Back to Journals » Infection and Drug Resistance » Volume 14
Predictive Role of Targeted, Active Surveillance Cultures for Detection of Methicillin-Resistant Staphylococcus aureus
Authors Al Musawi S, Alkhaleefa Q, Alnassri S , Alamri A , Alnimr A
Received 4 October 2021
Accepted for publication 4 November 2021
Published 12 November 2021 Volume 2021:14 Pages 4757—4764
DOI https://doi.org/10.2147/IDR.S340871
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Safiya Al Musawi,1 Qassim Alkhaleefa,2 Samia Alnassri,3 Aisha Alamri,4 Amani Alnimr2
1Department of Pathology, Salmaniya Medical Complex, Ministry of Health, Manama, Kingdom of Bahrain; 2Department of Microbiology, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia; 3Department of Infection Control, King Fahad Hospital of the University, Dammam, Kingdom of Saudi Arabia; 4Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Imam Abdulrahman Bin Faisal University, Dammam, Kingdom of Saudi Arabia
Correspondence: Amani Alnimr
Department of Microbiology, College of Medicine, Imam Abdul Rahman Bin Faisal University, P.O. Box 1982, Dammam, 31441, Kingdom of Saudi Arabia
Tel +966 56 318 1019
Email [email protected]
Aisha Alamri
Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Imam Abdul Rahman Bin Faisal University, Dammam, 3144, Kingdom of Saudi Arabia
Tel +966 538187263
Email [email protected]
Introduction: Methicillin-resistant Staphylococcus aureus (MRSA) colonisation is an important source of healthcare-acquired infections. Reliable screening strategies for MRSA colonisation are essential for the timely implementation of infection control measures.
Aim: This study determined reliable MRSA screening sites to predict colonisation in resource-limited settings and estimated the impact of missed MRSA cases when shifting from multi- to single-site screening.
Methodology: A cross-sectional study was conducted in patients with positive MRSA surveillance cultures from the routinely screened sites (nasal, axillary, groin, and throat) from January 2009 to December 2019.
Results: A total of 1906 screening tests were positive for MRSA cultures (n = 1345 patients). As a single site, the nasal cavity showed the highest MRSA detection, with a sensitivity of 66.8% (95% CI = 64– 69) with 277.9 missed isolation days. Screening three or more anatomical sites detected 97– 100% of MRSA cases, with 0– 24.5 missed isolation days. Screening the axilla and groin separately or in combination showed a good clinical utility index (CUI) of > 0.6 to < 0.8, while an excellent CUI was obtained upon screening other site samples (> 0.8). The combined nasal and throat cultures demonstrated a sensitivity of 93.2 (95% CI = 91– 94) with 57.2 missed isolation days.
Conclusion: Multi-site screening is the optimal strategy for minimising MRSA exposure within a healthcare facility. For active MRSA surveillance, a combination of nasal and throat cultures can provide a practical approach in low-resource settings compared to nasal sampling alone.
Keywords: methicillin-resistant Staphylococcus aureus, screening sites, infection control, Saudi Arabia
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has been one of the leading causes of healthcare-acquired infections since its identification in the 1960s.1,2 The prevalence of community- and hospital-acquired MRSA has increased over time, with a wide diversity of strains isolated from a carrier state.3 MRSA colonisation in humans increases the risk of subsequent clinical infections.4,5 Nasal screening of 758 patients on hospital admission demonstrated significantly higher MRSA infection rates (19% versus 2%) in the following 12-month period among colonised individuals,6 the anatomical colonisation site may be a key factor in MRSA infection.7 Despite the anterior nares being the primary S. aureus colonisation site,4 certain strains, such as the MRSA-PFGE type USA300, preferentially colonise the groin, leading to infections in adjacent areas.7 It was shown that MRSA infection could be predicted by a preceding MRSA carriage at multiple sites, which can be detected by bacteriological cultures or molecular tools.8,9 A recent meta-analysis of 52,642 MRSA cases by Chipolombwe et al highlighted the need for multi-site sampling in MRSA-colonisation detection.10 The optimal site or combination of sites for predicting MRSA colonisation is inconsistent among various studies,10 and the diagnostic cultures have been found to miss ~35–84% of the colonisation status.11,12 These findings reflect the controversy regarding the optimal anatomical site for MRSA sampling.
