Back to Journals » Infection and Drug Resistance » Volume 15
Prevalence and Molecular Characterization of Extended Spectrum β-Lactamase and Carbapenemase-Producing Enterobacteriaceae Isolates from Bloodstream Infection Suspected Patients in Addis Ababa, Ethiopia
Authors Seman A , Mihret A , Sebre S , Awoke T , Yeshitela B , Yitayew B , Aseffa A , Asrat D , Abebe T
Received 19 November 2021
Accepted for publication 11 March 2022
Published 29 March 2022 Volume 2022:15 Pages 1367—1382
DOI https://doi.org/10.2147/IDR.S349566
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Aminu Seman,1,2 Adane Mihret,1,2 Shemse Sebre,1,2 Tewachew Awoke,3 Biruk Yeshitela,2 Berhanu Yitayew,4 Abraham Aseffa,2 Daniel Asrat,1 Tamrat Abebe1
1Department of Microbiology, Immunology, and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia; 2Bacterial and Viral Disease Research Directorate, Armauer Hansen Research Institute, Addis Ababa, Ethiopia; 3Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Bahir Dar University, Bahir Dar, Ethiopia; 4Department of Medical Laboratory Science, College of Medicine and Health Sciences, Debre Berhan University, Debre Berhan, Ethiopia
Correspondence: Aminu Seman, Department of Microbiology, Immunology, and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University, P.O. Box: 9086, Addis Ababa, Ethiopia, Tel +251 920 747 176, Email [email protected]; [email protected]
Background: Production of Extended spectrum beta-lactamase (ESBL) and Carbapenemase is the most common strategy for drug resistance in clinical isolates of Enterobacteriaceae. This study was conducted to determine the magnitude of ESBL and Carbapenemase production (CPE) among clinical isolates of Enterobacteriaceae causing bloodstream infections (BSI) in Ethiopia.
Methods: A cross-sectional study was performed from September 2018 to January 2019 in Ethiopia. A total of 2397 BSI suspected patients were enrolled and blood culture was performed using a BacT/Alert instrument in combination with conventional methods for identification. After antimicrobial susceptibility test, phenotypic confirmation of ESBLs was done by combined disc-diffusion. Meanwhile carbapenemase production was done by modified carbapenem inactivation method. Multiplex PCR was conducted to detect the presence of blaCTX-M,blaSHV, blaTEM, blaKPC and blaNDM genes.
Results: A total of 104 (4.3%) Enterobacteriaceae were isolated from 2397 BSI suspected patients. Klebsiella pneumoniae (55/104, 52%) was the predominant isolate followed by E. coli, (19.2%, 20/104) and K.oxytoca (17.3%, 18/104). ESBL and carbapenemase production were observed from 70 (67.3%, 57.4 − 76.2% at 95% CI) and 8 (7.7%, 3.4– 14.6% at 95% CI) isolates respectively. The highest frequency of ESBL and carbapenemase production was observed in K. pneumoniae 78.2% (43/55) and 9.1% (5/55), respectively. All the 70 isolates confirmed as ESBL producers harbored at least one of the ESBL genes and the majority of them carried multiple beta-lactamase genes (84.3%), where blaCTX-M, type was the most predominant (67.3%). Similarly, the entire eight isolates positive for carbapenemase carried blaNDM but none of them carried blaKPC.
Conclusion: In our study, the rate of ESBL production among BSI-causing Enterobacteriaceae was alarming and most of the isolates carried multiple types of ESBL genes. A significant magnitude of CPE isolates causing BSI was recorded.
Keywords: CTX-M, TEM, SHV, NDM, KPC, Ethiopia
Introduction
Extended-spectrum beta-lactamases (ESBLs) are plasmid-encoded enzymes, found frequently in Enterobacteriaceae, which hydrolyze third-generation cephalosporins, penicillin and monobactams.1,2 The first report of ESBL was reported in Germany in the year 1983 from Enterobacteriaceae and since then, it has been observed that ESBL-producing Enterobacteriaceae (ESBL-E) are a real threat to human health.3
These enzymes are commonly detected in the members of the Enterobacteriaceae like Klebsiella pneumoniae and Escherichia coli4 and less commonly in Enterobacter, Serratia, Klebsiella oxytoca, Salmonella, Morganella morganii, Proteus spp.5–7 So far, more than 500 β-lactamases have been reported in Enterobacteriaceae strains of which the CTX-M, TEM, and SHV beta-lactamases are the most common types and are proved to be the most successful in terms of promiscuity and dissemination across various epidemiological niches.8,9
In Ethiopia phenotypic reports are showing a high prevalence of ESBL among Enterobacteriaceae isolates by Teklu et al,10 Moges et al11 and Legese et al12 and current reports are coming showing the genotypes in the region but there is still no detailed reports on the distribution and dominance of ESBL genes.
ESBLs are emerging both in the community and in hospitals and have increasingly been described worldwide since their introduction.1 The global threat posed by ESBL producing Enterobacteriaceae is mainly because ESBL producing bacteria do not show resistance only to penicillins, most cephalosporins, and aztreonam but also to other classes of antibiotics such as aminoglycosides, cotrimoxazole, tetracycline, fluoroquinolones, and chloramphenicol, which results in narrow treatment options.4,13,14
Therefore, the production of ESBL by Enterobacteriaceae has forced clinicians to a better alternative antibiotic for the treatment of invasive infections by these bacteria. So, carbapenems were the first choice and the last-resort antibiotics for therapy of invasive bacterial infections producing ESBL. However, the more frequent use and misuse of carbapenems have in turn led to increased resistance to carbapenems.8,15–17
Carbapenem-resistant Enterobacteriaceae (CRE) have been reported worldwide mainly due to (i) acquisition of carbapenemase genes via mobile genetic elements, or (ii) a combination of extended-spectrum beta-lactamase and/or cephalosporinase, with a decreased outer-membrane permeability or efflux overexpression.18,19 Carbapenemase-producing isolates are much more important from a public health perspective and are by far the most current clinical issue in antibiotics resistance Enterobacteriaceae.20
The most frequently identified carbapenemase genes are the Klebsiella pneumoniae carbapenemase (KPC), followed by Metallo-beta-lactamases (MBLs) such as New Delhi MBL (NDM), and OXA-type genes. Certain carbapenemases dominate in specific regions and countries.21,22 In Ethiopia, only a few phenotypic reports are available on the magnitude of CPE in the study site Legese et al12 and Tadesse et al23 but still, there are no reports on the genotypic level.
