Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Pre- and postmortem imaging of transplanted cells
Authors Andrzejewska A, Nowakowski A, Janowski M, Bulte J, Gilad A, Walczak P, Lukomska B
Received 26 February 2015
Accepted for publication 28 April 2015
Published 2 September 2015 Volume 2015:10(1) Pages 5543—5559
DOI https://doi.org/10.2147/IJN.S83557
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Anna Andrzejewska,1 Adam Nowakowski,1 Miroslaw Janowski,1–4 Jeff WM Bulte,3–7 Assaf A Gilad,3,4 Piotr Walczak,3,4,8 Barbara Lukomska1
1NeuroRepair Department, 2Department of Neurosurgery, Mossakowski Medical Research Centre, Polish Academy of Sciences, Warsaw, Poland; 3Russell H Morgan Department of Radiology and Radiological Science, Division of MR Research, 4Cellular Imaging Section and Vascular Biology Program, Institute for Cell Engineering, 5Department of Biomedical Engineering, 6Department of Chemical & Biomolecular Engineering, 7Department of Oncology, The Johns Hopkins University School of Medicine, Baltimore, MD, USA; 8Department of Radiology, Faculty of Medical Sciences, University of Warmia and Mazury, Olsztyn, Poland
Abstract: Therapeutic interventions based on the transplantation of stem and progenitor cells have garnered increasing interest. This interest is fueled by successful preclinical studies for indications in many diseases, including the cardiovascular, central nervous, and musculoskeletal system. Further progress in this field is contingent upon access to techniques that facilitate an unambiguous identification and characterization of grafted cells. Such methods are invaluable for optimization of cell delivery, improvement of cell survival, and assessment of the functional integration of grafted cells. Following is a focused overview of the currently available cell detection and tracking methodologies that covers the entire spectrum from pre- to postmortem cell identification.
Keywords: stem cells, transplantation, SPECT, MRI, bioluminescence, cell labeling
Introduction
Cell transplantation has been explored as a new clinical approach to repair injured tissue. Recently, the therapeutic properties of a variety of cell types have been studied extensively, and it is now clear that cells delivered to remote sites home to the area of injury and stimulate repair and regeneration of the damaged tissue. However, to gain some insight into exogenous cell migration, tissue localization, and the level of engraftment, the cells require labeling and subsequent tracking. Moreover, cell-tracking studies require a label that it is uniquely distinguishable and biologically stable for the quantitative analysis of dynamic processes in living cells. A variety of methods have been developed to identify transplanted cells premortem (in vivo) and postmortem (ex vivo). These methods rely on various contrast mechanisms and most require some premodification of the cells of interest. In this review, we describe a broad range of techniques that are suitable for cell-tracking studies, from methods applicable solely for postmortem microscopy assessment to those that facilitate longitudinal cell-tracking in live animals and from physical cell labeling methods to the use of reporter genes or strategies that do not require exogenous labeling, but are based on the detection of inherent marker labels, such as the Y chromosome, newly replicating DNA, or natural mutations.
Premortem cell detection techniques
SPECT and PET imaging
Radioactive substances have been employed in medical practice for many years. While the resolution of single-photon emission computed tomography (SPECT) and positron emission tomography (PET) is relatively low, nuclear medicine compensates with an outstanding sensitivity at the whole-body level. One of the main differences between the SPECT and PET techniques is the type of radioisotopes. In SPECT, radioisotopes directly emit gamma waves, whereas in PET imaging, the nuclei of the radioactive element first generate a positron, which annihilates after contact with the electron from the body, producing two collinear gamma rays that are then detected coincidently. Technetium-99m, Indium-111, and Gallium-67 are the most commonly used radiometal isotopes in SPECT, while Galium-68, Copper-64, Yttrium-86, and Zirconium-89 are usually applied in PET. Moreover, there are several nonmetallic radionuclides that could be utilized for PET, including 18F, 11C, 13N, 15O, and 124I. Most of the radioisotopes mentioned earlier have been adapted for oncological diagnosis and therapy. Before application, they are conjugated with an adequate chelator (eg, DOTA, DTPA, TETA) and/or a targeting molecule (antibody or small protein) in order to obtain thermodynamic stability, chemical inertness, and optimal biodistribution in the body.1,2 The labeling of cells with radioisotopes is a simple and very efficient procedure, but preparation of radioisotopes is complex and costly, including access to highly specialized equipment at the site of application, since, due to rapid decay, the labels cannot be prepared in advance. While the compounds bearing radioisotopes are easily internalized by cells from the high-concentration incubation medium, the radioisotopes can leak.3 Kim et al4 used a direct labeling method for rat adipose-derived stem cells, with a radioactive 124I-HIB-label. PET imaging combined with computed tomography enabled the identification of cells after transplantation in the myocardium for more than 9 days. This technique also provided additional information about the survival rate of transplanted cells. The radioactivity level was inversely proportional to that of apoptosis of transplanted cells.4 However, in general, the fast decay of radioactivity is another obstacle allowing for cell imaging only acutely after transplantation. The half-life of SPECT radioisotopes ranges from hours to a few days, and it is even shorter for the 18F used for PET imaging. In addition, the diagnostic/therapeutic centers must be equipped with highly specialized instruments. The doses applied clinically spread throughout the body, thus carrying minimal risk for individual cells. In contrast, the accumulation of radioisotopes in labeled cells can be potentially destructive, as it is for thyroid cells treated with radioiodine.5,6 Despite the drawbacks, radioisotopes are actively involved in stem cell tracking both in the preclinical setting and in clinical trials.7,8
Radioisotopes can be used for direct labeling of cells, but also can be conjugated with ligands of specific receptors present in a particular type of cell. A properly constructed compound can identify the transplanted stem cells inside the host body. Tarantal et al,9 showed that injection of 89Zr conjugated to human-specific CD45 antibody into the circulatory system of Rhesus monkeys facilitated imaging of the biodistribution of grafted human stem cells with PET. Another interesting application of noninvasive PET imaging was geared at differentiation of cancer stem cells from healthy brain cells. The CD133 epitope being a promising cancer stem cell marker was used for that purpose. Radioactive tracer 64Cu-NOTA conjugated to CD133 antibody was shown to successfully detect cancer stem cells in both subcutaneous and intracranial tumors.10 Another example of that system consists of a somatostatin receptor and a derivative of the somatostatin inhibitor, [111In]DTPA-octreotide (Octreoscan), which is approved in the United States and Europe for the diagnosis of tumors with high endogenous somatostatin receptor expression, such as breast cancers, pituitary tumors, or pheochromocytomas, but such technology has not been used for the detection of transplanted cells as yet.11
A PET reporter gene/probe system was described for the first time by Tjuvajev et al12 in 1995. It was based on the use of a reporter gene together with a reporter probe. There are three main classes of reporter genes: enzymes, receptors, and transporters. The reporter gene can be introduced in an expressive vector into transplanted cells, and its product accumulates inside the cells. Then, the product of the reporter gene interacts selectively with radioactive molecules that are delivered in the reporter probe, which results in the selective accumulation of the probe in transplanted cells. These cells can then be detected using PET. An example of this kind of system is an in vivo tracing of transplanted rat bone marrow mesenchymal stem cells (MSCs) transfected with the recombinant adenovirus vector, Ad5-hERL-IRES-VEGF (Ad-EIV), in the muscle of a rat host. Ad-EIV carries a reporter gene (hERL) coding the human estrogen receptor ligand-binding domain (hERL). After intravenous tail vein injection of a radiolabeled, biologically inactive, estrogen analog, 16α-[18F] fluoro-17β-estradiol (18F-FES), the radioactive signal originating from the grafted cells was visible with PET.13 The engraftment of human and rat hematopoietic stem cells in the rat was successfully observed in vivo using 1-(2-deoxy-2-18fluoro-β-L-arabinofuranosyl)-5-methyluracil and deoxycytidine kinase as a reporter gene. This kinase carries a mutation within the active site.14 The use of another kinase – the herpes simplex virus thymidine kinase (sr39TK) – as a reporter gene was shown to be very beneficial in PET imaging of transplanted, genetically modified human hematopoietic stem cells in rats. The radioactive signal from stem cells engineered to express sr39TK could be recorded after the infusion of [18F]-FHBG. Moreover, sr39TK is also a suicide gene, and, after administration of gancyclovir, the cells that express its active form undergo apoptosis. The possibility to induce fast and selective destruction of a transplanted, genetically modified stem cell population is quite important in the context of possible clinical application, alleviating to some extent safety concerns related to insertional mutagenesis and cell transformation.15 The human sodium iodide symporter (hNIS) is another reporter gene that is increasingly used. It has demonstrated success in monitoring adenoviral-based gene delivery.16 The expression of hNIS has been easily achieved in tumor cells, and the subsequent application of the radioactive 188Re probe proved to be theranostic.17 Moreover, the tumor-homing property of MSCs has been used for tumor-selective radionuclide accumulation via hNIS expression, with positive therapeutic effects.18 The hNIS has been also used in the field of regenerative medicine to determine the viability of transplanted cells, and has shown less variable results, and thus, a superior profile compared to eGFP (enhanced green fluorescent protein) for in vivo imaging.19,20 The observation of stem cells labeled with radiotracers can not only provide researchers with information about the fate of these cells, but can also enable optimization of the delivery route and technique.21 The increasing requirement for a detailed visualization of stem cells often leads to the development of multimodal approaches. Some newly introduced radioisotopes, such as 52Mn, can also be visualized by both PET and magnetic resonance imaging (MRI) scanners.22 However, the application of radioactive tracers for regenerative medicine carries the risk of not only radiation-induced cell death, but also mutagenesis, which could potentially result in tumor formation, a very grim complication even in a very delayed fashion. In addition, the crossing of radionuclides through the intact blood–brain barrier, as present in restorative neurotransplantation, has not been studied as yet.