Nosocomial MRSA infections are associated with a significant increase in the length of hospital stay, expense, mortality and morbidity,13,14 which can be prevented through infection control interventions.2 Huang et al determined that MRSA identification, followed by contact isolation precautions, effectively reduces MRSA cross-transmission in intensive care settings.15 In addition, a multi-centre initiative in the United States reduced MRSA infection rates by 55% between 2005 and 2017 by implementing the interventions recommended by the Centers for Disease Control and Prevention (CDC), including screening patients for MRSA upon admission.2 Nevertheless, this approach can significantly impact healthcare resources, especially when they are limited. Various strategies have been applied to maintain a cost-effective diagnostic MRSA screening yield. For instance, targeted active MRSA surveillance is a cost-effective strategy in an academic institution, unlike universal screening for routine admission or prior to surgery.16–18 Institutions may consider single-site screening for practicality and cost reduction. This study compared the predictive role of single-site versus multi-site MRSA screening in a low-prevalence setting to estimate the MRSA cases that could be missed by single-site screening.
Methods
Study Settings
A retrospective cross-sectional study was conducted from January 2009 to December 2019 at the King Fahd Hospital of the University Al-Khobar (KFHU) Khobar, Dhahran, Saudi Arabia (550-bed secondary care and academic training facility). The institution follows an active, targeted MRSA surveillance protocol in which all patients with MRSA-acquisition risk factors are routinely screened upon admission.19 Patients with positive MRSA-culture results from screening sites (nasal, axillary, groin, and throat) were included in this study. MRSA colonized patients were defined as those who tested positive for MRSA screening in one or more of these sampling sites (nose, throat, axilla or groin) in the absence of a concurrent local infection such as an abscess. The data were obtained from electronic hospital records, including gender, age, screening location, and antimicrobial susceptibility results. Patients with incomplete screening in any anatomical sites, missing susceptibility results, or repeated MRSA-positive results within six months were excluded from this study.
Microbiological Analysis
The swabs received at the laboratory were plated on mannitol salt agar or CHROMagar MRSA chromogenic medium (SPML, Riyadh, Saudi Arabia) and incubated overnight at 37 °C. Suspected MRSA growth was confirmed by catalase, coagulase or DNAse tests followed by automated microbial identification using the fully automated VITEK 2 system (bioMérieux Inc., Durham, NC, USA) in 2009–2016 and the VITEK MS (bioMérieux Inc.) in 2017–2019, following the manufacturer’s instructions. MRSA screening was performed using cefoxitin discs (30 µg) on Muller-Hinton agar (SPML, Dammam, Saudi Arabia), followed by antibiotic susceptibility testing using the VITEK 2 automated system. D-tests were performed to detect inducible clindamycin resistance in all strains showing macrolide resistance and lincosamide susceptibility.20 The Clinical and Laboratory Standards Institute (CLSI) breakpoints were used to interpret the susceptibility results.20 The Xpert® MRSA molecular assay (Cepheid, Sunnyvale, USA) was used to resolve any discrepancy between the cefoxitin disc diffusion and the VITEK 2 system.
Statistical Analysis
The analysis was performed with the Statistical Package for Social Sciences (IBM Corp., Armonk, NY, USA) version 26.0 using a patient-based approach. The 95% confidence interval (CI) of the sensitivities were calculated, and the proportion’s difference of p-value < 0.05 was considered statistically significant. The number needed to diagnose (NND) was calculated as NND = NND = 1/[Sensitivity – (1 – Specificity)] where the smaller the NND, the more useful the assay. The clinical utility index (CUI) was calculated as previously described.21 The missed isolation days were estimated based on the historical institution data of isolation days per confirmed MRSA case, considering that any positive MRSA culture signifies a true carrier state in routine clinical practice.
Ethical Approval
This study was approved by the Institutional Review Board of Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia (IRB number 2020-01-368). This retrospective, sample-based, surveillance study was exempt from the requirement of informed consent, as it involved data collected from laboratory databases and infection control records for the purpose of research.