Carbapenemase producing Enterobacteriaceae isolates are usually resistant to many other beta-lactam and non-beta-lactam antibiotics, leading to multi-resistant isolates.18 Even if there are some phenotypic data about the magnitude of ESBL and Carbapenemase production among Enterobacteriaceae isolates responsible for BSI in Ethiopia. There is no genotypic data about the magnitude of ESBL and Carbapenemase from Enterobacteriaceae associated with bloodstream infection. Therefore, this study aimed to determine the magnitude of Carbapenemase-producing Enterobacteriaceae (CPE), ESBL producing Enterobacteriaceae (ESBL-PE) causing bloodstream infection and to characterize the types of carbapenemase and ESBL genes produced by Enterobacteriaceae.
Materials and Methodology
Study Setting
This hospital-based cross-sectional study was conducted among bloodstream infection suspected patients attending Tikur Anbessa Specialized Hospital (TASH) during five months period from September 2018 to January 2019. TASH is one of the largest hospitals in Ethiopia located in the capital city of Ethiopia established in 1972, with over 800 beds and provides clinical services to 370,000–400,000 patients a year.
Sample and Demographic Data Collection
Demographic data were collected from every patient using stanDardized questionnaires and patient record forms. During the febrile period, a volume of 5–10 mL or 1–3 mL of the peripheral blood sample was collected from adults and pediatric patients, respectively.
Bacterial Isolation and Identification
The blood sample was inoculated into BacT/ALERT® 3D (bioMeriéux-France) culture bottles and incubated in an automated BacT/ALERT® 3D machine at 35°±2°C in 5% CO2.24,25 Bacterial identification was carried out by sub-culturing of the sample on MacConkey agar (Mac, Oxoid UK), 5% sheep blood agar plate (BAP, Oxoid UK) and chocolate agar plate (CHA, Oxoid UK). Identification of the bacterial isolates was performed using panels of standard biochemical tests including, indole, urea, triple sugar iron, citrate, lysine decarboxylase, motility and Malonate24 (S1 Table 1).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility test was carried out by the Kirby–Bauer disc diffusion method on Muller Hinton agar (Oxoid, UK) according to Clinical and Laboratory Standards Institute (CLSI) guidelines.26 Antibiotics tested in this study were, ampicillin (10 μg), gentamicin (10 μg), amikacin (30 μg), tobramycin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), ciprofloxacin (5 μg), trimethoprim/sulphamethoxazol (25 μg), cefepime (30 μg), cefoxitin (30 μg), meropenem (10 μg), Imipenem (10 μg) amoxicillin/clavulanic acid (20/10 μg), piperacillin-tazobactam (100/10 μg) and cefuroxime (30 μg). All the antibiotics tested were supplied by BD USA.
Pure colony of isolates with turbidity equal to 0.5 McFarland standards were lawn cultured on Mueller-Hinton agar (MHA) plates. Then the antimicrobial discs were placed on the MHA plates and incubated at 35°C for 24h. After overnight incubation, the zone of inhibition was measured and interpreted as per the recommendation of CLSI.26
Phenotypic Detection of ESBLs
The Enterobacteriaceae isolates that were non-susceptible to at least one of the third-generation cephalosporin were screened for ESBL production by combination disc-diffusion test (CDT) on MHA by using ceftazidime (CAZ) and cefotaxime (CTX) alone and with ceftazidime + clavulanic acid (CAZ/CLA) and cefotaxime + clavulanic acid (CTX/CLA) as recommended by CLSI 2018. The increase in zone size diameter by ≥5 mm for CTX/CLA and CAZ/CLA, when compared with that CTX and CAZ alone, was confirmed as the presence of ESBL.
Phenotypic Detection of Carbapenemase
Isolates that were resistant and intermediate to either meropenem or imipenem were tested for production of carbapenemase by using the modified carbapenem inactivation method (CIM) as described by CLSI;26. Briefly; 1µL loop full colony of test isolate from overnight blood agar plate was suspended in 2 mL of nutrient broth (Oxoid UK) and 10µg of meropenem disk was added to the nutrient broth and fully immersed. The tubes were incubated at 37°C in ambient air without agitation for 4 h ± 15 min. Subsequently, the meropenem disks were removed using a 10 µL inoculation loop and applied to Mueller-Hinton agar plates (Oxoid, UK) freshly inoculated with a 0.5 McFarland suspension of a carbapenem-susceptible strain (Escherichia coli ATCC® 25922) after overnight incubation results were interpreted as described by CLSI.26,27
Molecular Characterization of ESBLs and Carbapenemase Genes
DNA Extraction
The DNA was extracted from fresh colonies of Enterobacteriaceae isolates by the boiling method as described previously.8 Briefly, 3 to 5 colonies of an overnight growth of each isolate on nutrient agar (Oxoid, UK) were suspended in 500 µL of nuclease-free water. The suspension was boiled at 94 °C for 10 min in a dry block incubator (Thermo-fisher scientific, California) and placed in a freezer at −20°C for 10 minutes, then placed at room temperature for one minute and centrifuged at 14,000 g for 5 min. Finally, 150 µL of the supernatant was transferred into nuclease free Eppendorf tube and measured using Nano drop (Thermo Scientific) for quality and quantity of DNA prior to storage at −20°C until analysis.