X-ray and US imaging
X-ray-based fluoroscopy and computer tomography, as well as ultrasonography, are extensively exploited modalities in clinical imaging. X-ray-based methods of cellular imaging work by the absorption of X-rays by contrast agents, which are detected by various 2D and 3D detectors. Ultrasonography depends on the recording of echoes of ultrasonic waves. Heavy elements are the preferred cellular labels for X-ray imaging, while bubbles are the most frequently used contrast agents for ultrasonography. Unfortunately, even heavy elements and bubbles in cell-loadable quantities are difficult to detect with current state-of-the-art detectors. Thus, indirect approaches have been tested to support cell transplantation with these modalities, including, for example, coencapsulation of cells with bromine compounds.23,24 Another proposed option is the suspension of cells within a tantalum-labeled scaffold (hydrogel).25 Microbubbles can be easily internalized by stem cells, thus enabling their localization within internal organs, but such an approach is not useful for cell imaging within the central nervous system due to the low bone permeability of ultrasonic pulses.26 What is of interest is the current use of extracellular bubbles to facilitate cell homing to injured tissues after intravascular delivery.27,28
Relaxation-based MR contrast agents
In vivo tracking of stem cells with MRI based on relaxation requires prelabeling of cells with special compounds that can change the water relaxation time and/or magnetic susceptibility, and then, determining the location of these compounds based on the image intensity. MRI contrast agents can be divided into two main groups: exogenous and endogenous.
Metal-based compounds are primary among the exogenous-based labeling strategies. Metallic marker tags can be primarily based on iron, manganese, and gadolinium. They can be divided into two main groups. The first group includes MRI contrast agents that affect the longitudinal relaxation time, T1, where the spin lattice relaxation time is generated. T1-weighted contrast agents involve gadolinium (Gd3+) and manganese (Mn3+) chelates, where the mode of action is based on the decrease of the T1 relaxation time. In practice, highly intense T1-weighted images are produced with positive contrast. Contrast agents for stem cell labeling based on Gd elements include chelated Gd–lipid nanoparticles, Gd-chelated dextran nanoparticles, and Gd-DOTA–peptide complexes, and gadolinium oxide nanoparticles.29–32 However, the use of gadolinium nanoparticles in stem cell tracking is plagued by significant risk of cytotoxicity, which may compromise cell function or survival. It is also unclear how these compounds are metabolized in labeled cells.33 There is a potential threat that the toxic, free inorganic Gd3+ could be released and influence cell metabolism of the labeled cells as was reported in the case of the rat spinal cord cultured neurons, where free Gd3+ distorted the activity of K+–Cl− membrane cotransporter.34
Among the contrast agents with manganese compounds as an MRI signal source, MnCl2, and mesoporous and hollow MnO nanoparticles have been employed.35–37 An interesting study was performed with the systemic administration of MnCl2 that made possible simultaneous monitoring and elimination of human embryonic stem cell-derived teratoma cells.38 While a T1-positive contrast agent is more desirable, it is also characterized by lower sensitivity than T2* contrast agents, which capitalize on the magnetic field inhomogeneity produced by iron-oxide-based compounds. The second main group of the metallic contrast agents are those related to transverse relaxation time, T2, where the spin–spin relaxation time is reduced. T2-weighted contrast agents are primarily superparamagnetic iron nanoparticles (SPIOs). The most common use for stem cell labeling is related to Fe3O4 magnetite nanoparticles, which are composed of nonstoichiometric Fe2+ and Fe3+ oxides.39,40
Several types of SPIOs can be used for stem cell imaging, such as citrate-coated or dextran-coated SPIOs and ferumoxides.41,42 Ferumoxides are composed of iron particles of approximately 5 nm in size, but with a hydrodynamic diameter of approximately 80–150 nm, where the iron crystals are covered with a dextran layer. Some ferumoxides are combined with poly-L-lysine.43 Ultrasmall SPIOs (USPIOs), whose diameter size fluctuates approximately 35 nm, are another type of SPIOs. This type of contrast agent has been used successfully for adult rat neural stem cell labeling and tracking.44
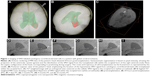
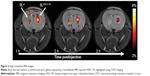
A commercially available Feraheme® (ferumoxytol, AMAG Pharmaceuticals, Waltham, MA, USA) is an iron oxide with a hydrophilic carboxydextran coat.45 The overall colloidal particle size is approximately 17–31 nm in diameter. Ferumoxytol was found to be useful in human neural stem cell tracking.46 For instance, the meso-2,3-dimercaptosuccinic acid-coated iron nanoparticles were used for efficient adipose-derived stem cell labeling, thereby increasing the spectrum of possible uses for different coating materials.47 An elegant method of simultaneous MRI and fluorescence imaging is achieved by using Molday ION Rhodamine B nanoparticles, where 50 nm magnetite-based nanocolloidals are also labeled with the fluorochrome Rhodamine B, with a 565–620 nm emission wavelength.48 A similar approach was employed in bone marrow-derived stem progenitor cells, where cells were labeled with the T2 contrast agent ferumoxide and a fluorescent tissue marker.49 Ferumoxide has also been successfully used in intraventricular delivery in the pediatric patient with global ischemia (Figure 1), and long-term observation did not reveal negative consequences.50 Microsized paramagnetic iron oxide nanoparticles are another type of MRI contrast agent, in which the diameter of nanoparticles is approximately 1 μm.51 For instance, iron nanoparticles were covered with a divinylbenzene polymer to detect the migration of bone marrow-derived stromal cells in a stroke model.52 Despite the numerous examples of its use, similar to Gd-based contrast agents, SPIO labeling does not seem to be neutral for labeled cells. SPIO-based stem cell labeling as a method is presently not FDA (US Food and Drug Administration) approved, because the molecular mechanisms of SPIO actions are not clearly understood as yet. So, there are still concerns about the uncertain side effects on cell function, as well as the possible negative influence on the fate of the labeled cells.53 For instance, labeling with SPIOs could impair functional properties like migration capacity and colony formation abilities of labeled MSCs.54 However, there are FDA-approved SPIO formulations, but each clinical application needs Institutional Review Board’s approval for specific SPIO formulation and cell type to be used. In summary, despite the fact that several different MRI contrast agents exist and safety issues are not still resolved, the most popular T2 and T1 MRI-based stem cell labeling and in vivo tracking agents are SPIOs and gadolinium oxide nanoparticles.33 There are some data that indicate that the use of dual T1 and T2 MRI contrast agents could improve cell detection accuracy.31 SPIO-based agents are the only MR contrast agents that have so far been used for clinical (stem) cell tracking.55
Apart from the metallic elements approach for MRI, stem cell labeling with the use of nonmetallic elements, such as fluorine, can be efficiently implemented, despite the lower sensitivity compared to iron-based nanoparticles.56–58 In addition to exogenous substances for MR labeling, endogenous MR tracer tags can be specified. An interesting technical solution for the MR labeling is a genetic-based strategy. Bengtsson et al59 reported that in β-galactosidase-engineered bone marrow cells, β-galactosidase activity in the presence of a special compound – S-Gal™ (Sigma-Aldrich, St Louis, MO, USA) – resulted in enhanced T2 MR contrast imaging. In another biological MRI approach, cells of interest were transfected with genes encoding proteins that were, in turn, able to bind iron particles, such as ferritin, transferrin receptors, and iron transporters derived from magnetotactic bacteria for the biomineralization of iron.60–63 In addition, stem cell surfaces could be engineered by artificial antigens with subsequent detection by SPIO-coupled antibodies.64
Chemical exchange saturation transfer (CEST) MRI
MRI has been used extensively over the last 3 decades for anatomical, functional, and dynamic imaging. Rapid improvements in MRI instrumentation and techniques have led to increased spatial resolution.65 The MRI contrast can be further enhanced by the expression of certain proteins (encoded by genes, termed reporter genes) that increase the MRI contrast. For instance, these proteins can be involved in iron metabolism and storage, and can act as enzymes that convert certain compounds to paramagnetic contrast agents.66–71
(Paramagnetic) chemical exchange saturation transfer ([PARA]CEST) is a novel approach for generating MRI contrast, in which the dynamic exchange process between an exchangeable proton and the surrounding water protons is used to amplify the desired contrast.72–74 One advantage of CEST is that the magnetization of different protons can be specifically saturated at different resonance frequencies, enabling the in vivo detection of multiple targets simultaneously.75 This rapidly evolving approach to creating contrast has been applied to detect temperature changes, pH, enzyme activity, metal ions, and metabolites like glycogen and glucose, and glycosaminoglycan.76–87
One potential application of such reporter genes is in regenerative medicine, where cells (eg, stem, progenitors, or immune cells) are transplanted into patients to repair a damaged tissue. Since the fate of these cells after transplantation is mostly unknown, it would be greatly beneficial if the cells could be tagged. Ideally, the “tag” should be a protein or an enzyme that is expressed only in the transplanted cells and for as long as the cells are viable. Therefore, reporter genes based on CEST MRI have an advantage, since they are bioorganic and biocompatible and are constitutively and continuously expressed by the cell. The first generation of a CEST-based reporter gene was a synthetic gene that encodes a lysine-rich protein. This reporter was used to distinguish glioma cells that overexpress the transgene from control cells in vivo in an animal model.88 Along the same lines, a synthetic gene was used to encode to an artificial protein that could sense cellular signaling.89 These studies were followed by a reengineering of the human protamine 1 (hPRM-1) gene as a CEST-based reporter. Since the hPRM-1 is a human protein, which is normally only expressed in sperm cells, it has a very low background signal in the body. Moreover, this is a human protein, and, therefore, would not be expected to trigger an immune reaction. Protamine was also used to monitor sustained drug release. Recently, CEST was applied to detect the activity of the theranostic enzymes, cytosine deaminase, carboxypeptidase G2, and herpes simplex virus type-1 thymidine kinase (HSV1-TK) (Figure 2).80,90–93 In these cases, the reporter can be used both for tagging the cells, if an imaging probe is used, and also as a suicide gene if a different compound (a prodrug) is used. Thus, such a theranostic gene has an additive value, especially where cells are needed to eradicate tumors. Unlike other labeling techniques, which have a very limited selection of compounds that can be used as imaging agents, CEST MRI that relies on bioorganic compounds allows almost endless possibilities for probe selection and design. Therefore, this technology holds great promise for the next generation of diagnostic imaging tools.