Results
Our retrospective study from 2009 to 2019 included 1906 non-replicate, MRSA-positive cultures obtained from four sites (nose, throat, axillary and groin) of each screened patient (n = 1345) with a median age of 23 years (SD = 24.61), representing any site of colonisation. The cases included 51% males and 48.3% females with the highest frequency of MRSA isolation noted in the age group >50 years and 45.9% of strains cumulatively isolated from patients <20 years. The detailed demographic data are shown in Table 1.
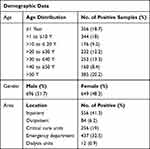 |
Table 1 Epidemiological Characteristics of Study Population (n = 1345, Median Age = 23 Years, SD = 24.61) |
MRSA was recovered from 889 nasal, 619 throat, 181 groin, and 101 axillary swabs. As a single site, the nasal cavity exhibited the highest MRSA detection sensitivity of 66.8% (95% CI = 64–69). A three-site combination improved the detection sensitivity, with the optimal combination being nasal, throat and groin, followed by nasal, throat, and axilla. The nasal, throat, groin, and axilla showed 100% sensitivity with a 95% CI = 98–100. The exclusive single-site carriage was 225 (16.7%), 155 (11.5%), 41 (3%), and 2 (0.15%) for the nasal, throat, groin, and axilla, respectively.
Based on the calculated CUI, screening the axilla and groin separately or in combination showed good utility, while other screening strategies showed excellent utility. The highest CUI was achieved by screening three or more sites. The predictions of the various screening sites for culture-based MRSA detection from single and combination sites are illustrated in Table 2.
 |
Table 2 Prediction of Different Screening Sites for Culture-Based MRSA Detection |
The MRSA isolates showed 38.98% erythromycin, 32.95% clindamycin, 24.46% trimethoprim-sulfamethoxazole, and 1.6% rifampin resistance rates. No vancomycin- or linezolid-resistant isolates were detected. Over the eleven-year period, there was a significant reduction in the clindamycin (p = 0.017) and rifampin (p = 0.000) resistance rates. However, no significant difference was observed in the erythromycin resistance (p = 0.313). There is an increasing tendency over years for trimethoprim-sulfamethoxazole resistance with p value of 0.062. The trend of antimicrobial resistance among the MRSA screening cultures is shown in Figure 1.
 |
Figure 1 Trend of antimicrobial resistance among MRSA screening cultures. |
Discussion
The growing number of MRSA infections has become a complex health problem, in which individuals with mucosa or skin MRSA colonisation can be a reservoir for ongoing hospital transmission.2,11 Reported MRSA carriage risk factors included exposure to antibiotics, previous hospitalisation, major surgery, chronic comorbidities and, in some studies, male gender.22 However, our cohort did not show a significant male-factor predominance (Table 1). A previous study from the same region reported a 20–40 age group female predominance for MRSA nasal colonisation.23 Notably, our study found that >45% of MRSA colonisation occurred in young patients and children (Table 1). These MRSA isolation frequencies may reflect more active surveillance in pediatric wards over other hospital units, although outbreak investigation is warranted in such a case. In a meta-analysis of 18 studies conducted in the United States from 1999 to 2011, the MRSA colonisation prevalence and pooled acquisition rate in neonatal and pediatric intensive care units were approximately 1.9% (95% CI = 1.3–2.6) and 4.1% (95% CI = 1.2–8.6), respectively.24 The colonisation rate in that analysis was higher in neonates who were admitted after discharge from the birth hospital (5.8 versus 0.2%), and the risk of MRSA infection during hospitalisation was amplified among colonized neonates (Relative Risk = 24.2, 95% CI 8.9–66.0). Detection of MRSA carriage in young age group necessitates implementing effective infection control intervention and, in some cases, decontamination.