PCR Protocol
ESBL genes (blaCTX-M, blaTEM, and blaSHV) and carbapenem resistance determining genes (blaKPC and blaNDM) were detected using conventional PCR by following a previous protocol.28,29 Table 1 shows sets of specific primers used for the detection of ESBL and carbapenemase genes.
 |
Table 1 Primer Sequences Used for Detection Extended-Spectrum Beta-Lactamase and Carbapenemase Genes |
Detection of ESBL and carbapenemase genes were performed in two separate PCR reactions by multiplexing the three ESBL genes (blaCTX-M, blaTEM, and blaSHV) in one reaction tube and the two carbapenemase genes (blaKPC and blaNDM) in another tube. For ESBL genes the PCR was performed in the T3000 Biometra thermocycler in a final volume of 25 µL containing 12.5 µL 2 x HotStarTaq multiplex PCR Master Mix (QIAGEN), 1.5 µL of each primer (0.2 μM), 1.5 µL of template DNA (300 ng), and 9.5 µL of nuclease-free water. Whereas carbapenemase gene detection was performed in the T3000 Biometra thermocycler in a final volume of 15 µL containing 7.5 µL 2 x HotStarTaq multiplex PCR Master Mix (QIAGEN), 1 µL of each primer (0.2 μM), 1 µL of sample DNA (300 ng), and 5.5 µL of nuclease-free water. The PCR cycling parameters for both reactions were: initial denaturation at 95°C for 15 minutes followed by 35 cycles each of denaturation at 94°C for 30s, annealing at 58 °C for 90s, extension at 72 °C for 90s, and final extension at 72 °C for 10 minutes. The PCR products were visualized by performing gel-electrophoresis in 1.5% agarose gel after staining in ethidium bromide with the aid of a gel imaging system, GelDoc (Bio-Rad). A 100bp ladder molecular weight marker (Promega) was used to measure the molecular weight of amplified products. The molecular characterization of ESBLs and carbapenemase genes were conducted at the Armauer Hansen Research Institute, Addis Ababa, Ethiopia.
Data Quality Assurance
The reliability of the study findings was guaranteed by implementing quality control measures throughout the whole process of the laboratory work. The quality of microbiological methods used for bacterial identification was controlled by running K.pneumoniae ATCC® 700603 and Escherichia coli ATCC® 25922 strains for every new batch. Escherichia coli ATCC® 25922 and ATCC® 35218 standard strains were used to check the quality and effectiveness of antibiotics. For the ESBL confirmatory test, Klebsiella pneumoniae ATCC® 700603 (ESBLs positive) and Escherichia coli ATCC® 25922 (ESBLs negative) control strains were used. For carbapenemase confirmatory tests control strains Klebsiella pneumoniae ATCC® BAA-1705 (positive) and Escherichia coli ATCC® 25922 (negative) were used. For optimization of the multiplex PCR known control strains were used as a positive control. DNA samples from reference blaTEM, blaSHV, and blaCTX-M positive strains were used as positive controls during ESBL detection and known Klebsiella pneumoniae isolates carrying blaKPC and blaNDM were used as a positive control along with the carbapenemase detection. Before multiplexing of the primers for ESBL and carbapenemase genes, each primer was tested by a monoplex PCR reaction.
Data Analysis
The statistical analyses were performed using SPSS version 26.0 (IBM Corporation, USA). The comparison of variables was carried out by the chi-square test and Fischer’s exact test where appropriate. A p-value of < 0.05 was considered statistically significant.
Ethical Consideration
The study protocol, including consent procedure and ethical issues, was approved by Addis Ababa University, College of Health Sciences, Department of Microbiology, Immunology, and Parasitology Research Ethical Review committee (DRERC) (Ref. no. DERC/17/18/02-M) and Armauer Hansen Research Institute/All Africa Leprosy TB Rehabilitation and Training center (AAERC) research ethics Committee (AAERC) (Ref. no. P011/18). A support letter was obtained from the study site. Written informed consent was obtained from both adult study participants and from parents or guardians on the behalf of the children and newborn infants who participated in the study.
Result
Socio-Demographic Characteristics of Study Participants
A total of 597 (24.9%) patients had a positive culture from 2397 blood culture tests performed for patients suspected of bloodstream infection. The median ages of the patients were 9 years (1 day–75 years) and a total of 41 (39.4%) were females Table 2.
 |
Table 2 Sociodemographic Characteristics of Study Participants and Magnitude of Isolated Enterobacteriaceae from BSI Patients at TASH, 2019 |
Distribution of Enterobacteriaceae Isolates
Out of the total 597 culture-positive bacterial isolates, 104 (17.4%) were Enterobacteriaceae. K. pneumoniae, (52.9%, 55/104), was the major isolate followed by E. coli, (19.2%, 20/104) and K. oxytoca (17.3%, 18/104). The majority of the bacteria were isolated from inpatients (99%, 103/104) and mainly from pediatrics wards 39.4%, (n=41/104) and ICU wards (35.6%, 35/104) of the hospital as shown in Table 2.
Among the total Enterobacteriaceae isolates, 63 (60.6%) were isolated from males, and 40 (38.5%) of the isolates were from patients under the age of one month. A larger proportion of the Enterobacteriaceae isolates (66/104, 63.5%) were identified from patients with underlining medical conditions (Table 2). Of those patients with underlining medical conditions, 22 (33.3%) and 18 (27.7%) were patients with solid organ cancer and genetic disorders respectively.
Antibiotics Resistance Pattern of Enterobacteriaceae Isolates
Generally, In this study the resistance patterns of Enterobacteriaceae isolated from a blood culture was evaluated for 16 antibiotics (S2 Table 2). The overall resistance patterns of the isolates were shown in Figure 1. A high rate of resistance was observed to ampicillin 103 (99%), cefuroxime 98 (94.2%), and amoxicillin/clavulanic acid 94 (90.4%). Out of those tested antibiotics meropenem, imipenem and amikacin showed better in vitro activity against the isolated Enterobacteriaceae 82.7% (86), 84.6% (88) and 91.3% (95) sensitivity respectively. Generally, Enterobacteriaceae isolates in this study had a relatively lower resistance rate to the aminoglycosides (amikacin 8.7% and tobramycin 45.2%) and carbapenems (imipenem 15.4% and meropenem 17.3%).
Prevalence of ESBL Producing Enterobacteriaceae Isolates
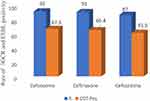
From the total 104 non-duplicate isolates of Enterobacteriaceae, 92 (88.5%), 91 (87.5%), and 87 (83.7%) were resistant to cefotaxime, ceftriaxone and ceftazidime respectively and confirmed for ESBL production by combined disc-diffusion (CDT). As shown in Figure 2, from the isolates tested for ESBL production, 70 (67.3%; 57.4–76.2% at 95% CI)) were ESBL producers which makes the prevalence of ESBL producing Enterobacteriaceae causing bloodstream infection 2.9%, (70/2397) in one of the largest teaching hospitals in Ethiopia. The most common species presenting ESBL activity were K. pneumoniae (61.4%, 43/70) and E coli (18.6%, 13/70).