Bioluminescent imaging (BLI)
BLI is an attractive approach for tracking transplanted cells, as it specifically, and with relatively high sensitivity, reports on the viability of cells expressing a BLI reporter gene. In addition, this technique with the use of specific promoters, eg, Oct4 could potentially be used to observe alterations of gene expression in cultured or transplanted stem cells.94 This imaging modality relies on stable expression of the reporter gene in the target cells, and such expression can be induced using one of the well-established molecular biology techniques, eg, lentiviral transduction, or with the use of primary cells isolated from transgenic, light-producing animals.95–100 Several BLI reporter genes have been isolated, including the North American firefly (Photinus pyralis; FLuc), jellyfish (Aequorea), sea pansy (Renilla; RLuc), corals (Tenilla), the click beetle (Pyrophorus plagiophthalamus), and several bacterial species (Vibrio fischeri, Vibrio harveyi); however, for cell tracking purposes, FLuc is, by far, most widely used.101 The variety of currently available luciferase types with different substrate specificities opens a new possibility to observe two or more cell lines simultaneously. Leng et al102 adapted this idea to show the therapeutic effect of transplanted MSC in a model of human breast cancer. In this study, MSCs engineered to express RLuc were injected in mice bearing Fluc-expressing tumors, and due to different enzyme substrates, it enabled simultaneous detection of both MSCs and tumor cells. That imaging paradigm revealed the inhibition of cancer progression induced by the presence of MSC. This example showed that dual luciferase imaging enables investigation of the interaction between cell populations.102
The principle of BLI is based on the luciferase reaction that requires an enzyme (firefly luciferase), its substrate (D-luciferin or coelenterazine), ATP, and oxygen. This enzyme-catalyzed oxidation results in oxyluciferin, a product that, when decaying, emits photons. Photons are detected by specialized charge-coupled device (CCD) cameras that convert photons into electrons. The noise of the system is reduced by supercooling the CCD camera and mounting the camera in a light-tight chamber (Figure 3). The sensitivity of this imaging modality is dependent on several factors, including the optical properties and expression level of the reporter gene, the depth of labeled cells within the body, and the sensitivity of the detection device.103 The major challenge of in vivo BLI is that emitted photons must pass through the tissues and have to be detected outside the body. For that reason, detection of cells implanted superficially is much more effective than are deeper targets. As a general rule, the signal drops approximately tenfold for each 10 mm of tissue depth.104 In order to address the problem of tissue attenuation that limits sensitivity, significant effort has been directed toward improving the optical properties of imaging probes. The potential strategies include developing reporter genes with red-shifted emission profiles of photons known to have better tissue penetration, or developing new generations of substrates, such as CycLuc1, which offers significantly better pharmacokinetics and effectively results in an improved imaging signal.96,105 Another technique with the potential to improve detection of luciferase-expressing cells is based on the modification of ABC-transporter activity. These proteins are involved in the removal of xenobiotics from cells. The result of their inhibition is a higher accumulation of D-luciferin inside cells, and thus, an increased availability of substrate for the luciferase enzyme.106 An important advancement in BLI was the development of image processing algorithms for extracting spatial information from the imaging signal, referred to as optical tomography.107 Another quite interesting alternative is the use of bioluminescent luciferase from Gaussia princeps (GLuc), which is naturally secreted from cells in an active form. GLuc is present in body fluids. After the addition of the substrate to urine or serum, the measurement of the bioluminescent signal can provide information about the condition and viability of the transplanted cells.108
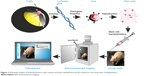  | Figure 3 Schematic outline of bioluminescence cells creation and their identification by BLI method in host body after transplantation. |
Overall, the advantages of BLI are robust reporter genes, effective and nontoxic substrates, and low cost and high throughput. These qualities make it an excellent and broadly used imaging modality for monitoring the survival of transplanted cells in small animals.109 The disadvantages of this technique include low spatial resolution, and susceptibility to the dynamic changes in light scattering and absorption properties of the tissue (bleeding, vascularization, pigmentation, etc), which can compromise the accuracy of the quantitative analysis of the photon signal.
Optoacoustic/photoacoustic imaging
Optoacoustic imaging is based on the phenomenon of a transverse wave of light (photons generated by a short laser pulse or less expensive high-power LED [light-emitting diode]) hitting specific molecules and causing their transient thermal expansion, which, in turn, generates an acoustic wave sensed by external detectors, such as microphones or piezoelectric tools (Figure 4).110 Thus, the stimulation is the same as for fluorescent imaging, but the detected signal is based on an acoustic wave, which can be a huge advantage. In fluorescent imaging, the energy of photons generated by a fluorescent lamp or a laser is sufficient for relatively deep penetration to tissues, and the limiting factor is the energy of reflected photons, which is insufficient to reach detectors. In optoacoustic imaging, this limitation is overcome by the detection of an acoustic wave, which has dramatically better penetration of tissues. The lack of tissue autofluorescence is an additional advantage of optoacoustic technology. While fluorescence provides a scattering contrast, optoacoustic technology provides an absorption contrast.111,112 Several systems have been developed for optoacoustic imaging. Portable, hand-held imaging probes can reach 1.5 cm in depth at a speed of 10 volumetric frames per second and a spatial resolution of 200 μm.113 Photoacoustic microscopy provides high-resolution images of tissue slices, can be combined with multiphoton microscopy, and can also be used for in vivo photoacoustic imaging.115 The recent development of optoacoustic devices with tomographic capabilities, based on multispectral immixing (multispectral optoacoustic tomography – MSOT), has fueled even more interest in this imaging modality.116–118 This was made possible because of the relatively high yield of the acoustic signal. An MSOT device has just come on the market (http://www.ithera-medical.com).
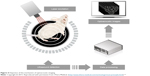  | Figure 4 Depiction of the mechanism of optoacoustic imaging. |
Optoacoustic imaging can be virtually always applied to fluorescence imaging, including reporter genes and probes.118,119 The resolution of images from infrared fluorescent protein-labeled cells has been found to be 10× higher (100 μm) compared to fluorescence molecular tomography.119 However, the applicability goes beyond fluorescence, with the recognition of gold as a strong generator of an optoacoustic signal.119–121 Gold nanoparticles, nanorods, and nanocages are widely available and biocompatible; thus, they have been increasingly used as an optoacoustic contrast agent.122–124 The utility of optoacoustic imaging has been shown to be advantageous not only for cell labeling, but also for the imaging of biomaterial scaffolds (Table 1).19
Postmortem cell identification
Immunohistochemical and genetic techniques
Thymidine analog markers
Various thymidine analogs can be used effectively to label proliferating cells. Among these, 5-bromo-2′-deoxyuridine (BrdU) is extensively used for labeling cells through its incorporation into newly synthesized DNA. BrdU applied to donor cells before transplantation could be used to trace their migration and identify the destination and accumulation in the host tissues.125–127 However, this method has a number of limitations and pitfalls. It has been shown that BrdU can be transmitted from the labeled cells into the neighboring cells of the recipient, leading to incorrect results by identifying the host cells as transplanted cells.128 Furthermore, due to proliferation of the originally labeled cells, the interpretation of a positive signal is often merely guesswork due to the label dilution when BrdU is transferred to the daughter cells.
