Back to Journals » Infection and Drug Resistance » Volume 15
Preliminary Investigation of Iron Acquisition in Hypervirulent Klebsiella pneumoniae Mediated by Outer Membrane Vesicles
Authors Lan Y, Zhou M, Li X, Liu X, Li J, Liu W
Received 1 October 2021
Accepted for publication 18 January 2022
Published 2 February 2022 Volume 2022:15 Pages 311—320
DOI https://doi.org/10.2147/IDR.S342368
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
You Lan, Mao Zhou, Xin Li, Xuan Liu, Jun Li, Wenen Liu
Department of Clinical Laboratory, Xiangya Hospital Central South University, Changsha, Hunan, 410008, People’s Republic of China
Correspondence: Wenen Liu
Department of Clinical Laboratory, Xiangya Hospital Central South University, No. 87 Xiangya Road, Changsha, Hunan, 410008, People’s Republic of China
, Tel +86 731 8432 7437
, Fax +86 731 8432 7332
, Email [email protected]
Objective: To investigate the role of outer membrane vesicles (OMVs) and related proteins in iron acquisition of hypervirulent Klebsiella pneumoniae (HVKP) and classic Klebsiella pneumoniae (cKP).
Methods: The OMVs of HVKP and cKP under iron-deficient and iron-sufficient media were extracted and purified by ultracentrifugation. Transmission electron microscopy (TEM) was used to identify OMVs. The quantitative proteomics were performed based on mass spectrometry.
Results: Four OMVs samples secreted by HVKP and cKP under iron-deficient and iron-sufficient environment were isolated and collected (HVKP OMVs under iron-deficient environment (A1), HVKP OMVs under iron-sufficient environment (A2), cKP OMVs under iron-deficient environment (B1), cKP OMVs under iron-sufficient environment (B2)). The amount of OMVs released by HVKP in iron-deficient medium was significantly larger than that in iron-sufficient medium (P < 0.05). HVKP secreted more OMVs than cKP in iron-deficient medium (P < 0.05). A total of 1074 kinds of proteins were identified in four samples. A comparison between the iron-deficient vs iron-sufficient environment showed that 61 proteins in HVKP OMVs were identified with a significant change in abundance under iron-deficient environment. Among them, 17 proteins were related to iron acquisition and transportation systems. While in cKP OMVs, 62 proteins significantly changed under iron-deficient environment in which 5 proteins were related to iron acquisition and transportation systems. Upon comparison of the HVKP vs cKP OMVs under iron deficiency, 81 proteins were detected with a significant change in which 8 proteins were related to iron acquisition and transportation systems.
Conclusion: Above all, the results of this study suggest a potential role for OMVs in iron acquisition of HVKP and provide evidence of potential connections between OMVs and strong iron-acquisition ability of HVKP during iron limitation.
Keywords: hypervirulent Klebsiella pneumoniae, iron acquisition, proteomics
Introduction
Klebsiella pneumoniae (KP) is one of the most important opportunistic pathogen responsible for community-acquired and hospital-acquired infections, which is second only to Escherichia coli.1,2 Hypervirulent Klebsiella pneumoniae (HVKP) is a new Klebsiella pneumoniae variant first discovered by Taiwanese scholars in the 1980s.3–5 The most important distinction between HVKP and classic Klebsiella pneumoniae(cKP) is that HVKP can cause severe invasive and metastatic diseases such as liver abscess, necrotizing fasciitis, meningitis and endophthalmitis in healthy and young individuals, followed by disastrous and even life-threatening sequelae, which is extremely harmful to society.6,7
A battery of virulence factors contribute to the hypervirulence of HVKP, including capsule, siderophores, lipopolysaccharide (LPS), fimbriae, outer membrane proteins, and type 6 secretion system (T6SS), of which the special capsule and extremely strong iron acquisition ability are the predominant virulence factor of HVKP.8 Iron is essential for the life metabolism of microorganisms by functioning as prosthetic group or activator for various enzymes. Iron-deficient environment is an important protective measure to the invasion of microorganisms which drives bacterial pathogens to evolve various iron acquisition virulence systems to cope with this situation.9,10
Several studies have shown that outer membrane vesicles (OMVs) are involved in bacterial virulence.11–13 Gram-negative bacteria naturally secreted OMVs which is spherical vesicles with diameters ranging from 20 to 250 nm.14,15 OMVs are membrane structure composed of phospholipids and bilayer. Increasing studies have shown that OMVs are particularly effective at delivering cargo to host cells which play an important role in the process of bacterial virulence factor transmission, biofilm formation, invasion and infection, natural self-defense, response to external pressure and nutrient intake. The OMVs isolated from Mycobacterium tuberculosis contain mycobactin, which can serve as an iron donor and supports replication of iron-starved mycobacteria.16 A new OMVs-associated iron acquisition mechanism was revealed in Pseudomonas aeruginosa.17 These studies indicated that the OMVs-associated mechanism of iron acquisition may be widely employed in bacterial pathogens.