Distribution of ESBL Producing Isolates Based on Ward Type, Sex, and Age Groups
Among the total ESBL producing isolates 40/70 (57.1%) were isolated from males. Of the total ESBL producing Enterobacteriaceae isolates (42.9%, 30/70) and (24.3%, 17/70) isolates were from patients with an age group of less than one month and between one to fifteen years respectively. Regarding ward type where ESBL producing Enterobacteriaceae isolates were identified, (41.4%, 29/70) and (37.1%, 26/70) of isolates were from pediatrics and Intensive care unit wards respectively. The majority of ESBL producing isolates were identified from patients with underlining medical conditions (65.7%, 46/70) and patients with genetic disorder and cancer had accounted for (34.8%, 16/46) and (26%, 12/46) respectively Table 3. The highest intra-species frequency of ESBL production was observed among K. pneumoniae (78.2%, 43/55) followed by E. coli (65%,13/20). ESBL production was significantly higher among K. pneumoniae (P=0.003, COR=9.556 (2.190–41.690), P=0.005, AOR=9.476 (1.988–45.175) at 95% CI) and E. coli (P=0.05, COR=4.952 (0.986–24.875), P=0.037, AOR=6.333 (1.121–35.795) than other Enterobacteriaceae isolates by univariate analysis and multivariate analysis respectively using a logistic regression model.
 |
Table 3 Magnitude and Distribution of ESBL Producing Enterobacteriaceae Isolates from Patients with BSI at TASH, 2019 |
Prevalence of Carbapenemase-Producing Enterobacteriaceae (CPE) Isolates
From the total 104 isolates of Enterobacteriaceae 18 (17.3%) isolates showed non-susceptibility to carbapenem and were tested for carbapenemase production using modified carbapenem inactivation method. Out of the carbapenem non-susceptibility isolates 8(44.4%) were carbapenemase positive with an overall magnitude of carbapenemase production among Enterobacteriaceae 7.7% (8/104) (3.4–14.6% at 95% CI). This makes the magnitude of CPE causing bloodstream infections (0.33%, 8/2397). K. pneumoniae was the major carbapenemase-producing isolate accounting for 62.5% (5/8) and one isolate of each K. oxytoca, Morganella
morganii and Serratia marcescens were carbapenemase producers. Table 4.
 |
Table 4 Magnitude and Distribution of Carbapenemase Producing Enterobacteriaceae Isolates from Patients with BSI at TASH, 2019 |
Distribution of Carbapenem-Resistant Enterobacteriaceae Based on Ward Type, Sex, and Age Groups
Out of the total 104 Enterobacteriaceae isolates only 18 (17.3%) were found to be CRE, which includes 10 (55.6%) K. pneumoniae, two isolates of each E. coli and K. oxytoca (11.1%) and one isolate each of Enterobacter species, M. morganii, Providencia rettgeri and Serratia marcescens. Out of the total CRE isolates (72.2%, 13) were isolated from males, 14 (77.8%) were from patients aged <15 years; and from those 18 CRE isolates 50%, were from the pediatric ward. Regarding the nature of patients in which CRE isolated (72.2%, n=13) had underline medical condition from those (53.8%, n=7) were solid cancer patients Table 5.
 |
Table 5 Distribution of Carbapenem-Resistant Enterobacteriaceae Isolates from Patients with BSI at TASH, 2019 |
Antimicrobial Resistance Pattern of ESBL-PE and CPE
Table 6 shows the antibiotics resistance profile of ESBL and carbapenemase-producing and none producing isolates against commonly prescribed antibiotics. Those Enterobacteriaceae isolates with ESBL activity showed higher resistance levels to all tested antibiotics than the non-ESBL producing isolates except to piperacillin-tazobactam (42.8% vs 55.9%), meropenem (4.3% vs 44.1%), imipenem (4.3% vs 38.2%) and amikacin (5.7% vs 14.7%). Production of ESBL is significantly associated with enhanced resistance to amoxicillin-clavulanate, trimethoprim-sulfamethoxazole, cefipeme and cefuroxime (p<0.05). Besides the enhanced resistance of ESBL producing isolates, ESBL producing isolates showed a higher level of MDR pattern (p<0.05).
 |
Table 6 Comparison of Antimicrobial Resistance Patterns Between ESBLand Carbapenemase Producing and Non-Producing Enterobacteriaceae Isolates at TASH, 2019 |
Additionally, carbapenemase-producing isolates were found more resistant to all antibiotics included in this study except to trimethoprim/sulfamethoxazole (62.5% vs 90.6%) than the non-CPE (p<0.05). CPE are found 100% resistant to the antibiotics ampicillin, gentamicin, cefotaxime, ceftriaxone, ceftazidime, cefoxitin, cefipeme and cefuroxime Table 6. Furthermore, carbapenemase-producing Enterobacteriaceae isolates had a 100% MDR pattern which was significantly higher than the non-producer 90.6%, (p<0.05).