A new thymidine analog, 5-ethynyl-2-deoxyuridine (EdU), has been introduced for tracking transplanted cells.129 While BrdU leaks from the labeled cells, EdU overcomes this difficulty since it is covalently bound to DNA and persists in the nucleus of tracked cells.130 It was shown that EdU labeling did not interfere with in vitro cell proliferation, differentiation, cytokine secretion, or migratory response.131 EdU-tracked cells are detected by a chemical reaction using an azide-conjugated fluor (red or green Alexa Fluors) that binds to the EdU alkyne moiety. However, sometimes “false-positive” staining occurs since certain types of unlabeled cells, eg, bone marrow cells, reveal a positive reaction with Alexa-azide fluor in the absence of an alkyne label.132
Species-specific markers
In the context of preclinical studies in xenogeneic transplantation, it is possible to use species-specific markers to distinguish donor cells from surrounding host cells. Thus, exogenous cells of human origin, engrafted within rodent tissues, could be identified using antibodies specific for human antigens. Recently, successful attempts at species-specific immunolabeling include antibodies to anti-human mitochondria, anti-human nuclei, or anti-human neuron-specific enolase.133–137
Y chromosome markers
Localization of donor-derived cells in the sex-mismatched transplant recipients can be detected by chromosome fluorescence in situ hybridization (FISH). Y chromosome-specific probes for FISH have been used to determine male cells transplanted into female recipients in both animals and humans.138,139 This method could be used to identify transplanted cells for a long period of time, regardless of their differentiation into different cell phenotypes.140,141 Double staining of tissue sections by Y chromosome FISH and immunohistochemistry enables monitoring of the path taken by transplanted male cells and their fate toward maturation.142,143 When FISH is combined with nuclear dyes, eg, DAPI (4′6-diamidino-2-phenylindole) or ethidium bromide counterstaining to highlight the nucleus, the nuclear localization of the Y chromosome probe could be confirmed, thus eliminating false-positive results.
Fluorescent dyes
The widespread use of fluorescence-based microscopy resulted from the many available fluorophores that could be used to label cells. Fluorophores are divided into exogenous dyes that interact with different cellular components or endogenous fluorescent proteins constitutively produced by cells. The density of staining and localization of fluorophores is influenced by their properties and the target cells being labeled. Fluorescent dyes are suitable for attaching to the cell structures of interest and for controlled localization and expression levels.
Cell membrane dyes
Lipophilic carbocyanine fluorescent dyes that bind to the cell membrane lipid bilayer have been widely used due to their low cytotoxicity. Membrane dyes can be used to visualize the donor cells in the host tissues or to assess the proliferation of transplanted cells after their division. Commonly used are PKH lipophilic dyes, eg, PKH26 used to track stem cells derived from the different sources.134,144–146 These dyes stain the whole plasma membrane of cells through lateral diffusion, then spread into intracellular compartments.147 Recently, PKH dyes have been validated for their applicability to stain microvesicles isolated from MSC.148
Other types of carbocyanine lipophilic membrane dyes, such as the chloromethyl-dialky-carbocyanine (CM-DiI) series, have been recently used to track multiple cell populations.149,150 These dyes have a number of advantages. They heavily intercalate within the lipid layer of the cell membrane, providing strong, highly photostable and long-lasting staining.151 Most importantly, these dyes have not been reported to be cytotoxic, even at high concentrations. However, CM-DiI dyes reveal decreased detection after mitotic cell division when the fluorescence of daughter cells appears to be halved compared to that of the mother cells.152 More importantly, the most significant problem with lipophilic membrane dyes is their transfer between the cells. The acquisition of PKH and CM-DiI dyes by neighboring cells, mostly tissue macrophages, has been shown, particularly if the labeled cells were dead.153,154 Although, even if there are no dead cells among the transplanted cell population, the noncovalently bound DiI can dissociate from labeled donor cells and get picked up by the host cells.155
Cytoplasmic and nuclear dyes
Cytoplasmic and nuclear dyes seem to be very efficient for cell tracking; however, they can affect different cellular functions. There are several fluorescent dyes permeable through the cell membrane. Among these, CFDA-SE (carboxyfluorescein diacetate succinimidyl ester) and CFSE (carboxyfluorescein succinimidyl ester) are frequently used.156–159 These dyes penetrate the cells where they are metabolized to amine-reactive chemicals, which then covalently bind to cytosolic cell components and are retained within the cytoplasm for a long period of time in vitro.147 However, CFDA-SE and CFSE are easily photobleached and their fluorescence decreases rapidly within the tissue. For this reason, they could be used for cell labeling only in short-term studies.
The other fluorescent dyes, eg, Hoechst and DAPI, reveal a high affinity for double-stranded DNA. They are mostly used to stain the nuclei of fixed cells in immunofluorescent studies, but they also could be applied to track cells before transplantation.160 However, despite the high labeling efficiency of dead cells after their fixation, DAPI and Hoechst are poor dyes for live cells due to inefficient penetration of the intact cell membrane. Moreover, since binding to DNA is noncovalent, the label can dissociate from DNA and it can be released from donor cells and taken up by the host cells, giving false-positive results.
All the abovementioned methods of staining are primarily based on immunohistological evaluation, which provides only “snapshot” evidence rather than a comprehensive study of transplanted cells over time. Such limited techniques partially reflect the conflicting observation of exogenous cells in the host. To address these limitations, the most reliable methods for assessing transplanted cells in vivo were determined. Among these methods, different reporter genes that are introduced into cells in vectors or integrated into the DNA of transplanted cells are highly accurate tools for following cell graft fate. Stable expression of fluorescent proteins in cells can be obtained through transfection and enables the long-term detection of genetically engineered cells in vitro and in vivo.
Reporter genes
Fluorescent proteins
GFP, first isolated from the jellyfish Aequorea victoria, is a widely used fluorescent reporter in various in vivo studies. The emission of light occurs after absorption of radiation in the ultraviolet spectrum. GFP-transgenic animals expressing this fluorescent protein in all tissues have been extensively maintained, and the isolated cells were applied in a suitable host.161–163 However, the level of GFP expression in transgenic animals is highly variable among animals and even among the cells within the same animal.164 Harting et al165 reported that only 50% of MSCs isolated from GFP-transgenic rodents expressed GFP. Due to insufficient GFP expression and the complexity and cost of development of transgenic animals, vector-mediating GFP transfection in vitro is still the preferred technique for GFP labeling.166–168 The generation of various colors of blue/yellow-shifted GFP variants, or the mutation-based development of red fluorescent proteins from anemones and tropical corals, have enlarged the reporter strategies available to track cells. The transfected GFP is densely localized and tightly packed in the nucleus, making the fluorescent signal in a small area easy to detect in the deep tissues after transplantation of GFP+ cells. However, the GFP gene is also problematic because many mammalian tissues are endogenously fluorescent. Another limitation of GFP usage is the oxygen accessibility that is necessary for proper fluorophore formation.169 This type of fluorescent dye may also interfere with DNA replication or transcription, and thus, may affect cell function. While fluorescence can potentially also be visualized in vivo, the amount of fluorescent signal is very low due to light absorption by the host tissues. In addition, because of the additional autofluorescence, this method is used for postmortem identification of transplanted cells. Even in that case, the signal from GFP must be enhanced by the application of specific antibodies.
β-galactosidase
β-galactosidase is a hydrolase enzyme encoded by the LacZ gene of Escherichia coli, and has been applied to convert a colorless substrate, eg, Blue-gal or X-gal, into a blue reaction product. Various cells transfected with the LacZ gene or isolated from LacZ-transgenic rodents have been visualized histochemically after their infusion into the host.126,170,171 However, the β-gal gene as a label is problematic because many mammalian cells demonstrate endogenous β-gal activity. While there are reports about the difference between bacterial and mammalian β-gal activity, it has been challenging for LacZ gene users to overcome the recipient tissue background problem.
Alkaline phosphatase
Alkaline phosphatase (AP) is a hydrolytic enzyme (EC 3.1.3.1) that catalyzes the nonspecific transestrification of phosphoryl ester optimally in alkaline environment. In in vitro research, E. coli AP gene (phoA) is commonly used as a reporter gene fused with gene of interest. Its product is naturally secreted to culture medium and can be easily detected by colorimetric and luminescent methods.172 Moreover, it may work as sensor of subcellular localization of particular proteins.173 The research with murine AP injected into the tail vein of mouse shows that this enzyme causes no immunogenic responses, which is a huge advantage in in vivo studies.174 Currently secreted AP (SEAP) originating from human placenta is broadly used. The removal of some of amino acids from the carboxy end of this protein inhibits the secretion process, and AP protein is accumulated inside the cell. Furthermore, modified AP is stable at high temperature contrary to the endogenous expressing enzyme which is produced naturally by some cells. Due to this feature, the researchers can dispose of nonspecific background signal from endogenous AP.175 SEAP fused with continuously expressing protein was successfully used to identify postmortem the transplanted neural cells in central nervous system by immunohistochemistry and fluorescence-activated cell sorting. Moreover, this technique enables assessment of percentage of cells which survive in host body after transplantation (Table 2).176
Conclusion
Cell therapy is a rapidly growing field of medicine. Cell imaging dramatically increases our understanding of the therapeutic effects. There are a variety of options to visualize transplanted cells, with a major division into premortem (in vivo) and postmortem (ex vivo) techniques (Figure 5). Noninvasive, premortem techniques that permit evaluation of the efficiency of transplantation and the migration of transplanted cells are essential for animal model studies and human clinical therapeutic approaches. Postmortem techniques allow to see the transplanted cells within the context of surrounding tissue in detail due to the possibility of imaging at high magnification. Driven by the urgent need, there has been continuous progress in cell labeling approaches, which we have presented in our review. This progress includes novel nucleotide analogs for postmortem stem cell identification, such as EdU; novel MRI reporter genes, such as MagA; novel MR contrast techniques, such as CEST; as well as entirely novel methods such as optoacoustic imaging. The breadth of options allows for fitting the cell labeling and detection methods to the specific needs of particular applications on the preclinical and clinical level, which will allow for a more comprehensive application of regenerative medicine.