There has been little research focusing on the role of OMVs in the HVKP virulence mechanism. In order to understand the correlation between OMV and iron acquisition ability, differential proteomics analysis were performed on OMVs secreted by HVKP and cKP culture under iron-deficiency and iron-sufficient conditions in this study. Stimulation of OMVs release, influences on the protein patterns of OMVs and comparison of HVKP and cKP OMVs under iron limitation were observed in this study. It may provide ideas for the virulence mechanism related to HVKP iron acquisition, and lay the scientific foundation for the prevention and treatment of HVKP infection.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains used in this study were HVKP and cKP. HVKP is NTUH-K2044 originated from a gift from Taiwan University, which isolated from a Taiwanese liver abscess and was widely used as a model strain for HVKP. cKP is Klebsiella pneumoniae CMCC46117 from National Center for Clinical Laboratory. The iron-deficient (Fe-) media were prepared with M9 Minimal Medium (MM, Sangon Biotech, China). MM contains 6.78g/L Na2HPO4,3.0g/L KH2PO4, 0.5g/L NaCl, 1.0g/L NH4Cl, 0.493g/L MgSO4·7H2O, 0.011g/L CaCl2 and 4g/L glucose (Sinopharm, China). The iron-containing media with concentrations of 0.02μg/mL FeCl3 MM and 2.0μg/mL FeCl3 MM were prepared by adding 5μL and 500μL 200 μg/mL FeCl3(0.02g FeCl3·6H2O(Sinopharm, China) added to 100mL H2O, PH2.53) to the 50mL Minimal Medium respectively. Ultra-pure water were used for all media preparation. Frozen HVKP and cKP strains were resuscitated and purified by growing after 2 times of passages on Columbia Blood Agar at 37°C, 5% CO2. Pure colonies were cultured in iron-deficient and iron-sufficient media, respectively.
Growth Curve Analysis
Bacterial suspension of 0.5 McFarland Standard (McF) was made by picking a pure colony of bacterial into sterile saline. For growth analyses, 50μL bacterial suspension were diluted in 50ml of Fe- media, 0.02μg/mL and 2.0μg/mL FeCl3 MM which were preheat at 37°C. The culture media were placed in a constant temperature shaker (BILON cos-100c), and the conditions were set at 37 ° C and 200rpm/min. Optical density (OD600nm, [OD600]) was measured and recorded every 1h by Infinite M200 PRO.
Bacterial OMVs Extraction, Purification and Identification
250μL of bacterial suspension was added to Fe- media and 2.0μg/mL FeCl3 MM prepared in advance, each of them was 250mL. These media were put on a constant temperature shaker at 37 °C and 200rpm/min until the logarithmic phase of bacterial growth, which was determined according to the growth curve in the previous step. Bacterial culture solution was centrifuged at 4000×g 4°C (Eppendorf 5702) for 15 min to remove impurities such as large intact bacterial cells. Then the supernatants were filtered by 0.45μm Millipore (Billerica, MA) to remove small cell debris. Two runs of ultracentrifugation (J6MI, Beckman Coulter, USA) to remove residual impurities. The filtered supernatants were pelleted by centrifugation at 40,000 g for 3h, 4°C. Then the pellet was resuspended in sterile PBS buffer. Centrifugation and resuspension were repeated once. Finally, the bacterial OMVs were suspended in 500ul sterile PBS buffer. The morphology and size of bacterial OMVs were observed under a transmission electron microscope (Hitachi H-7500, Japan) using negative staining.