Molecular Characterization of ESBL and Carbapenemase Genes
The PCR reaction resulted in the occurrence of multiple genes from different species of Enterobacteriaceae as shown in Figure 3A. All 70 CDT ESBL positive isolates were positive for at least one of the ESBL genes. The beta-lactamase genes blaSHV, blaTEM and blaCTX-M were detected in 62 (59.2%), 66 (64%) and 70 (67.3%) of isolates, respectively (S3 Table 3). The majority of the ESBL-producing isolates (63.6%, 45/70) carried blaCTX-M+TEM+ SHV and 17.1% (12/70) carried blaTEM+ SHV. Out of 70 (67.3%) of ESBL producing isolates, 13 (12.5%) carried only blaCTX-M. Only two isolates (1.9%) carried blaSHV and no isolates of Enterobacteriaceae carried a single gene of blaTEM. K. pneumoniae was the species most commonly found to carry a combination of ESBL genes where 37 (86%) isolates carried more than one type of ESBL genes followed by E coli (69%, 9/13) and K oxytoca (63%, 7/11) as presented in Table 7.
 |
Table 7 Distribution of ESBL and Carbapenemase Genes Among Enterobacteriaceae Isolates from Patients with BSI at TASH, 2019 |
The CRE strains were further tested for their ability to produce carbapenemase and carriage of carbapenemase genes. A total of 8 out of 18 CRE showed a positive band to at least one of the carbapenemase encoding genes tested. All of these carbapenemase-producing Enterobacteriaceae isolates carried a single gene of the universal Metallo-beta-lactamase NDM (100%, 8/8) (S4 Table 4). Among the carbapenemase genes targeted KPC was not identified in any of the CRE isolates as shown in Figure 3B. Out of the 18 CRE isolates three Enterobacteriaceae carried ESBL genes; one isolates had blaCTX-M and blaTEM and two isolates carried all the three ESBL genes. And also, from the CPE isolates two had at least one ESBL gene; one isolate with blaCTX-M and blaTEM and the other one was carried blaCTX-M, blaSHV and blaTEM.
Discussion
Prevalence of ESBL and Carbapenemase-Producing Enterobacteriaceae
ESBL producing Enterobacteriaceae and Carbapenem-resistant Enterobacteriaceae (CRE) have become a serious worldwide problem. These multidrug-resistant organisms cause infections associated with high mortality and limited treatment options.10,31 In our study, the magnitude of ESBL and Carbapenemase-producing Enterobacteriaceae (CPE) were 67.3% and 7.7%. In different parts of the world varying figures of ESBL and CPE have been reported from bloodstream infection suspected patients, lower reports of ESBL were made in Brazil 21.3%,32 in Mexico 30.7% ESBL33 and 50% ESBL,34 in Antananarivo, Madagascar 26.3%,35 in Nigeria 41.7%,36 in Ethiopia 38.4%,2 in Iran 42.8%,8 in Egypt 48.93%;37 whereas higher figures of ESBL production was reported in Ethiopia 70.9%11 and 78.57%,12 in Germany 83.6%,38 in Cambodia 93.4%.39
The probable reason for this wide variation in the prevalence of ESBL from different geographic regions could be due to differences in the risk factors, such as the excessive use of broad-spectrum antibiotics, a high rate of patient transfer from the peripheral centers who received prior multiple antimicrobial treatments and high rate of GI colonization by ESBL producing Enterobacteriaceae in the study site.40,41 Therefore, these dissimilarities in the magnitude of ESBL production among Enterobacteriaceae in different regions indicate the need to recognize and take appropriate measures for the timely containment of these resistant microorganisms from further global spread.
In this study the major ESBL producing Enterobacteriaceae isolates were Klebsiella pneumoniae (78.2%) and Escherichia coli (65%) which is in agreement with other reports from Ethiopia K. pneumoniae (78.6%) and E. coli (52.2%),10 K. pneumoniae (89.8%) E. coli (75%)11 and India K. pneumoniae (74%), and E. coli (62%).7 Additionally, both K. pneumoniae and E. coli isolates in our study were observed the significant ESBL producer among the other Enterobacteriaceae this may be due to the phenomenon of easily acquiring antimicrobial resistance determining genes via horizontal gene transfer mechanism by both K. pneumoniae and E. coli.42,43
In recent years, there is an increased resistance to carbapenem and carbapenemase production among Enterobacteriaceae worldwide. These isolates were resistant to several antibiotic families and were associated with high morbidity and mortality.44 The prevalence of carbapenemase production in our study was 7.7% which is higher than previous reports from Kuwait 5.2%,45 in Kathmandu Nepal 4%4 but a comparable magnitude of carbapenemase production was reported from India 8%,46 Taiwan 8.6%,47 and the Czech Republic 6.6%.48
However, the prevalence of CPE in our study is lower than two previous reports from Ethiopia 12.12%12 and 16.2%,11 and much lower than report from Uganda 28.6%,49 France 36.2%,19 Germany 53.3%50 and multicenter observational study from seven Latin American countries 20.8%.51 From those CPE isolates Klebsiella pneumoniae was the major carbapenemase-producing isolate in our study and a similar magnitude was reported from Czech Republic,48 Nepal,4 Uganda49 and Pakistan52. The fact that K. pneumonia is the major organism responsible for ESBL and carbapenemase production could be associated with its notorious ability to accumulate and transfer resistance determinants and this has made it a leading causative agent of hospital-acquired infections.25 Therefore, rapid detection of CPE producing K. pneumonia in clinical laboratory is essential for timely diagnosis, easy control of spreading and administration of systems for tracking those super resistant organisms.
As expected ESBL and carbapenemase-producing isolates presented a higher in vitro resistance profile to most of the antibiotics tested. ESBL producing isolates exhibited greater resistance rate to the the following antibiotic groups: among the beta-lactams to ampicillin, cefotaxime, ceftazidime, ceftriaxone, cefepime and cefuroxime; the beta-lactam/inhibitor to amoxicillin-clavulanate; the aminoglycosides to gentamicin and tobramycin; the quinolone to ciprofloxacin; and trimethoprim/sulphamethoxazol than the non-ESBL producing isolates. A similar finding was reported by other researchers from Ethiopia,10 Burkina Faso,53 Saudi Arabia.54 This could be due to genes encoding ESBLs are often associated with determinants of resistance to other antimicrobial agents, including aminoglycosides, fluoroquinolones and trimethoprim-sulfamethoxazole.35 However, ESBL producing isolates were less resistant to the antibiotics meropenem (4.3%), imipenem (4.3%) and amikacin (5.7%) which is in agreement with other studies in Ethiopia, Addis Ababa10 and Togo.55
In our study, relatively carbapenemase-producing Enterobacteriaceae isolates showed a lower resistance rate to amikacin which is comparable with a study from the Republic of Korea 23%56 and from seven Latin American countries 18%51. Therefore, amikacin could be used as a treatment option for the CPE isolates causing bloodstream infections. However, in our study CPE isolates were found completely non-susceptible to ampicillin, gentamicin, cefotaxime, ceftriaxone, ceftazidime, piperacillin-tazobactam, amoxicillin-clavulanic acid, cefoxitin and cefepime which is in agreement with a study in the Republic of Korea.56
To this end, there is no published report on the genotypic characterization of ESBL and carbapenemase genes from Enterobacteriaceae causing bloodstream infection from the study site. Therefore, this study gives a first generalized picture of the problem in Ethiopia. In our study, the majority of the phenotypic ESBL positive isolates carried multiple bla genes (84.3%), where blaCTX-M -type was the most predominant. A similar report of blaCTX-M predominance in ESBL genes was reported worldwide37,54,57 and a significant number of blaTEM and blaSHV genes were detected besides the blaCTX-M type of ESBL.