Acknowledgments
The study was supported by a National Centre for Research and Development grant number 101 in ERA-NET NEURON project: “MEMS-IRBI”, MSCRE-0178-00, R21 NS08154, R01 NS045062, R01 NS076573, and NMSS RG 4994-A-3. We would like to thank Mary McAllister for editorial assistance and I-Hsun Wu for her contribution to Figure 5.
Disclosure
The authors report no conflicts of interest in this work.
References
Bhattacharyya S, Dixit M. Metallic radionuclides in the development of diagnostic and therapeutic radiopharmaceuticals. Dalton Trans. 2011;40(23):6112–6128. | ||
Zeglis MB, Houghton JL, Evans MJ, Viola-Villegas N, Lewis JS. Underscoring the influence of inorganic chemistry on nuclear imaging with radiometals. Inorg Chem. 2014;53(4):1880–1899. | ||
Mathiasen AB, Kastrup J. Non-invasive in-vivo imaging of stem cells after transplantation in cardiovascular tissue. Theranostics. 2013;3(8):561–572. | ||
Kim MH, Wooa SK, Lee KC, et al. Longitudinal monitoring adipose-derived stem cell survival by PET imaging hexadecyl-4-124I-iodobenzoate in rat myocardial infarction model. Biochem Biophys Res Commun. 2015;456(1):13–19. | ||
Janowski M, Bulte JW, Walczak P. Personalized nanomedicine advancements for stem cell tracking. Adv Drug Deliv Rev. 2012;64(13):1488–1507. | ||
Bonnema SJ, Fast S, Hegedüs L. The role of radioiodine therapy in benign nodular goitre. Best Pract Res Clin Endocrinol Metab. 2014;28(4):619–631. | ||
Khabbal J, Kerkelä E, Mitkari B, et al. Differential clearance of rat and human bone marrow-derived mesenchymal stem cells from the brain after intra-arterial infusion in rats. Cell Transplant. 2015;24(5):819–828. | ||
Barbosa da Fonseca LM, Battistella V, de Freitas GR, et al. Early tissue distribution of bone marrow mononuclear cells after intra-arterial delivery in a patient with chronic stroke. Circulation. 2009;120(6):539–541. | ||
Tarantal AF, Lee CC, Kukis DL, Cherry SR. Radiolabeling human peripheral blood stem cells for positron emission tomography (PET) imaging in young rhesus monkeys. PLoS One. 2013;8(10):e77148. | ||
Gaedickea S, Braunb F, Prasada S, et al. Noninvasive positron emission tomography and fluorescence imaging of CD133+ tumor stem cells. Proc Natl Acad Sci U S A. 2014;111(6):E692–E701. | ||
van der Lely AJ, de Herder WW, Krenning EP, Kwekkeboom DJ. Octreoscan radioreceptor imaging. Endocrine. 2003;20(3):307–311. | ||
Tjuvajev JG, Stockhammer G, Desai R, et al. Imaging the expression of transfected genes in vivo. Cancer Res. 1995;55(24):6126–6132. | ||
Qin C, Lan X, He J, et al. An in vitro and in vivo evaluation of a reporter gene/probe system hERL/(18)F-FES. PLoS One. 2013;8(4):e61911. | ||
McCracken MN, Gschweng EH, Nair-Gill E, et al. Long-term in vivo monitoring of mouse and human hematopoietic stem cell engraftment with a human positron emission tomography reporter gene. Proc Natl Acad Sci U S A. 2013;110(5):1857–1862. | ||
Gschweng EH, McCracken MN, Kaufman ML, et al. HSV-sr39TK positron emission tomography and suicide gene elimination of human hematopoietic stem cells and their progeny in humanized mice. Cancer Res. 2014;74(18):5173–5183. | ||
Varma NR, Barton KN, Janic B, et al. Monitoring adenoviral based gene delivery in rat glioma by molecular imaging. World J Clin Oncol. 2013;4(4):91–101. | ||
Guo R, Zhang M, Xi Y, et al. Theranostic studies of human sodium iodide symporter imaging and therapy using 188Re: a human glioma study in mice. PLoS One. 2014;9(7):e102011. | ||
Knoop K, Schwenk N, Dolp P, et al. Stromal targeting of sodium iodide symporter using mesenchymal stem cells allows enhanced imaging and therapy of hepatocellular carcinoma. Hum Gene Ther. 2013;24(3):306–316. | ||
Hwang do W, Jang SJ, Kim YH, et al. Real-time in vivo monitoring of viable stem cells implanted on biocompatible scaffolds. Eur J Nucl Med Mol Imaging. 2008;35(10):1887–1898. | ||
Quach CH, Jung KH, Paik JY, Park JW, Lee EJ, Lee KH. Quantification of early adipose-derived stem cell survival: comparison between sodium iodide symporter and enhanced green fluorescence protein imaging. Nucl Med Biol. 2012;39(8):1251–1260. | ||
Pantin JM, Hoyt RF, Aras O, et al. Optimization of intrabone delivery of hematopoietic progenitor cells in a swine model using cell radiolabeling with [89]zirconium. Am J Transplant. 2015;15(3):606–617. | ||
Lewis CM, Graves AS, Hernandez R, et al. 52Mn production for PET/MRI tracking of human stem cells expressing divalent metal transporter 1 (DMT1). Theranostics. 2015;5(3):227–239. | ||
Barnett BP, Arepally A, Stuber M, Arifin DR, Kraitchman DL, Bulte JW. Synthesis of magnetic resonance-, X-ray- and ultrasound-visible alginate microcapsules for immunoisolation and noninvasive imaging of cellular therapeutics. Nat Protoc. 2011;6(8):1142–1151. | ||
Kedziorek DA, Solaiyappan M, Walczak P, et al. Using C-arm x-ray imaging to guide local reporter probe delivery for tracking stem cell engraftment. Theranostics. 2013;3(11):916–926. | ||
Mohandas G, Oskolkov N, McMahon MT, Walczak P, Janowski M. Porous tantalum and tantalum oxide nanoparticles for regenerative medicine. Acta Neurobiol Exp (Wars). 2014;74(2):188–196. | ||
Cui W, Tavri S, Benchimol MJ, et al. Neural progenitor cells labeling with microbubble contrast agent for ultrasound imaging in vivo. Biomaterials. 2013;34(21):4926–4935. | ||
Tong J, Ding J, Shen X, et al. Mesenchymal stem cell transplantation enhancement in myocardial infarction rat model under ultrasound combined with nitric oxide microbubbles. PLoS One. 2013;8(11):e80186. | ||
Kokhuis TJ, Skachkov I, Naaijkens BA, et al. Intravital microscopy of localized stem cell delivery using microbubbles and acoustic radiation force. Biotechnol Bioeng. 2015;112(1):220–227. | ||
Nejadnik H, Henning TD, Do T, et al. MR imaging features of gadofluorine-labeled matrix-associated stem cell implants in cartilage defects. PLoS One. 2012;7(12):e49971. | ||
Brekke C, Williams SC, Price J, Thorsen F, Modo M. Cellular multiparametric MRI of neural stem cell therapy in a rat glioma model. Neuroimage. 2007;37(3):769–782. | ||
Cao L, Li B, Yi P, et al. The interplay of T1- and T2-relaxation on T1-weighted MRI of hMSCs induced by Gd-DOTA-peptides. Biomaterials. 2014;35(13):4168–4174. | ||
Shen Y, Shao Y, He H, et al. Gadolinium(3+)-doped mesoporous silica nanoparticles as a potential magnetic resonance tracer for monitoring the migration of stem cells in vivo. Int J Nanomedicine. 2013;8:119–127. | ||
Wang Y, Xu C, Ow H. Commercial nanoparticles for stem cell labeling and tracking. Theranostics. 2013;3(8):544–560. | ||
Ishibashi H, Hirao K, Yamaguchi J, et al. Inhibition of chloride outward transport by gadolinium in cultured rat spinal cord neurons. Neurotoxicology. 2009;30(1):155–159. | ||
Aoki I, Takahashi Y, Chuang KH, et al. Cell labeling for magnetic resonance imaging with the T1 agent manganese chloride. NMR Biomed. 2006;19(1):50–59. | ||
Yamada M, Gurney PT, Chung J, et al. Manganese-guided cellular MRI of human embryonic stem cell and human bone marrow stromal cell viability. Magn Reson Med. 2009;62(4):1047–1054. | ||
Shin J, Anisur RM, Ko MK, Im GH, Lee JH, Lee IS. Hollow manganese oxide nanoparticles as multifunctional agents for magnetic resonance imaging and drug delivery. Angew Chem Int Ed Engl. 2009;48(2):321–324. | ||
Kim T, Momin E, Choi J, et al. Mesoporous silica-coated hollow manganese oxide nanoparticles as positive T1 contrast agents for labeling and MRI tracking of adipose-derived mesenchymal stem cells. J Am Chem Soc. 2011;133(9):2955–2961. | ||
Chung J, Dash R, Kee K, et al. Theranostic effect of serial manganese-enhanced magnetic resonance imaging of human embryonic stem cell derived teratoma. Magn Reson Med. 2012;68(2):595–599. | ||
Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17(7):484–499. | ||