Relative Quantification of OMVs
Stewart assay was used to quantify bacterial OMVs.18 Similar with OMVs extraction steps, the pellet after the first ultracentrifugation was resuspended in MV buffer, which contains 50mM Tris, 5mM NaCl and 1mM MgSO4, pH 7.4. The pellet after the second ultracentrifugation was suspended in 1mL MV buffer. At the same time an equal volume of chloroform was added and then centrifuged at 4000×g for 15 min. The lower organic layer was left, add an equal volume of ferric thiocyanate solution, which contains 27.03g/L FeCl3·6H2O and 30.4g/L NH4SCN, centrifuged at 4000g for 15 min. Took the lower organic layer and dried it with nitrogen. The dried sample was resuspended in 1mL chloroform, and its absorbance at 470nm was measured three times repeatedly. The resulting absorbance value was then normalized to the OD600 of the extracted culture.
Quantitative Proteomics Analysis of OMVs
OMVs secreted by HVKP and cKP in Fe- media and 2.0μg/mL FeCl3 MM were extracted for proteomics analysis. Comparative proteomics analysis was carried out by mass spectrometry. The lysis solution (2.5%SDS/100mMTris-HCL, pH=8.5) was added to the OMVs precipitate after two runs of ultracentrifugation. After separation of protein and enzymatic hydrolysis, Mass spectrometry analysis was processed by using TripleTOF5600 LC/MS system (SCIEX). Data generated by TripleTOF5600 was searched in MaxQuant (V1.6.2.10). The database search algorithm was performed by using MaxLFQ. The Proteome Reference Database of Klebsiella pneumoniae in UniProt was used in this study (Uniprot_Proteome_Klebsiellapneumoniae IS43_20181210.fasta). The proteins identified by mass spectrometry were screened with the standard Unused≥1.3 (confidence interval>95%). After deletion of the entries searched in the reverse library and contaminated proteins, the remaining proteins were used for subsequent analysis. LFQ intensities were converted to a log scale (log2). Missing values were imputed from a normal distribution. Mean±1.96SD was the threshold to identify proteins with significant changes in abundance.
Bioinformatics analysis mainly uses the Gene Ontology database (http://geneontology.org/) to perform biological Process, cellular component and molecular function on a single sample.
Statistical Analysis
All data analysis was performed by IBM SPSS 21.0 software. Comparison between the two groups was performed using paired sample t-test or two independent sample t-test. A P value < 0.05 was considered statistically significant.
Results
Growth Curve Analysis
The growth of HVKP in Fe- media (ODmax 0.54), 0.02μg/mL FeCl3 MM (ODmax 0.66) and 2.0μg/mL FeCl3 MM (ODmax 0.67) was shown in Figure 1. The growth of HVKP in the iron-containing media was similar, while in the Fe- media it was lower. Also, cKP could grow in iron-containing medium, but the growth rate was related to the concentration of iron. In the Fe- media, it was no growth of cKP within the first 17 hours, as we can see in Figure 2. The growth of cKP was susceptible to the concentration of iron in the media. At the same time, the strong iron acquisition ability of HVKP made it more suitable for iron-deficient environments.
 |
Figure 1 The growth of HVKP in the iron-containing and iron-deficient media. |
 |
Figure 2 The growth of cKP in the iron-containing and iron-deficient media. |
According to the growth curve analysis, we extracted OMVs from culture media when its absorbance value reached 0.5 (OD600) in the logarithmic growth phase to obtain a larger amount of OMVs. And the extracted OMVs were used for subsequent electron microscope observation and quantitative comparison. In addition, MM and 2.0μg/mL FeCl3 MM were used to simulate iron-deficiency and iron-sufficient environment in this study.
Observation of OMVs
Under the transmission electron microscope, it can be observed from Figures 3 and 4 that the morphology of OMVs secreted by HVKP and cKP are mostly spherical vesicle structure, and part of them are irregular in shape. Multiple vesicles can be seen to gather into clusters. The size of OMVs is between 20–250nm, which were similar to OMVs secreted by other Gram-negative bacteria.
 |
Figure 3 Observation of OMVs secreted by HVKP under the transmission electron microscope. |
 |
Figure 4 Observation of OMVs secreted by cKP under the transmission electron microscope. |
Quantitative Comparison of OMVs
As shown in Figure 5, compared with the iron-sufficient environment, HVKP secreted more OMVs when cultured in an iron-deficient environment, the difference was statistically significant (P <0.05). However, the number of OMVs secreted by cKP when grown in Fe- media or 2.0μg/mL FeCl3 MM was not significantly different. Furthermore, the difference between the OMVs released by HVKP and cKP in the same environment was compared. The results showed that HVKP secreted more OMVs than the cKP in the Fe- media, and the difference was statistically significant (P <0.05). There was no significant difference between HVKP and cKP in the 2.0μg/mL FeCl3 MM.