In the present study the all ESBL producing E. coli isolates expressed blaCTX-M, and more than 60% expressed blaTEM and blaSHV gene. Whereas all of ESBL producing Klebsiella pneumoniae isolates carried blaCTX-M and blaTEM genes and about 93% carried blaSHV gene.
Additionally, about 44.4% of carbapenem-resistant Enterobacteriaceae isolates were found to carry the carbapenemase gene. The metallo-β-lactamase, blaNDM was the predominant carbapenemase gene detected in our study which is in agreement with other reports from Pakistan52 Turkey58 and United Kingdom.59 Therefore, strains producing the blaNDM are spreading widely across the globe and posing series public health threats mainly because of their wider range of resistance to clinically available antibiotic58 and the plasmid associated with blaNDM is capable of wide rearrangement, widespread horizontal transmission and flexibility among bacterial species.60
Klebsiella pneumoniae is an important nosocomial pathogen causing wide range of clinical infections in hospitalized and immune-compromised patients. Currently, the bacterium is acquiring/developing multi-resistance determining genes like blaNDM and becoming worldwide threats17 as it was reported elsewhere in Netherland,61 Thailand,62 Pakistan52 and Turkey58 Klebsiella pneumoniae was the predominant Enterobacteriaceae producing blaNDM.
Conclusion
The prevalence of ESBL production among Enterobacteriaceae causing bloodstream infection is quite alarming and, in most cases, the isolates carried multiple types of ESBL genes. The rate of multidrug resistance pattern and co-resistance to other non-beta-lactam antibiotics was higher among ESBL and carbapenemase producing Enterobacteriaceae. The antibiotic drug amikacin was the most effective drug against ESBL-PE and CPE isolates tested in our study. A significant magnitude of CPE isolates causing bloodstream infection were recorded. The metallo-beta lactamase blaNDM was the only carbapenemase gene detected from those CPE isolates. K. pneumoniae was the most dominant ESBL and carbapenemase-producing Enterobacteriaceae.
Limitation of the Study
Due to resource issues we are unable to determine the level of ESBL/AmpC co-expression because getting AmpC confirmatory disk was impossible to us and also, we are unable to confirm the methallo-beta lactamase by eCIM phenotypic confirmation method due to unavailability of EDTA-Imipenem/meropenem disks in Ethiopia.
Acknowledgments
The authors hereby thank Addis Ababa University (AAU) and Armauer Hansen Research Institute (AHRI) for their financial and material support and Tikur Anbessa Specialized Hospital Laboratory staff.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Nijhuis R, van Zwet A, Stuart JC, Weijers T, Savelkoul P. Rapid molecular detection of extended-spectrum β-lactamase gene variants with a novel ligation-mediated real-time PCR. J Med Microbiol. 2012;61(11):1563–1567. doi:10.1099/jmm.0.047910-0
2. Siraj SM, Ali S, Wondafrash B. Extended-spectrum β-lactamase production in Klebsiella pneumoniae and Escherichia coli at Jimma University specialized hospital, south-west, Ethiopia. Mol Microbiol. 2015;5. doi:10.1186/s13756-019-0488-4
3. Palmeira JD, Ferreira HMN. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production–a threat around the world. Heliyon. 2020;6(1):e03206. doi:10.1016/j.heliyon.2020.e03206
4. Nepal K, Pant ND, Neupane B, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):1–7. doi:10.1186/s12941-017-0236-7
5. Nedjai S, Barguigua A, Djahmi N, et al. Prevalence and characterization of extended spectrum beta-lactamases in Klebsiella-Enterobacter-Serratia group bacteria, in Algeria. Med Mal Infect. 2012;42(1):20–29. doi:10.1016/j.medmal.2011.10.001
6. Tokajian S, Eisen JA, Jospin G, Farra A, Coil DA. Whole genome sequencing of extended-spectrum beta-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front Cell Infect Microbiol. 2015;5:32. doi:10.3389/fcimb.2015.00032
7. Siddiqui N, Bhakre J, Damle A, Bajaj J. Prevalence of extended-spectrum beta-lactamase (ESBL) producing gram-negative bacilli from various clinical isolates. IOSR J Dent Med Sci. 2014;13:186–189.