The duration of MRSA colonisation in asymptomatic carriers is uncertain and can vary from a few months to several years.25–27 Previous studies have suggested that previously colonised patients are at risk of continued carriage for at least four years.28 Thus, patients who have been MRSA carriers may require frequent screening upon each admission. In addition, the evidence suggested that active enhanced MRSA surveillance led to earlier, more frequent discontinuation of contact precautions when they were no longer needed (relative risk = 2.5, 95% CI = 1.5–4.7), as shown in a randomised clinical trial of >600 patients, resulting in reduced hospital costs associated with the reduced need for isolation.29 In consideration of frequent testing, sampling fewer sites for MRSA carriage without a negative impact on the diagnostic yield is preferred. Our study highlighted the limitation of nasal culture-based screening only, which failed to detect 33.2% of the cases, resulting in 277.9 missed isolation days (95% CI = 253–294) (Table 2). It also showed that neither the groin nor the axilla alone or in combination is adequate for screening. Although the anterior nares are considered the primary site of MRSA carriage, several studies have shown that the groin can be a frequent reservoir in patients with skin and soft tissue infections; this was not evident in our study.30,31 Combining throat and nasal samples resulted in missing only 57.2 days (95% CI = 49–73), with high sensitivity, negative predictive value and CUI. This approach can potentially be used as minimal screening if comprehensive multi-site MRSA surveillance testing is not practical owing to cost or other considerations. Local epidemiological factors have to be taken into consideration when adopting such an approach since a systemic review of 52,642 patients from centres (mostly in Europe and the United States) found that throat and groin was the optimal two-swab combination for MRSA detection (89.6%; 62.5–100%), with an increase in the detection yield by the three swab combination of nose, throat and groin to 94.2% (81–100%).10 This review also showed that MRSA detection in the nasal cavity alone exhibited 68% (34–91%) sensitivity, and the throat and nose exhibited the highest yields as single anatomical sites, consistent with our findings. It should be noted that the nose is the most frequent MRSA colonisation site, and the shedding of nasally carried MRSA occurs more readily with concomitant respiratory tract infections, resulting in increased cross-transmission in hospitals.32 The cumulative sensitivity of MRSA detection from various sample combinations (Table 2) is also similar to the findings of Senn et al, in which the sensitivity of nasal swab culture alone was 48%, increasing to 79% when combined with groin swabs and 96% when groin and throat swabs were combined.33 Several factors may contribute to the variation in the MRSA-colonisation detection rate, such as laboratory testing tools, usage of selective and broth-enrichment media, and previous decolonisation procedures.34,35 Other possible approaches to reduce the cost of frequent screening include pooling various swabs in one culture broth or transport medium,36 which has not been systematically evaluated.
Lautenbach et al found the throat to be the second most frequent reservoir for MRSA carriage, consistent with our finding.37 An exclusive throat carriage was previously reported. A European study of 3464 patients found that 12% of MRSA cases were detected only from throat cultures, confirming nasal screening alone to be inadequate.38 Similarly, in this study, we found 11.5% of exclusive MRSA throat carriers. The anterior nares alone were the exclusive site for 16.7% of the patients, while 3% and 0.15% of the cases were exclusive carriers at the groin or axilla, consistent with previous reports of most carriers being colonised at multiple sites.39,40 MRSA colonisation can occur in other anatomical sites such as chronic or surgical wounds, sputum, intravascular catheter sites, stool and the genitourinary tract. A study of 71 hospitalised colonised patients showed a 67% carriage rate in the gastrointestinal tract associated with skin colonisation.41 Whether routine stool screening for MRSA is clinically useful in detecting more MRSA cases is unknown. Notably, the resistance patterns of the asymptomatic MRSA strains in our study showed reduced resistance to some antimicrobial classes over time (Figure 1), suggesting an increase in community origin. However, the distinction between hospital- and community-acquired MRSA is no longer certain.42 In addition, our data showed an overall increase in trimethoprim-sulfamethoxazole resistance in MRSA over years, similar to published studies from United States and Germany.43,44 Trimethoprim-sulfamethoxazole is considered as alternative agents for the treatment of certain MRSA infection and is sometimes used in salvage therapy.45
In this study, the missed isolation days for nasal MRSA screening culture only were 277.9 days. A published study evaluating multiple anatomical site MRSA screening upon admission at a teaching hospital in France found that nasal cultures alone missed 27.0% of MRSA, corresponding to 560 theoretical isolation days.40 The extent of missed MRSA isolation days depends on the patient population, local infection-prevention measures for colonised patients, comorbidities that require longer stays, among other factors. The optimal CUI and NND results were achieved by screening three or more anatomical sites (Table 2). The data on optimal and cost-effective sampling sites for MRSA screening are obscure.12 Because multifaceted approaches are often followed to reduce the incidence of MRSA outbreaks, the individualised role of screening remains uncertain. Further investigation is required to assess the cost-efficacy of the diagnostic yield of MRSA screening tools in high and low MRSA prevalence.