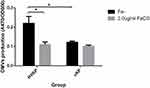 |
Figure 5 Quantitative comparison of OMVs secreted by HVKP and cKP in the iron-containing (2.0μg/mL FeCl3) and iron-deficient media (Fe-). *P<0.05. Abbreviation: A470, absorbance at 470 nm. |
Proteomics Analysis of OMVs
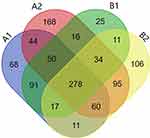
After mass spectrometry, 1074 kinds of proteins were identified from the purified OMVs in four samples. Among them, 619 proteins were identified from HVKP OMVs under iron-deficient environment (A1). 745 proteins were identified from HVKP OMVs in the iron-sufficient environment (A2). A total of 522 proteins were identified from cKP OMVs under iron-deficient environment (B1) and 612 proteins under iron-sufficient environment (B2). The number of proteins identified in each sample and their cross-relationship were shown in Figure 6. The entire list of proteins is provided in the Supplementary Materials.
GO (Gene ontology) analysis of OMVs proteome demonstrate the overlap among four samples. Overall, analysis of cellular component showed that the OMVs proteins mostly enfold 8 kinds of cell components, which are protein-containing complex, organelle part, organelle, membrane, cell, extracellular region, membrane part, nucleoid. Besides the above, A1, B1 enfold membrane-enclosed lumen. A2, B2 enfold cell part. These results suggest that OMVs content is not simply bacterial metabolites, but also many cell-derived components. OMVs could be important vehicles to transmit and communicate cellular information. The OMVs proteins from four samples primarily participate in 11 biological processes, which are cellular process, cellular component organization or biogenesis, localization, metabolic process, multi-organism process, biological regulation, reproductive process, biological adhesion, response to stimulate, developmental process, carbon utilization. Besides the above, A1 participates in locomotion. A2, B1 participate in detoxification. The OMVs proteins from four samples were mainly involved in 9 kinds of molecular functions, which are molecular transducer activity, molecular function regulator, antioxidant activity, translation regulator activity, transcription regulator activity, catalytic activity, binding, transporter activity, structural molecule activity. Besides the above, A2, B1 involved in molecular carrier function.
A comparison between the iron-deficient vs iron-sufficient environment showed that 61 proteins were identified with a significant change in abundance: 34 proteins significantly increased and 27 proteins significantly decreased in HVKP OMVs under iron-deficient environment. Among them, 17 proteins were related to iron acquisition and transportation systems. While in cKP OMVs, 37 proteins significantly increased and 25 proteins significantly decreased under iron-deficient environment. Among them, 5 proteins were related to iron acquisition and transportation systems. Upon comparison of the HVKP vs cKP OMVs in the absence of iron, 45 proteins significantly increased and 36 proteins significantly decreased in HVKP OMVs. Among them, 8 proteins were related to iron acquisition and transportation systems. Notably, the increased proteins under iron-deficient environment were mainly involved in receptors of iron transport, TonB-ExbB-ExbD energy conversion systems, and ATP binding cassette (ABC) protein receptors. The specific iron-acquisition related proteins and their functions were shown in Table 1. These results highlight the abundance of iron-associated proteins in OMVs under iron-deficient environment and demonstrate a novel connection between OMVs and iron-acquisition.
 |
Table 1 The OMVs Proteins Up-Regulated in Iron-Deficient Medium and Functions |
Discussion
OMVs are particularly effective at delivering cargo to host cells. The diverse contents incorporated into OMVs including bacterial virulence factors, active substances and genetic materials so that OMVs get involved in many important biological functions such as inducing immune response, promoting cell-to-cell information exchange, bacterial biofilm formation, and nutrition competition.19–23 In this study, mass spectrometry was used for quantitative proteomics analysis of OMVs secreted by HVKP and cKP under iron-deficient and iron-sufficient environment. The aim of this study was to screen out potential iron acquisition-related proteins and help us further understand the iron acquisition mechanism of HVKP.