8. Montazeri EA, Khosravi AD, Saki M, Sirous M, Keikhaei B, Seyed-Mohammadi S. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae causing bloodstream infections in cancer patients from southwest of Iran. Infect Drug Resist. 2020;13:1319. doi:10.2147/IDR.S254357
9. Ali T, Ali I, Khan NA, Han B, Gao J. The growing genetic and functional diversity of extended spectrum beta-lactamases. Biomed Res Int. 2018;2018. doi:10.1155/2018/9519718
10. Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019;8(1):39. doi:10.1186/s13756-019-0488-4
11. Moges F, Setegn Eshetie WA, Mekonnen F, et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One. 2019;14(4):e0215177. doi:10.1371/journal.pone.0215177
12. Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase-and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27. doi:10.2147/IDR.S127177
13. Nakai H, Hagihara M, Kato H, et al. Prevalence and risk factors of infections caused by extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. J Infect Chemother. 2016;22(5):319–326. doi:10.1016/j.jiac.2016.02.004
14. Hijazi SM, Fawzi MA, Ali FM, Abd el galil KH. Multidrug-resistant ESBL-producing Enterobacteriaceae and associated risk factors in community infants in Lebanon. JIDC. 2016;10(09):947–955. doi:10.3855/jidc.7593
15. Van Aken S, Lund N, Ahl J, Odenholt I, Tham J. Risk factors, outcome and impact of empirical antimicrobial treatment in extended-spectrum β-lactamase-producing Escherichia coli bacteraemia. Scand J Infect Dis. 2014;46(11):753–762. doi:10.3109/00365548.2014.937454
16. Hijazi S, Fawzi M, Ali F, Abd el galil K. Prevalence and characterization of extended-spectrum beta-lactamases producing Enterobacteriaceae in healthy children and associated risk factors. Ann Clin Microbiol Antimicrob. 2016;15(1):1–9. doi:10.1186/s12941-016-0121-9
17. Sheikh AF, Khoshnood S, Saki M, et al. Prevalence of carbapenemases and ESBL encoding genes among K. pneumoniae isolates obtained from an educational hospital in Ahvaz, Southwestern Iran. Gene Rep. 2021;23:101128. doi:10.1016/j.genrep.2021.101128
18. Manageiro V, Romão R, Moura IB, et al. Molecular epidemiology and risk factors of carbapenemase-producing Enterobacteriaceae isolates in Portuguese hospitals: results from European survey on carbapenemase-producing Enterobacteriaceae (EuSCAPE). Front Microbiol. 2018;9:2834. doi:10.3389/fmicb.2018.02834
19. Dortet L, Cuzon G, Ponties V, Nordmann P. Trends in carbapenemase-producing Enterobacteriaceae, France, 2012 to 2014. Eurosurveillance. 2017;22(6):30461. doi:10.2807/1560-7917.ES.2017.22.6.30461
20. Nordmann P, Gniadkowski M, Giske C, et al. Identification and screening of carbapenemase-producing Enterobacteriaceae. CMI. 2012;18(5):432–438. doi:10.1111/j.1469-0691.2012.03815.x
21. Wilson H, Török ME. Extended-spectrum β-lactamase-producing and carbapenemase-producing Enterobacteriaceae. Microbial Genom. 2018;4:7.
22. Samuelsen Ø, Overballe-Petersen S, Bjørnholt JV, et al. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS One. 2017;12(11):e0187832. doi:10.1371/journal.pone.0187832
23. Tadesse S, Mulu W, Genet C, Kibret M, Belete MA, Mascellino MT. Emergence of high prevalence of extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae species among patients in Northwestern Ethiopia Region. Biomed Res Int. 2022;2022:1–9. doi:10.1155/2022/5727638
24. Onken A, Said AK, Jørstad M, Jenum PA, Blomberg B, Galdiero M. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS One. 2015;10(12):12. doi:10.1371/journal.pone.0145632
25. Leal HF, Azevedo J, Silva GEO, et al. Bloodstream infections caused by multidrug-resistant gram-negative bacteria: epidemiological, clinical and microbiological features. BMC Infect Dis. 2019;19(1):609. doi:10.1186/s12879-019-4265-z
26. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
27. Sfeir M, Satlin M, Fauntleroy K, Jenkins S, Westblade L. Blood-modified carbapenem inactivation method: a phenotypic method for detecting carbapenemase-producing Enterobacteriaceae directly from positive blood culture broths. J Clin Microbiol. 2020;58(2):e01377–01319. doi:10.1128/JCM.01377-19
28. Mohammed Y, Gadzama GB, Zailani SB, Aboderin AO. Characterization of extended-spectrum beta-lactamase from Escherichia coli and Klebsiella species from North Eastern Nigeria. J Clin Diagnostic Res. 2016;10(2):DC07.
29. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
30. Darweesh MF. Molecular characterization of ESBL gene in Citrobacter spp. and antibacterial activity of omega-3 against resistant isolates. Curr Issues Pharm Med Sci. 2017;30(3):156–161. doi:10.1515/cipms-2017-0029
31. Kakru DK, Khurshid S, Arshi S. Prevalence of carbapenemase producing genes among carbapenem resistant enterobacteriaceae isolated from blood in a Tertiary Care Hospital, Kashmir. Int J Curr Microbiol App Sci. 2019;8(4):2859–2865. doi:10.20546/ijcmas.2019.804.333
32. KdS N, Conte D, Maia FV, Dalla-Costa LM. Distribution of extended-spectrum β-lactamase types in a Brazilian tertiary hospital. Rev Soc Bras Med Trop. 2015;48(2):162–169. doi:10.1590/0037-8682-0009-2015
33. Garza-Gonzalez E, Ibarra SIM, Llaca-Díaz JM, Gonzalez GM. Molecular characterization and antimicrobial susceptibility of extended-spectrum β-lactamase-producing Enterobacteriaceae isolates at a tertiary-care centre in Monterrey, Mexico. J Med Microbiol. 2011;60(1):84–90. doi:10.1099/jmm.0.022970-0
34. Muro S, Garza-González E, Camacho-Ortiz A, et al. Risk factors associated with extended-spectrum β-lactamase-producing Enterobacteriaceae nosocomial bloodstream infections in a tertiary care hospital: a clinical and molecular analysis. Chemotherapy. 2012;58(3):217–224. doi:10.1159/000339483
35. Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multidrug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013;13(1):85. doi:10.1186/1471-2180-13-85
36. Ibadin EE, Omoregie R, Anogie NA, Igbarumah IO, Ogefere HO. Prevalence of extended spectrum β-lactamase, ampC β-lactamase and metallo-β-lactamase among gram negative bacilli recovered from clinical specimens in Benin city, Nigeria. Int J Enteric Pathog. 2017;5(3):85–91. doi:10.15171/ijep.2017.20
37. Abdallah H, Wintermans B, Reuland E, et al. Extended-spectrum β-lactamase-and carbapenemase-producing Enterobacteriaceae isolated from Egyptian patients with suspected blood stream infection. PLoS One. 2015;10(5):e0128120. doi:10.1371/journal.pone.0128120
38. Schmiedel J, Falgenhauer L, Domann E, et al. Multiresistant extended-spectrum β-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol. 2014;14(1):187. doi:10.1186/1471-2180-14-187
39. Vlieghe ER, Huang T-D, Phe T, et al. Prevalence and distribution of beta-lactamase coding genes in third-generation cephalosporin-resistant Enterobacteriaceae from bloodstream infections in Cambodia. Eur J Clin Microbiol Infect Dis. 2015;34(6):1223–1229. doi:10.1007/s10096-015-2350-9
40. Rani DS, Jahnavi DI, Nagamani DK. Phenotypic and molecular characterization of ESBL producing Enterobacteriaceaein A Tertiary Care Hospital. IOSR-JDMS. 2016;15(08):27–34. doi:10.9790/0853-1508092734
41. Desta K, Woldeamanuel Y, Azazh A, et al. High gastrointestinal colonization rate with extended-spectrum beta-lactamase-producing enterobacteriaceae in hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS One. 2016;11(8):e0161685. doi:10.1371/journal.pone.0161685
42. Khaertynov KS, Anokhin VA, Rizvanov AA, et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front Med. 2018;5:225. doi:10.3389/fmed.2018.00225
43. Poirel L, Madec J-Y, Lupo A, et al. Antimicrobial resistance in Escherichia coli. Microbiol Spectr. 2018;6(4):
44. Wartitima EL, Bahmani F-Z, Elouennass M, Benouda A. Prevalence of carbapenemase-producing Enterobacteriaceae in a University Hospital in Rabat, Morocco: a 19-months prospective study. Int Arab J Antimicrob Agents. 2012;2(3):1–6.