Conclusion
This study concluded the inadequacy of nasal sampling alone for MRSA detection because it missed a significant proportion of colonised cases. We proposed combined nasal and throat screening as the optimal alternative to comprehensive multiple-site culture surveillance when resources are limited. The combination approach of all anatomical sites is the MRSA screening strategy with the highest yield for institutions seeking no missed isolation days. In large populations, in which young patients represented a majority, no gender preference was noted for MRSA colonization. Further prospective studies are required to determine the cost-effectiveness of various infection control intervention methods and screening approaches, including comparing molecular versus routine testing in different prevalence settings.
Abbreviations
CDC, Centers for Disease Control and Prevention; CI, confidence intervals; CLSI, Clinical and Laboratory Standards Institute; CUI, clinical utility index; DNAse, deoxyribonuclease; KFHU, King Fahd Hospital of the University Al-Khobar; MRSA, methicillin-resistant Staphylococcus aureus; NND, number needed to diagnose; PFGE, pulsed-field gel electrophoresis; SD, standard deviation.
Acknowledgments
The authors would like to thank all members of the Diagnostic Microbiology Laboratory and Infection Control department at KFHU who were involved in the routine processing of data related to this work.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Funding
The authors received no financial support for this work.
Disclosure
The authors declare they have no conflicts of interest to disclose that are relevant to this study.
References
1. Jevons MP. “Celbenin” - resistant Staphylococci. Br Med J. 1961;1(5219):124–125. doi:10.1136/bmj.1.5219.124-a
2. Antibiotic resistance threats in the United States, 2019. Centers for Disease Control and Prevention; 2019 [cited August 2, 2021]. doi:10.15620/cdc:82532.
3. Alkharsah K, Rehman S, Alkhamis F, Alnimr A, Diab A, Al-Ali A. Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann Clin Microbiol Antimicrob. 2018;17(1). doi:10.1186/s12941-018-0260-2
4. Wertheim H, Melles D, Vos M, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–762. doi:10.1016/S1473-3099(05)70295-4
5. Ramarathnam V, De Marco B, Ortegon A, Kemp D, Luby J, Sreeramoju P. Risk factors for development of methicillin-resistant Staphylococcus aureus infection among colonized patients. Am J Infect Control. 2013;41(7):625–628. doi:10.1016/j.ajic.2012.08.005
6. National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004;32(8):470–485. doi:10.1016/j.ajic.2004.10.001
7. Szumowski J, Wener K, Gold H, et al. Methicillin‐Resistant Staphylococcus aureus colonization, behavioral risk factors, and skin and soft‐tissue infection at an ambulatory clinic serving a large population of HIV‐infected men who have sex with men. Clin Infect Dis. 2009;49(1):118–121. doi:10.1086/599608
8. Kluytmans J, Wertheim H. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005;33(1):3–8. doi:10.1007/s15010-005-4012-9
9. Davis K, Stewart J, Crouch H, Florez C, Hospenthal D. Methicillin-resistant Staphylococcus aureus (MRSA) Nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis. 2004;39(6):776–782. doi:10.1086/422997
10. Chipolombwe J, Török M, Mbelle N, Nyasulu P. Methicillin-resistant Staphylococcus aureus multiple sites surveillance: a systemic review of the literature. Infect Drug Resist. 2016;9:35–42.
11. Grundmann H, Aires-de-sousa M, Boyce J, Tiemersma E. Emergence and resurgence of methicillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368(9538):874–885. doi:10.1016/S0140-6736(06)68853-3
12. Struelens M, Hawkey P, French G, Witte W, Tacconelli E. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin Microbiol Infect. 2009;15(2):112–119. doi:10.1111/j.1469-0691.2009.02698.x
13. Pada S, Ding Y, Ling M, et al. Economic and clinical impact of nosocomial methicillin-resistant Staphylococcus aureus infections in Singapore: a matched case–control study. J Hosp Infect. 2011;78(1):36–40. doi:10.1016/j.jhin.2010.10.016