In this study, Stewart assay was used to quantify the amount of OMVs released by HVKP and cKP under iron-deficient and iron-sufficient environments. Significant increase in HVKP OMVs production was detected under iron-deficient environment, indicating that the higher production of OMVs could be a part of the adaptive response of HVKP to iron limitation. This result is consistent with previous reports of increased vesicle production by bacterial pathogens when cultured under nutrient-deficient environments. Previous studies demonstrate that the pathogenic bacteria usually release more OMVs than their nonpathogenic counterparts.24,25 A higher rate of OMVs production of HVKP than cKP under iron-deficient environment was also detected in this work, displaying a stronger response of HVKP to iron limitation. This phenomenon provides evidence for the idea that OMVs production may be a part of the adaptive response that promotes acquisition of iron and survival under iron limitation.
To cope with iron limitation, pathogenic bacteria have evolved many sophisticated mechanisms to sequestrate metal iron from the environment. The iron sequestration mechanisms including secretion of siderophores, high-affinity iron-chelated molecules, expression of iron-binding proteins. Iron availability is essential for the bacterial virulence and infection. Study in Mycobacterium tuberculosis has shown that OMVs of M. tuberculosis containing mycobactin siderophore can serve as an iron donor and supports replication of iron-starved mycobacteria.16 Vesicular proteomes of Campylobacter jejuni identified 134 OMVs proteins and found a variety of virulence factors and proteins for bacterial survival.26 Comprehensive proteomic analysis of OMVs from Shigella flexneri identified 148 proteins including a variety of important virulence factors, such as lpa proteins, TolC family, murein hydrolases.27 In Acinetobacter baumannii, it was found that the proteins incorporated in OMVs can enhance the invasive ability of pathogenic bacteria.28 To investigate the connection between iron availability and OMVs proteomes, quantitative proteomics were employed to identify and screen proteins with significant change in abundance under iron limitation. A variety of proteins associated with iron acquisition, binding and transportation were identified in OMVs proteomes. In HVKP OMVs, Iron(III) dicitrate transport protein FecA and 1-phosphofructokinase were found a>9-fold increase in abundance under iron deficiency. CirA (Catecholate siderophore receptor), FeoB (iron transport protein), IroN (Outer Membrane Siderophore receptor) were found a >8-fold increase. These data indicates the strong adaptive response of HVKP under iron deficiency. Previous study by Muselius’ et al reported that Ent siderophores were observed in cellular proteome rather than in OMVs proteomes.29 In agreement with their findings, siderophores were not observed in HVKP and cKP OMVs proteomes. Taken together, Klebsiella pneumoniae may employ different iron acquisition mechanisms from the M. tuberculosis which incorporated mycobactin siderophore into OMVs serving as an iron donor for iron-starved mycobacteria.
A recent study revealed that low iron concentrations up-regulate the expression of the locus I of the T6SS which is encoding essential components known to be necessary to assemble a T6SS.30 In addition to the proteins related to the iron acquisition and transport system, the proteomic analysis also found 6 bacterial T6SS related proteins incorporated in OMVs. VgrG forming a cell-puncturing tip, it was not only a directly interact device, but also a secreted protein of T6SS, which exerted virulent infections. ClpB is an energy-providing ATPase that mediates the formation of T6SS secretion channels and the release of effector proteins, while tssK interacts with the membrane structure to form a T6SS membrane complex.31 It has been reported that the Pseudomonas aeruginosa H3-T6SS system secretes the effector protein TseF to recognize the signaling molecule PQS in OMVs, and cooperates with the iron transporters in outer membrane FptA and OprF to recruit bacterial OMVs.17 Based on these, we speculated that there may be a T6SS-OMVs-Fe interaction network mode for HVKP iron acquisition mechanism. These three factors cooperate together to help HVKP compete for iron. Further investigations are needed to verify this notion.
In summary, this study showed the stimulation of OMVs release, increase of proteins associated with iron acquisition in OMVs and comparison of HVKP and cKP OMVs under iron-limitation. The possible mechanism of iron acquisition by HVKP was preliminarily discussed, which provided candidate proteins for follow-up research, and laid the foundation for further study of OMV-mediated HVKP virulence regulation.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: [iProX AND IPX0003384000].
Acknowledgments
We are thankful to the Taiwan University for donation of NTUH-K2044.