Riegler J, Liew A, Hynes SO, et al. Superparamagnetic iron oxide nanoparticle targeting of MSCs in vascular injury. Biomaterials. 2013;34(8):1987–1994. | ||
Andreas K, Georgieva R, Ladwig M, et al. Highly efficient magnetic stem cell labeling with citrate-coated superparamagnetic iron oxide nanoparticles for MRI tracking. Biomaterials. 2012;33(18):4515–4525. | ||
Chen XW, Zhu DJ, Ju YL, Zhou SF. Therapeutic effect of transplanting magnetically labeled bone marrow stromal stem cells in a liver injury rat model with 70%-hepatectomy. Med Sci Monit. 2012;18(10):BR375–BR382. | ||
Albukhaty S, Naderi-Manesh H, Tiraihi T. In vitro labeling of neural stem cells with poly-L-lysine coated super paramagnetic nanoparticles for green fluorescent protein transfection. Iran Biomed J. 2013;17(2):71–76. | ||
Bakhru SH, Altiok E, Highley C, et al. Enhanced cellular uptake and long-term retention of chitosan-modified iron-oxide nanoparticles for MRI-based cell tracking. Int J Nanomedicine. 2012;7:4613–4623. | ||
Khurana A, Nejadnik H, Chapelin F, et al. Ferumoxytol: a new, clinically applicable label for stem-cell tracking in arthritic joints with MRI. Nanomedicine (Lond). 2013;8(12):1969–1983. | ||
Gutova M, Frank JA, D’Apuzzo M, et al. Magnetic resonance imaging tracking of ferumoxytol-labeled human neural stem cells: studies leading to clinical use. Stem Cells Transl Med. 2013;2(10):766–775. | ||
Qin J, Li K, Peng C, et al. MRI of iron oxide nanoparticle-labeled ADSCs in a model of hindlimb ischemia. Biomaterials. 2013;34(21):4914–4925. | ||
Addicott B, Willman M, Rodriguez J, et al. Mesenchymal stem cell labeling and in vitro MR characterization at 1.5 T of new SPIO contrast agent: Molday ION Rhodamine-B™. Contrast Media Mol Imaging. 2011;6(1):7–18. | ||
Meng Y, Zhang F, Blair T, et al. MRI of auto-transplantation of bone marrow-derived stem-progenitor cells for potential repair of injured arteries. PLoS One. 2012;7(2):e31137. | ||
Janowski M, Walczak P, Kropiwnicki T, et al. Long-term MRI cell tracking after intraventricular delivery in a patient with global cerebral ischemia and prospects for magnetic navigation of stem cells within the CSF. PLoS One. 2014;9(2):e97631. | ||
Roose D, Leroux F, De Vocht N, et al. Multimodal imaging of micron-sized iron oxide particles following in vitro and in vivo uptake by stem cells: down to the nanometer scale. Contrast Media Mol Imaging. 2014;9(6):400–408. | ||
Tarulli E, Chaudhuri JD, Gretka V, Hoyles A, Morshead CM, Stanisz GJ. Effectiveness of micron-sized superparamagnetic iron oxide particles as markers for detection of migration of bone marrow-derived mesenchymal stromal cells in a stroke model. J Magn Reson Imaging. 2013;37(6):1409–1418. | ||
Schäfer R, Kehlbach R, Müller M, et al. Labeling of human mesenchymal stromal cells with superparamagnetic iron oxide leads to a decrease in migration capacity and colony formation ability. Cytotherapy. 2009;11(1):68–78. | ||
Bulte JW. In vivo MRI cell tracking: clinical studies. AJR Am J Roentgenol. 2009;193(2):314–325. | ||
Ahrens ET, Bulte JW. Tracking immune cells in vivo using magnetic resonance imaging. Nat Rev Immunol. 2013;13(10):755–763. | ||
Ruiz-Cabello J, Barnett BP, Bottomley PA, Bulte JW. Fluorine (19F) MRS and MRI in biomedicine. NMR Biomed. 2011;24(2):114–129. | ||
Ribot EJ, Gaudet JM, Chen Y, Gilbert KM, Foster PJ. In vivo MR detection of fluorine-labeled human MSC using the bSSFP sequence. Int J Nanomedicine. 2014;9:1731–1739. | ||
Bengtsson NE, Brown G, Scott EW, Walter GA. lacZ as a genetic reporter for real-time MRI. Magn Reson Med. 2010;63(3):745–753. | ||
Campan M, Lionetti V, Aquaro GD, et al. Ferritin as a reporter gene for in vivo tracking of stem cells by 1.5-T cardiac MRI in a rat model of myocardial infarction. Am J Physiol Heart Circ Physiol. 2011;300(6):H2238–H2250. | ||
Iordanova B, Ahrens ET. In vivo magnetic resonance imaging of ferritin-based reporter visualizes native neuroblast migration. Neuroimage. 2012;59(2):1004–1012. | ||
Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med. 2006;56(1):51–59. | ||
Goldhawk DE, Lemaire C, McCreary CR, et al. Magnetic resonance imaging of cells overexpressing MagA, an endogenous contrast agent for live cell imaging. Mol Imaging. 2009;8(3):129–139. | ||
Chung J, Kee K, Barral JK, et al. In vivo molecular MRI of cell survival and teratoma formation following embryonic stem cell transplantation into the injured murine myocardium. Magn Reson Med. 2011;66(5):1374–1381. | ||
Duyn JH, Koretsky AP. Novel frontiers in ultra-structural and molecular MRI of the brain. Curr Opin Neurol. 2011;24(4):386–393. | ||
Alfke H, Stöppler H, Nocken F, et al. In vitro MR imaging of regulated gene expression. Radiology. 2003;228(2):488–492. | ||
Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59(6):1225–1231. | ||
Genove G, DeMarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005;11(4):450–454. | ||
Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7(2):109–117. | ||
Louie AY, Hüber MM, Ahrens ET, et al. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18(3):321–325. | ||
Kodibagkar VD, Yu J, Liu L, Hetherington HP, Mason RP. Imaging β-galactosidase activity using 19F chemical shift imaging of LacZ gene-reporter molecule 2-fluoro-4-nitrophenol-beta-D-galactopyranoside. Magn Reson Imaging. 2006;24(7):959–962. | ||
Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson. 2000;143(1):79–87. | ||
van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn Reson Med. 2011;65(4):927–948. | ||
Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391–411. | ||
McMahon MT, Gilad AA, DeLiso MA, Berman SM, Bulte JW, van Zijl PC. New “multicolor” polypeptide diamagnetic chemical exchange saturation transfer (DIACEST) contrast agents for MRI. Magn Reson Med. 2008;60(4):803–812. | ||
Liu G, Qin Q, Chan KW, et al. Non-invasive temperature mapping using temperature-responsive water saturation shift referencing (T-WASSR) MRI. NMR Biomed. 2014;27(3):320–331. | ||
Wu Y, Soesbe TC, Kiefer GE, Zhao P, Sherry AD. A responsive europium(III) chelate that provides a direct readout of pH by MRI. J Am Chem Soc. 2010;132(40):14002–14003. | ||
Chan KW, Liu G, Song X, et al. MRI-detectable pH nanosensors incorporated into hydrogels for in vivo sensing of transplanted-cell viability. Nat Mater. 2013;12(3):268–275. | ||
Yoo B, Pagel MD. A PARACEST MRI contrast agent to detect enzyme activity. J Am Chem Soc. 2006;128(43):14032–14033. | ||
Liu G, Liang Y, Bar-Shir A, et al. Monitoring enzyme activity using a diamagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. J Am Chem Soc. 2011;133(41):16326–16329. | ||
Hingorani DV, Randtke EA, Pagel MD. A catalyCEST MRI contrast agent that detects the enzyme-catalyzed creation of a covalent bond. J Am Chem Soc. 2013;135(17):6396–6398. | ||
Trokowski R, Ren J, Kálmán FK, Sherry AD. Selective sensing of zinc ions with a PARACEST contrast agent. Angew Chem Int Ed Engl. 2005;44(42):6920–6923. | ||
Haris M, Cai K, Singh A, Hariharan H, Reddy R. In vivo mapping of brain myo-inositol. Neuroimage. 2011;54(3):2079–2085. | ||
van Zijl PC, Jones CK, Ren J, Malloy CR, Sherry AD. MRI detection of glycogen in vivo by using chemical exchange saturation transfer imaging (glycoCEST). Proc Natl Acad Sci U S A. 2007;104(11):4359–4364. | ||
Chan KW, McMahon MT, Kato Y, et al. Natural D-glucose as a biodegradable MRI contrast agent for detecting cancer. Magn Reson Med. 2012;68(6):1764–1773. | ||
Walker-Samuel S, Ramasawmy R, Torrealdea F, et al. In vivo imaging of glucose uptake and metabolism in tumors. Nat Med. 2013;19(8):1067–1072. | ||
Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc Natl Acad Sci U S A. 2008;105(7):2266–2270. | ||
Gilad AA, McMahon MT, Walczak P, et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. | ||
Airan RD, Bar-Shir A, Liu G, et al. MRI biosensor for protein kinase A encoded by a single synthetic gene. Magn Reson Med. 2012;68(6):1919–1923. | ||
