescia M, et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J Antimicrob Chemother. 2011;66(6):1231–1235. doi:10.1093/jac/dkr115
27. Gharsa H, Ben Slama K, Lozano C, et al. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet Microbiol. 2012;156(3–4):367–373. doi:10.1016/j.vetmic.2011.11.009
28. Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol. 2011;301(8):630–634. doi:10.1016/j.ijmm.2011.09.004
29. van Cleef BA, Van Benthem BHB, Verkade EJM, et al. Dynamics of methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus carriage in pig farmers: a prospective cohort study. Clin Microbiol Infect. 2014;20(10):764–771. doi:10.1111/1469-0691.12516
30. Mascaro V, Leonetti M, Nobile CGA, et al. Prevalence of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) among farm and slaughterhouse workers in Italy. J Occup Environ Med. 2018;60(8):e416–e425. doi:10.1097/JOM.0000000000001385
31. Santman- Berends I, Luttikholt S, Den Brom RV, et al. Estimation of the use of antibiotics in the small ruminant industry in the Netherlands in 2011 and 2012. PLoS One. 2014;9(8):e105052. doi:10.1371/journal.pone.0105052
32. Zanelli G, Sansoni A, Zanchi A, et al. Staphylococcus aureus nasal carriage in the community: a survey from central Italy. Epidemiol Infect. 2002;129(2):417–420. doi:10.1017/s0950268802007434
33. Van Bijnen EM, Paget J, de Lange-de Klerk ES, et al. Antibiotic exposure and other risk factors for antimicrobial resistance in nasal commensal Staphylococcus aureus: an ecological study in 8 European countries. PLoS One. 2015;10(8):e0135094. doi:10.1371/journal.pone.0135094
34. Ariza-Miguel J, Hernández M, Fernández-Natal I, Rodríguez-Lázaro D. Methicillin-resistant Staphylococcus aureus harboring mecC in livestock in Spain. J Clin Microbiol. 2014;52(11):4067–4069. doi:10.1128/JCM.01815-14
35. Pexara A, Solomakos N, Sergelidis D, Angelidis AS, Govaris A. Occurrence and antibiotic resistance of enterotoxigenic Staphylococcus aureus in raw ovine and caprine milk in Greece. Dairy Sci Technol. 2016;96:345–347. doi:10.1007/s13594-015-0272-z
36. Carfora V, Caprioli A, Marri N, et al. Enterotoxin genes, enterotoxin production, and methicillin resistance in Staphylococcus aureus isolated from milk and dairy products in Central Italy. Int Dairy J. 2015;42:12–15. doi:10.1016/j.idairyj.2014.10.009
37. Basanisi MG, Nobili G, La Bella G, et al. Molecular characterization of Staphylococcus aureus isolated from sheep and goat cheeses in southern Italy. Small Rumin Res. 2016;135:17–19. doi:10.1016/j.smallrumres.2015.12.024
38. Yan X, Schouls LM, Pluister GN, et al. The population structure of Staphylococcus aureus in China and Europe assessed by multiple-locus variable number tandem repeat analysis; clues to geographical origins of emergence and dissemination. Clin Microbiol Infect. 2016;22(1):
39. Baldwin LN, Lowe AD. Panton-Valentine Leukocidin associated with community acquired methicillin resistant Staphylococcus aureus: a case report and review of interim guidelines. Anaesthesia. 2008;63(7):764–766. doi:10.1111/j.1365-2044.2008.05482.x
40. Gonzalez BE, Rueda AM, Shelburne SA
41. Goerge T, Lorenz MB, van Alen S, Hübner NO, Becker K, Köck R. MRSA colonization and infection among persons with occupational livestock exposure in Europe: prevalence, preventive options and evidence. Vet Microbiol. 2017;200:6–12. doi:10.1016/j.vetmic.2015.10.027
42. Liu W, Liu Z, Yao Z, Fan Y, Ye X, Chen S. The prevalence and influencing factors of methicillin-resistant Staphylococcus aureus carriage in people in contact with livestock: a systematic review. Am J Infect Control. 2015;43(5):469–475. doi:10.1016/j.ajic.2014.12.009
43. Den Heijer CD, van Bijnen EM, Paget WJ, et al. Prevalence and resistance of commensal Staphylococcus aureus, including meticillin-resistant S. aureus, in nine European countries: a cross-sectional study. Lancet Infect Dis. 2013;13(5):409–415. doi:10.1016/S1473-3099(13)70036-7
44. Oppliger A, Moreillon P, Charrière N, Giddey M, Morisset D, Sakwinska O. Antimicrobial resistance of Staphylococcus aureus strains acquired by pig farmers from pigs. Appl Environ Microbiol. 2012;78(22):8010–8014. doi:10.1128/AEM.01902-12
45. Jamali H, Paydar M, Radmehr B, Dadrasnia A, Ismail S. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control. 2015;54:383–388. doi:10.1016/j.foodcont.2015.02.013
46. Patel M. Community-associated meticillin-resistant Staphylococcus aureus infections: epidemiology, recognition and management. Drugs. 2009;69(6):693–716. doi:10.2165/00003495-200969060-00004
47. Mendes RE, Sader HS, Deshpande LM, Diep BA, Chambers HF, Jones RN. Characterization of baseline methicillin-resistant Staphylococcus aureus isolates recovered from Phase IV clinical trial for linezolid. J Clin Microbiol. 2010;48(2):568–574. doi:10.1128/JCM.01384-09
48. Chua K, Laurent F, Coombs G, Grayson ML, Howden BP. Antimicrobial resistance: not community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA)! A clinician’s guide to community MRSA - its evolving antimicrobial resistance and implications for therapy. Clin Infect Dis. 2011;52(1):99–114. doi:10.1093/cid/ciq067
49. Swartz MN. Clinical practice. Cellulitis. N Engl J Med. 2004;350(9):904–912. doi:10.1056/NEJMcp031807
50. Tinelli M, Monaco M, Vimercati M, Ceraminiello A, Pantosti A. Methicillin-susceptible Staphylococcus aureus in skin and soft tissue infections, Northern Italy. Emerg Infect Dis. 2009;15(2):250–257. doi:10.3201/eid1502.080010
51. Köck R, Becker K, Cookson B, et al. Systematic literature analysis and review of targeted preventive measures to limit healthcare-associated infections by meticillin-resistant Staphylococcus aureus. Euro Surveill. 2014;19(29):pii20860.
52. Jayaweera JAAS, Kumbukgolla WW. Antibiotic resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolated from livestock and associated farmers in Anuradhapura, Sri Lanka. Germs. 2017;7(3):132–139. doi:10.18683/germs.2017.1118
53. Schauer B, Krametter-Frötscher R, Knauer F, et al. Diversity of methicillin-resistant Staphylococcus aureus (MRSA) isolated from Austrian ruminants and New World camelids. Vet Microbiol. 2018;215:77–82. doi:10.1016/j.vetmic.2018.01.006
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