Funding
There is no funding to report.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hawser S, Hoban DJ, Badal RE, et al. Epidemiology and antimicrobial susceptibility of gram-negative aerobic bacteria causing intra-abdominal infections during 2010–2011. J Chemother. 2015;27(2):67–73. doi:10.1179/1973947814Y.0000000164
2. Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185–188.
3. Cheng DL, Liu YC, Yen MY, et al. Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch Intern Med. 1991;151:1557–1559.
4. Liu YC, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146:1913–1916.
5. Wang JH, Liu YC, Lee SS, et al. Primary liver abscess due to Klebsiella pneumoniae inTaiwan. Clin Infect Dis. 1998;26:1434–1438.
6. Fang CT, Lai SY, Yi WC, et al. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi:10.1086/519262
7. Jae BJ. Klebsiella pneumoniae liver abscess. Infect Chemother. 2018;50(3):210–218. doi:10.3947/ic.2018.50.3.210
8. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a dangerous breed. Virulence. 2013;4(2):107–118. doi:10.4161/viru.22718
9. Andrews SC, Robinson K, Rodríguez-Quiñones F, et al. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237.
10. Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi:10.1038/nrmicro1046
11. Rivera J, Cordero RJ, Nakouzi AS, et al. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A. 2010;107:19002–19007. doi:10.1073/pnas.1008843107
12. Chatterjee D, Chaudhuri K. Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 2011;585:1357–1362. doi:10.1016/j.febslet.2011.04.017
13. Vanhove AS, Duperthuy M, Charrie`re GM, et al. Outer membrane vesicles are vehicles for the delivery of vibrio tasmaniensis virulence factors to oyster immune cells. Environ Microbiol. 2015;17:1152–1165.
14. Haurat MF, Aduse-Opoku J, Rangarajan M, et al. Selective sorting of cargo proteins into bacterial membrane vesicles. J Biol Chem. 2011;286:1269–1276.
15. Bonnington KE, Kuehn MJ. Protein selection and export via outer membrane vesicles. Biochim Biophys Acta. 2014;1843(8):1612–1619.
16. Prados RR, Brian CW, Daniel GP, et al. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol. 2014;196:1250–1256. doi:10.1128/JB.01090-13
17. Lin J, Zhang W, Cheng JA, et al. Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8:1–11. doi:10.1038/ncomms14888
18. Stewart JC. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem. 1980;104(1):10–14.
19. Berleman J, Auer M. The role of bacterial outer membrane vesicles for intra-and interspecies delivery. Environ Microbiol. 2013;15:347–354.
20. Amano A, Takeuchi H, Furuta N. Outer membrane vesicles function as offensive weapons in host–parasite interactions. Microbes Infect. 2010;12:791–798.
21. Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94.
22. Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J Biol Chem. 2000;275:12489–12496. doi:10.1074/jbc.275.17.12489
23. Wai SN, Lindmark B, Söderblom T, et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi:10.1016/S0092-8674(03)00754-2
24. Lai CH, Listgarten MA, Hammond BF, et al. Comparative ultra structure of leukotoxic and non-leukotoxic strains of Actinobacillus actinomycetemcomitans. Periodontal Res. 1981;16:379–389. doi:10.1111/j.1600-0765.1981.tb00989.x
25. Wai SN, Takade A, Amako K, et al. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol Immunol. 1995;39:451–456. doi:10.1111/j.1348-0421.1995.tb02228.x
26. Jang KS, Sweredoski MJ, Graham RL, et al. Comprehensive proteomics profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics. 2014;98:90–98. doi:10.1016/j.jprot.2013.12.014
27. Chen Y, Liu L, Fu H, et al. Comparative proteomic analysis of outer membrane vesicles from Shigella flexneri under different culture conditions. Biochem Biophys Res Commun. 2014;453(4):696–702.
28. Li ZT. Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microb Pathog. 2015;8l:46–52.
29. Muselius B, Sukumaran A, Yeung J, Geddes-McAlister J. Iron limitation in Klebsiella pneumoniae defines new roles for lon protease in homeostasis and degradation by quantitative proteomics. Front Microbiol. 2020;24(11):546.
30. Storey D, McNally A, Åstrand M, et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16(3):e1007969. doi:10.1371/journal.ppat.1007969
31. Russell AB, Peterson SB, Mougous JD, et al. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12(2):137–148. doi:10.1038/nrmicro3185
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.