Choi J, Kim K, Kim T, et al. Multimodal imaging of sustained drug release from 3-D poly(propylene fumarate) (PPF) scaffolds. J Control Release. 2011;156(2):239–245. | ||
Jamin Y, Eykyn TR, Poon E, Springer CJ, Robinson SP. Detection of the prodrug-activating enzyme carboxypeptidase G2 activity with chemical exchange saturation transfer magnetic resonance. Mol Imaging Biol. 2014;16(2):152–157. | ||
Bar-Shir A, Liu G, Liang Y, et al. Transforming thymidine into a magnetic resonance imaging probe for monitoring gene expression. J Am Chem Soc. 2013;135(4):1617–1624. | ||
Bar-Shir A, Liu G, Greenberg MM, Bulte JW, Gilad AA. Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat Protoc. 2013;8(12): 2380–2391. | ||
Ahn B, Parashurama N, Patel M, et al. Noninvasive reporter gene imaging of human Oct4 (Pluripotency) dynamics during the differentiation of embryonic stem cells in living subjects. Mol Imaging Biol. 2014;16(6):865–876. | ||
Liang Y, Ågren L, Lyczek A, Walczak P, Bulte JW. Neural progenitor cell survival in mouse brain can be improved by co-transplantation of helper cells expressing bFGF under doxycycline control. Exp Neurol. 2013;247:73–79. | ||
Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17(1):016004. | ||
Liang Y, Walczak P, Bulte JW. The survival of engrafted neural stem cells within hyaluronic acid hydrogels. Biomaterials. 2013;34(22):5521–5529. | ||
Berman SC, Galpoththawela C, Gilad AA, Bulte JW, Walczak P. Long-term MR cell tracking of neural stem cells grafted in immunocompetent versus immunodeficient mice reveals distinct differences in contrast between live and dead cells. Magn Reson Med. 2011;65(2):564–574. | ||
Gorelik M, Janowski M, Galpoththawela C, et al. Noninvasive monitoring of immunosuppressive drug efficacy to prevent rejection of intracerebral glial precursor allografts. Cell Transplant. 2012;21(10):2149–2157. | ||
Murphy CT, Moloney G, Hall LJ, et al. Use of bioluminescence imaging to track neutrophil migration and its inhibition in experimental colitis. Clin Exp Immunol. 2010;162(1):188–196. | ||
Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173(1):5–11. | ||
Leng L, Wang Y, He N, et al. Molecular imaging for assessment of mesenchymal stem cells mediated breast cancer therapy. Biomaterials. 2014;35(19):5162–5170. | ||
Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6(4):432–440. | ||
Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19(4):316–317. | ||
Evans MS, Chaurette JP, Adams ST Jr, et al. A synthetic luciferin improves bioluminescence imaging in live mice. Nat Methods. 2014;11(4):393–395. | ||
Huang R, Vider J, Serganova I, Blasberg RG. ATP binding cassette transporters modulate both coelenterazine- and D-luciferinbased bioluminescence imaging. Mol Imaging. 2011;10(3):215–226. | ||
Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23(3):313–320. | ||
Lewandrowski GK, Magee CN, Mounayar M, Tannous BA, Azzi J. Simultaneous in vivo monitoring of regulatory and effector T lymphocytes using secreted gaussia luciferase, firefly luciferase, and secreted alkaline phosphatase. In: Badr CE, editor. Bioluminescent Imaging: Methods and Protocols, Methods in Molecular Biology. New York, NY: Springer; 2014:211–227. | ||
Janowski M, Engels C, Gorelik M, et al. Survival of neural progenitors allografted into the CNS of immunocompetent recipients is highly dependent on transplantation site. Cell Transplant. 2014;23(2): 253–262. | ||
Yao J, Wang LV. Photoacoustic microscopy. Laser Photon Rev. 2013;7(5):758–778. | ||
Deán-Ben XL, Razansky D. Portable spherical array probe for volumetric real-time optoacoustic imaging at centimeter-scale depths. Opt Express. 2013;21(23):28062–28071. | ||
Tserevelakis GJ, Soliman D, Omar M, Ntziachristos V. Hybrid multiphoton and optoacoustic microscope. Opt Lett. 2014;39(7):1819–1822. | ||
Kneipp M, Turner J, Hambauer S, et al. Functional real-time optoacoustic imaging of middle cerebral artery occlusion in mice. PLoS One. 2014;9(4):e96118. | ||
iThera Medical. MSOT principle. Available from: http://www.ithera-medical.com/technology/msot-principle.html. Accessed January 1, 2015. | ||
Taruttis A, Rosenthal A, Kacprowicz M, Burton NC, Ntziachristos V. Multiscale multispectral optoacoustic tomography by a stationary wavelet transform prior to unmixing. IEEE Trans Med Imaging. 2014;33(5):1194–1202. | ||
Wang K, Schoonover RW, Su R, Oraevsky A, Anastasio MA. Discrete imaging models for three-dimensional optoacoustic tomography using radially symmetric expansion functions. IEEE Trans Med Imaging. 2014;33(5):1180–1193. | ||
Kusy S, Contag CH. Reporter gene technologies for imaging cell fates in hematopoiesis. Methods Mol Biol. 2014;1109:1–22. | ||
Cheng K, Cheng Z. Near infrared receptor-targeted nanoprobes for early diagnosis of cancers. Curr Med Chem. 2012;19(28): 4767–4785. | ||
Deliolanis NC, Ale A, Morscher S, et al. Deep-tissue reporter-gene imaging with fluorescence and optoacoustic tomography: a performance overview. Mol Imaging Biol. 2014;16(5):652–660. | ||
Nam SY, Ricles LM, Suggs LJ, Emelianov SY. In vivo ultrasound and photoacoustic monitoring of mesenchymal stem cells labeled with gold nanotracers. PLoS One. 2012;7(5):e37267. | ||
Liopo AV, Conjusteau A, Chumakova OV, Ermilov SA, Su R, Oraevsky AA. Highly purified biocompatible gold nanorods for contrasted optoacoustic imaging of small animal models. Nanosci Nanotechnol Lett. 2012;4(7):681–686. | ||
Vonnemann J, Beziere N, Böttcher C, el al. Polyglycerolsulfate functionalized gold nanorods as optoacoustic signal nanoamplifiers for in vivo bioimaging of rheumatoid arthritis. Theranostics. 2014;4(6):629–641. | ||
Ricles LM, Nam SY, Sokolov K, Emelianov SY, Suggs LJ. Function of mesenchymal stem cells following loading of gold nanotracers. Int J Nanomedicine. 2011;6:407–416. | ||
Zhang YS, Wang Y, Wang L, et al. Labeling human mesenchymal stem cells with gold nanocages for in vitro and in vivo tracking by two-photon microscopy and photoacoustic microscopy. Theranostics. 2013;3(8):532–543. | ||
Fitzpatrick KM, Raschke J, Emborg ME. Cell-based therapies for Parkinson’s disease: past, present, and future. Antioxid Redox Signal. 2009;11(9):2189–2208. | ||
Fuoco C, Salvatori ML, Biondo A, et al. Injectable polyethylene glycol-fibrinogen hydrogel adjuvant improves survival and differentiation of transplanted mesoangioblasts in acute and chronic skeletal-muscle degeneration. Skelet Muscle. 2012;2(1):24. | ||
Hattiangady B, Shetty AK. Neural stem cell grafting counteracts hippocampal injury-mediated impairments in mood, memory, and neurogenesis. Stem Cells Transl Med. 2012;1(9):696–708. | ||
Coyne TM, Marcus AJ, Woodbury D, Black IB. Marrow stromal cells transplanted to the adult brain are rejected by an inflammatory response and transfer donor labels to host neurons and glia. Stem Cells. 2006;24(11):2483–2492. | ||
Lin G, Huang YC, Shindel AW, et al. Labeling and tracking of mesenchymal stromal cells with EdU. Cytotherapy. 2009;11(7):864–873. | ||
Lin CS, Xin ZC, Dai J, Lue TF. Commonly used mesenchymal stem cell markers and tracking labels: limitations and challenges. Histol Histopathol. 2013;28(9):1109–1116. | ||
Ning H, Albersen M, Lin G, Lue TF, Lin CS. Effects of EdU labeling on mesenchymal stem cells. Cytotherapy. 2013;15(1):57–63. | ||
Lin G, Ning H, Banie L, et al. Bone marrow cells stained by azide-conjugated Alexa fluors in the absence of an alkyne label. Stem Cells Dev. 2012;21(13):2552–2559. | ||
Walczak P, Chen N, Eve D, Hudson J, et al. Long-term cultured human umbilical cord neural-like cells transplanted into the striatum of NOD SCID mice. Brain Res Bull. 2007;74(1–3):155–163. | ||
Li P, Zhang R, Sun H, et al. PKH26 can transfer to host cells in vitro and vivo. Stem Cells Dev. 2013;22(2):340–344. | ||