14. Siddiqui AH, Koirala J. Methicillin Resistant Staphylococcus Aureus. In: StatPearls. StatPearls Publishing; 2021.
15. Huang S, Yokoe D, Hinrichsen V, et al. Impact of routine intensive care unit surveillance cultures and resultant barrier precautions on hospital‐wide Methicillin‐Resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2006;43(8):971–978. doi:10.1086/507636
16. Kang J, Mandsager P, Biddle A, Weber D. Cost-effectiveness analysis of active surveillance screening for Methicillin-resistant Staphylococcus aureus in an academic hospital setting. Infect Control Hosp Epidemiol. 2012;33(5):477–486. doi:10.1086/665315
17. Robotham J, Deeny S, Fuller C, Hopkins S, Cookson B, Stone S. Cost-effectiveness of national mandatory screening of all admissions to English National Health Service hospitals for methicillin-resistant Staphylococcus aureus: a mathematical modelling study. Lancet Infect Dis. 2016;16(3):348–356. doi:10.1016/S1473-3099(15)00417-X
18. Murthy A, De Angelis G, Pittet D, Schrenzel J, Uckay I, Harbarth S. Cost-effectiveness of universal MRSA screening on admission to surgery. Clin Microbiol Infect. 2010;16(12):1747–1753. doi:10.1111/j.1469-0691.2010.03220.x
19. Infection Control Policy and Procedures; INF 12-009. King Fahd Hospital of the University Al-Khobar, Methicillin-resistant Staphylococcus aureus Management; 2019.
20. CLSI. Performance standards for antimicrobial susceptibility testing; Nineteenth informational supplement. CLSI document M100-19. Wayne, PA: Clinical and Laboratory Standards Institute; 2009.
21. Mitchell A. Sensitivity × PPV is a recognized test called the clinical utility index (CUI+). Eur J Epidemiol. 2011;26(3):251–252. doi:10.1007/s10654-011-9561-x
22. Humphreys H, Fitzpatick F, Harvey B. Gender differences in rates of carriage and bloodstream infection caused by Methicillin-resistant Staphylococcus aureus: are they real, do they matter and why? Clin Infect Dis. 2015;61(11):1708–1714. doi:10.1093/cid/civ576
23. Panhotra B, Saxena A, Al Mulhim A. Prevalence of methicillin-resistant and methicillin-sensitive Staphylococcus aureus nasal colonization among patients at the time of admission to the hospital. Ann Saudi Med. 2005;25(4):304–308. doi:10.5144/0256-4947.2005.304
24. Zervou F, Zacharioudakis I, Ziakas P, Mylonakis E. MRSA colonization and risk of infection in the neonatal and pediatric ICU: a meta-analysis. Pediatrics. 2014;133(4):e1015–e1023. doi:10.1542/peds.2013-3413
25. Scanvic A, Denic L, Gaillon S, Giry P, Andremont A, Lucet J. Duration of colonization by Methicillin-resistant Staphylococcus aureus after hospital discharge and risk factors for prolonged carriage. Clin Infect Dis. 2001;32(10):1393–1398. doi:10.1086/320151
26. Shenoy E, Paras M, Noubary F, Walensky R, Hooper D. Natural history of colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE): a systematic review. BMC Infect Dis. 2014;14(1). doi:10.1186/1471-2334-14-177
27. Sanford M, Widmer A, Bale M, Jones R, Wenzel R. Efficient detection and long-term persistence of the carriage of Methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1994;19(6):1123–1128. doi:10.1093/clinids/19.6.1123
28. Robicsek A, Beaumont J, Peterson L. Duration of colonization with Methicillin‐Resistant Staphylococcus aureus. Clin Infect Dis. 2009;48(7):910–913. doi:10.1086/597296
29. Shenoy E, Kim J, Rosenberg E, et al. Discontinuation of contact precautions for Methicillin-resistant Staphylococcus aureus: a randomized controlled trial comparing passive and active screening with culture and polymerase chain reaction. Clin Infect Dis. 2013;57(2):176–184. doi:10.1093/cid/cit206
30. Kaplan S, Forbes A, Hammerman W, et al. Randomized trial of “Bleach Baths” plus routine hygienic measures vs routine hygienic measures alone for prevention of recurrent infections. Clin Infect Dis. 2013;58(5):679–682. doi:10.1093/cid/cit764