45. Taqi M, Jamal W, Rotimi V. The prevalence of Extended-Spectrum β-lactamase (ESBL)-and Carbapenem–Resistant Enterobacteriaceae (CRE) isolates in positive blood cultures of patients in a teaching hospital in Kuwait over a 2-year period.
46. Devi LS, Broor S, Rautela RS, Grover SS, Chakravarti A, Chattopadhya D. Increasing prevalence of Escherichia coli and Klebsiella pneumoniae producing CTX-M-type extended-spectrum beta-lactamase, carbapenemase, and NDM-1 in patients from a rural community with community acquired infections: a 3-year study. Int J Bppl Basic Med. 2020;10(3):156.
47. Lai -C-C, Wu U-I, Wang J-T, Chang S-C. Prevalence of carbapenemase-producing Enterobacteriaceae and its impact on clinical outcomes at a teaching hospital in Taiwan. JFMA. 2013;112(8):492–496.
48. Hrabák J, Študentová V, Jakubů V, et al. Prevalence study on carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolates in Czech hospitals–results from Czech Part of European Survey on Carbapenemase–Producing Enterobacteriaceae (EuSCAPE). Epidemiol Mikrobiol Imunol. 2015;64(2):87–91.
49. Okoche D, Asiimwe BB, Katabazi FA, Kato L, Najjuka CF. Prevalence and characterization of carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS One. 2015;10(8):e0135745. doi:10.1371/journal.pone.0135745
50. Kaase M, Schimanski S, Schiller R, et al. Multicentre investigation of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in German hospitals. Int J Med Microbiol. 2016;306(6):415–420. doi:10.1016/j.ijmm.2016.05.009
51. Villegas MV, Pallares CJ, Escandón-Vargas K, et al. Characterization and clinical impact of bloodstream infection caused by carbapenemase-producing Enterobacteriaceae in seven Latin American countries. PLoS One. 2016;11(4):e0154092. doi:10.1371/journal.pone.0154092
52. Uddin F, Imam SH, Khan S, et al. NDM production as a dominant feature in carbapenem-resistant Enterobacteriaceae isolates from a Tertiary Care Hospital. Antibiotics. 2022;11(1):48. doi:10.3390/antibiotics11010048
53. Ouedraogo AS, Sanou M, Kissou A, et al. High prevalence of extended-spectrum ss-lactamase producing enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect Dis. 2016;16(1):326. doi:10.1186/s12879-016-1655-3
54. Hassan H, Abdalhamid B. Molecular characterization of extended-spectrum beta-lactamase producing Enterobacteriaceae in a Saudi Arabian tertiary hospital. J Infect Dev Ctries. 2014;8(3):282–288. doi:10.3855/jidc.3809
55. Salah FD, Soubeiga ST, Ouattara AK, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control. 2019;8(1):1–8. doi:10.1186/s13756-019-0552-0
56. Park J-W, Lee H, Park SY, Kim TH. Epidemiological, clinical, and microbiological characteristics of carbapenemase-producing Enterobacteriaceae bloodstream infection in the Republic of Korea. Antimicrob Resist Infect Control. 2019;8(1):1–9. doi:10.1186/s13756-019-0497-3
57. Parajuli NP, Maharjan P, Joshi G, Khanal PR. Emerging perils of extended spectrum beta-lactamase producing enterobacteriaceae clinical isolates in a teaching hospital of Nepal. Biomed Res Int. 2016;2016:1782835. doi:10.1155/2016/1782835
58. Karabay O, Altindis M, Koroglu M, Karatuna O, Aydemir ÖA, Erdem AF. The carbapenem-resistant Enterobacteriaceae threat is growing: NDM-1 epidemic at a training hospital in Turkey. Ann. Clin. Microbiol. Antimicrob. 2016;15(1):1–6. doi:10.1186/s12941-016-0118-4
59. Jain A, Hopkins KL, Turton J, et al. NDM carbapenemases in the United Kingdom: an analysis of the first 250 cases. J Antimicrob Chemother. 2014;69(7):1777–1784. doi:10.1093/jac/dku084
60. Khan AU, Maryam L, Zarrilli R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): a threat to public health. BMC Microbiol. 2017;17(1):1–12. doi:10.1186/s12866-017-1012-8
61. Vlek A, Frentz D, Haenen A, et al. Detection and epidemiology of carbapenemase producing Enterobacteriaceae in the Netherlands in 2013–2014. Eur J Clin Microbiol Infect Dis. 2016;35(7):1089–1096. doi:10.1007/s10096-016-2636-6
62. Rimrang B, Chanawong A, Lulitanond A, et al. Emergence of NDM-1-and IMP-14a-producing Enterobacteriaceae in Thailand. J Antimicrob Chemother. 2012;67(11):2626–2630. doi:10.1093/jac/dks267
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.