Hovakimyan M, Müller J, Wree A, Ortinau S, Rolfs A, Schmitt O. Survival of transplanted human neural stem cell line (ReNcell VM) into the rat brain with and without immunosuppression. Ann Anat. 2012;194(5):429–435. | ||
Bae SH, Kong TH, Lee HS, et al. Long-lasting paracrine effects of human cord blood cells on damaged neocortex in an animal model of cerebral palsy. Cell Transplant. 2012;21(11):2497–2515. | ||
Zhang C, Saatman KE, Royo NC, et al. Delayed transplantation of human neurons following brain injury in rats: a long-term graft survival and behavior study. J Neurotrauma. 2005;22(12):1456–1474. | ||
Crain BJ, Tran SD, Mezey E. Transplanted human bone marrow cells generate new brain cells. J Neurol Sci. 2005;233(1–2):121–123. | ||
Bonilla C, Zurita M, Otero L, Aguayo C, Vaquero J. Delayed intralesional transplantation of bone marrow stromal cells increases endogenous neurogenesis and promotes functional recovery after severe traumatic brain injury. Brain Inj. 2009;23(9):760–769. | ||
Mezey E, Key S, Vogelsang G, Szalayova I, Lange GD, Crain B. Transplanted bone marrow generates new neurons in human brains. Proc Natl Acad Sci U S A. 2003;100(3):1364–1369. | ||
Amariglio N, Hirshberg A, Scheithauer BW, et al. Donor-derived brain tumor following neural stem cell transplantation in an ataxia telangiectasia patient. PLoS Med. 2009;6(2):e1000029. | ||
Tran SD, Redman RS, Barrett AJ, et al. Microchimerism in salivary glands after blood- and marrow-derived stem cell transplantation. Biol Blood Marrow Transplant. 2011;17(3):429–433. | ||
Verghese E, Johnson C, Bertram JF, Ricardo SD, Deane JA. The fate of bone marrow-derived cells carrying a polycystic kidney disease mutation in the genetically normal kidney. BMC Nephrol. 2012; 13:91. | ||
Scheibe F, Ladhoff J, Huck J, et al. Immune effects of mesenchymal stromal cells in experimental stroke. J Cereb Blood Flow Metab. 2012;32(8):1578–1588. | ||
Polzer H, Volkmer E, Saller MM, et al. Long-term detection of fluorescently labeled human mesenchymal stem cell in vitro and in vivo by semi-automated microscopy. Tissue Eng Part C Methods. 2012;18(2):156–165. | ||
Pan H, Lan J, Luo X, Gao J, Xie X, Guo H. Biologic properties of gadolinium diethylenetriaminepentaacetic acid-labeled and PKH26-labeled human umbilical cord mesenchymal stromal cells. Cytotherapy. 2014;16(1):74–83. | ||
Progatzky F, Dallman MJ, Lo Celso C. From seeing to believing: labelling strategies for in vivo cell-tracking experiments. Interface Focus. 2013;3(3):20130001. | ||
Fierabracci A, Del Fattore A, Luciano R, Muraca M, Teti A, Muraca M. Recent advances in mesenchymal stem cell immunomodulation. The role of microvesicles. Cell Transplant. 2015;24(2):133–149. | ||
Adams GB, Alley IR, Chung UI, et al. Haematopoietic stem cells depend on Galpha(s)-mediated signalling to engraft bone marrow. Nature. 2009;459(7243):103–107. | ||
Xu L, Mahairaki V, Koliatsos VE. Host induction by transplanted neural stem cells in the spinal cord: further evidence for an adult spinal cord neurogenic niche. Regen Med. 2012;7(6):785–797. | ||
Darkazalli A, Levenson CW. Tracking stem cell migration and survival in brain injury: current approaches and future prospects. Histol Histopathol. 2012;27(10):1255–1261. | ||
Ferrari A, Hannouche D, Oudina K, et al. In vivo tracking of bone marrow fibroblasts with fluorescent carbocyanine dye. J Biomed Mater Res. 2001;56(3):361–367. | ||
Lassailly F, Griessinger E, Bonnet D. “Microenvironmental contaminations” induced by fluorescent lipophilic dyes used for noninvasive in vitro and in vivo cell tracking. Blood. 2010;115(26):5347–5354. | ||
Pawelczyk E, Jordan EK, Balakumaran A, et al. In vivo transfer of intracellular labels from locally implanted bone marrow stromal cells to resident tissue macrophages. PLoS One. 2009;4(8):e6712. | ||
Schormann W, Hammersen FJ, Brulport M, et al. Tracking of human cells in mice. Histochem Cell Biol. 2008;130(2):329–338. | ||
Markiewicz I, Sypecka J, Domanska-Janik K, Wyszomirski T, Lukomska B. Cellular environment directs differentiation of human umbilical cord blood-derived neural stem cells in vitro. J Histochem Cytochem. 2011;59(3):289–301. | ||
Jablonska A, Janowski M, Lukomska B. Different methods of immunosuppresion do not prolong the survival of human cord blood-derived neural stem cells transplanted into focal brain-injured immunocompetent rats. Acta Neurobiol Exp (Wars). 2013;73(1):88–101. | ||
Cellot S, Krosl J, Chagraoui J, Meloche S, Humphries RK, Sauvageau G. Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1(10) hematopoietic stem cells. Exp Hematol. 2007;35(5):802–816. | ||
Kato K, Cui S, Kuick R, et al. Identification of stem cell transcriptional programs normally expressed in embryonic and neural stem cells in alloreactive CD8+ T cells mediating graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16(6):751–771. | ||
Castanheira P, Torquetti LT, Magalhãs DR, Nehemy MB, Goes AM. DAPI diffusion after intravitreal injection of mesenchymal stem cells in the injured retina of rats. Cell Transplant. 2009;18(4):423–431. | ||
Liu N, Deguchi K, Yamashita T, Liu W, Ikeda Y, Abe K. Intracerebral transplantation of bone marrow stromal cells ameliorates tissue plasminogen activator-induced brain damage after cerebral ischemia in mice detected by in vivo and ex vivo optical imaging. J Neurosci Res. 2012;90(11):2086–2093. | ||
Murakami T, Kobayashi E. GFP-transgenic animals for in vivo imaging: rats, rabbits, and pigs. Methods Mol Biol. 2012;872:177–189. | ||
Singh V, Jaini R, Torricelli AA, Tuohy VK, Wilson SE. A method to generate enhanced GFP+ chimeric mice to study the role of bone marrow-derived cells in the eye. Exp Eye Res. 2013;116:366–370. | ||
Swenson ES, Price JG, Brazelton T, Krause DS. Limitations of green fluorescent protein as a cell lineage marker. Stem Cells. 2007;25(10):2593–2600. | ||
Harting MT, Jimenez F, Cox CS Jr. Isolation of mesenchymal stem cells (MSCs) from green fluorescent protein positive (GFP+) transgenic rodents: the grass is not always green(er). Stem Cells Dev. 2009;18(1):127–135. | ||
De Vocht N, Praet J, Reekmans K, et al. Tackling the physiological barriers for successful mesenchymal stem cell transplantation into the central nervous system. Stem Cell Res Ther. 2013;4(4):101. | ||
Czernik M, Fidanza A, Sardi M, et al. Differentiation potential and GFP labeling of sheep bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2013;114(1):134–143. | ||
Obokata H, Sasai Y, Niwa H, et al. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature. 2014;505(7485):676–680. | ||
Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91(26):12501–12504. | ||
Kopp HG, Hooper AT, Shmelkov SV, Rafii S. β-galactosidase staining on bone marrow. The osteoclast pitfall. Histol Histopathol. 2007;22(9):971–976. | ||
Komatsu K, Honmou O, Suzuki J, Houkin K, Hamada H, Kocsis JD. Therapeutic time window of mesenchymal stem cells derived from bone marrow after cerebral ischemia. Brain Res. 2010;1334:84–92. | ||
Lewis RE, Liao G, Young K, Douglas C, Kontoyiannis DP. Macrophage reporter cell assay for screening immunopharmacological activity of cell wall-active antifungals. Antimicrob Agents Chemother. 2014;58(3):1738–1743. | ||
Manoil C, Mekalanos JJ, Beckwith J. Alkaline phosphatase fusions: sensors of subcellular location. J Bacteriol. 1990;172(2):515–518. | ||
Maelandsmo GM, Ross PJ, Pavliv M, et al. Use of a murine secreted alkaline phosphatase as a non-immunogenic reporter gene in mice. J Gene Med. 2005;7(3):307–315. | ||
Schenborn E, Groskreutz D. Reporter gene vectors and assays. Mol Biotechnol. 1999;13(1):29–44. | ||
Mujtaba T, Han SS, Fischer I, Sandgren EP, Rao MS. Stable expression of the alkaline phosphatase marker gene by neural cells in culture and after transplantation into the CNS using cells derived from a transgenic rat. Exp Neurol. 2002;174(1):48–57. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.