31. Fritz S, Hogan P, Hayek G, et al. Household versus individual approaches to eradication of community-associated staphylococcus aureus in children: a randomized trial. Clin Infect Dis. 2011;54(6):743–751. doi:10.1093/cid/cir919
32. Sherertz RJ, Reagan DR, Hampton KD, et al. A cloud adult: the Staphylococcus aureus-virus interaction revisited. Ann Intern Med. 1996;124(6):539. doi:10.7326/0003-4819-124-6-199603150-00001
33. Senn L, Basset P, Nahimana I, Zanetti G, Blanc D. Which anatomical sites should be sampled for screening of methicillin-resistant Staphylococcus aureus carriage by culture or by rapid PCR test? Clin Microbiol Infect. 2012;18(2):E31–E33. doi:10.1111/j.1469-0691.2011.03724.x
34. Baker S, Brecher S, Robillard E, Strymish J, Lawler E, Gupta K. Extranasal Methicillin-resistant Staphylococcus aureus colonization at admission to an acute care veterans affairs hospital. Infect Control Hosp Epidemiol. 2010;31(1):42–46. doi:10.1086/649222
35. Van Heirstraeten L, Abrahantes J, Lammens C, et al. Impact of a short period of pre-enrichment on detection and bacterial loads of Methicillin-resistant Staphylococcus aureus from screening specimens. J Clin Microbiol. 2009;47(10):3326–3328. doi:10.1128/JCM.01088-09
36. Kerremans J, Maaskant J, Verbrugh H, van Leeuwen W, Vos M. Detection of methicillin-resistant Staphylococcus aureus in a low-prevalence setting by polymerase chain reaction with a selective enrichment broth. Diagn Microbiol Infect Dis. 2008;61(4):396–401. doi:10.1016/j.diagmicrobio.2008.04.004
37. Lautenbach E, Nachamkin I, Hu B, et al. surveillance cultures for detection of Methicillin-resistant Staphylococcus aureus: diagnostic yield of anatomic sites and comparison of provider- and patient-collected samples. Infect Control Hosp Epidemiol. 2009;30(4):380–382. doi:10.1086/596045
38. Mertz D, Frei R, Periat N, et al. Exclusive Staphylococcus aureus throat carriage. Arch Intern Med. 2009;169(2):172. doi:10.1001/archinternmed.2008.536
39. Kuehnert M, Kruszon‐Moran D, Hill H, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193(2):172–179. doi:10.1086/499632
40. Eveillard M, Lassence A, Lancien E, Barnaud G, Ricard J, Joly-Guillou M. Evaluation of a strategy of screening multiple anatomical sites for Methicillin-resistant Staphylococcus aureus at admission to a teaching hospital. Infect Control Hosp Epidemiol. 2006;27(2):181–184. doi:10.1086/500627
41. Bhalla A, Aron D, Donskey C. Staphylococcus aureus intestinal colonization is associated with increased frequency of S. aureus on skin of hospitalized patients. BMC Infect Dis. 2007;7(1). doi:10.1186/1471-2334-7-105
42. Miller L, Remington F, Bayer A, et al. Clinical and epidemiologic characteristics cannot distinguish community-associated Methicillin-resistant Staphylococcus aureus infection from Methicillin-Susceptible S. aureus infection: a prospective investigation. Clin Infect Dis. 2007;44(4):471–482. doi:10.1086/511033
43. Khamash D, Voskertchian A, Tamma P, Akinboyo I, Carroll K, Milstone A. Increasing clindamycin and trimethoprim-sulfamethoxazole resistance in pediatric Staphylococcus aureus infections. J Pediatric Infect Dis Soc. 2018;8(4):351–353. doi:10.1093/jac/dkab341
44. Nurjadi D, Klein S, Hannesen J, Heeg K, Boutin S, Zanger P. Molecular analysis of an increase in trimethoprim/sulfamethoxazole-resistant MRSA reveals multiple introductions into a tertiary care hospital, Germany 2012–19. J Antimicrob Chemother. 2021. doi:10.1093/jac/dkab341
45. Liu C, Bayer A, Cosgrove S, et al. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of Methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis. 2011;52(3):e18–e55. doi:10.1093/cid/ciq146
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
