Back to Journals » Drug Design, Development and Therapy » Volume 15
Potential Mechanism of S. baicalensis on Lipid Metabolism Explored via Network Pharmacology and Untargeted Lipidomics
Authors Ge PY , Qi YY, Qu SY, Zhao X, Ni S, Yao ZY, Guo R , Yang NY , Zhang QC , Zhu HX
Received 13 January 2021
Accepted for publication 31 March 2021
Published 4 May 2021 Volume 2021:15 Pages 1915—1930
DOI https://doi.org/10.2147/DDDT.S301679
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Georgios Panos
Ping-Yuan Ge,1 Yi-Yu Qi,1 Shu-Yue Qu,1 Xin Zhao,1 Sai-jia Ni,2 Zeng-Ying Yao,2 Rui Guo,3 Nian-Yun Yang,2 Qi-Chun Zhang,1,2 Hua-Xu Zhu2
1Jiangsu Key Laboratory for High Technology Research of TCM Formulae, Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China; 2Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China; 3School of Medicine and Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, People’s Republic of China
Correspondence: Qi-Chun Zhang; Hua-Xu Zhu Email [email protected]; [email protected]
Background: S. baicalensis, a traditional herb, has great potential in treating diseases associated with aberrant lipid metabolism, such as inflammation, hyperlipidemia, atherosclerosis and Alzheimer’s disease.
Aim of the Study: To elucidate the mechanism by which S. baicalensis modulates lipid metabolism and explore the medicinal effects of S. baicalensis at a holistic level.
Materials and Methods: The potential active ingredients of S. baicalensis and targets involved in regulating lipid metabolism were identified using a network pharmacology approach. Metabolomics was utilized to compare lipids that were altered after S. baicalensis treatment in order to identify significantly altered metabolites, and crucial targets and compounds were validated by molecular docking.
Results: Steroid biosynthesis, sphingolipid metabolism, the PPAR signaling pathway and glycerolipid metabolism were enriched and predicted to be potential pathways upon which S. baicalensis acts. Further metabolomics assays revealed 14 significantly different metabolites were identified as lipid metabolism-associated elements. After the pathway enrichment analysis of the metabolites, cholesterol metabolism and sphingolipid metabolism were identified as the most relevant pathways. Based on the results of the pathway analysis, sphingolipid and cholesterol biosynthesis and glycerophospholipid metabolism were regarded as key pathways in which S. baicalensis is involved to regulate lipid metabolism.
Conclusion: According to our metabolomics results, S. baicalensis may exert its therapeutic effects by regulating the cholesterol biosynthesis and sphingolipid metabolism pathways. Upon further analysis of the altered metabolites in certain pathways, agents downstream of squalene were significantly upregulated; however, the substrate of SQLE was surprisingly increased. By combining evidence from molecular docking, we speculated that baicalin, a major ingredient of S. baicalensis, may suppress cholesterol biosynthesis by inhibiting SQLE and LSS, which are important enzymes in the cholesterol biosynthesis pathway. In summary, this study provides new insights into the therapeutic effects of S. baicalensis on lipid metabolism using network pharmacology and lipidomics.
Keywords: S. baicalensis, network pharmacology, cortex metabolomics, lipid metabolism, molecular docking
Introduction
Lipid metabolism is an extremely complex process that not only is involved in constituting biological membranes but also functions as a signal and messenger, engaging in many physiological and pathological processes.1,2 Under normal physiological conditions, lipid homeostasis is tightly controlled; however, when disrupted or dysregulated by genetic and environmental factors, dyshomeostasis of lipid metabolism will lead to a high risk of developing lipid-associated metabolic diseases.3 As lipids play an essential role in the body, the disruption of several lipid metabolites may predispose individuals to many diseases, particularly because of the existence of the blood brain barrier (BBB) that blocks the transport of most lipids from the periphery.4 Lipid metabolism is tightly regulated, and as revealed by numerous studies, many diseases, including Alzheimer’s disease and cerebrovascular disease, may arise from altered lipid metabolism.5,6 For instance, accumulating genetic, clinical and modern pharmacological evidence has demonstrated that the pathogenesis of Alzheimer’s disease is closely associated with the dysregulation of cholesterol and sphingolipid metabolism.7,8 In addition, chronic inflammation has been reported to arise from dysregulation of endogenous bioactive lipids.9–12 Accordingly, pathways regulating lipid metabolism potentially represent an important druggable axis and therapeutic approach.
Traditional Chinese medicine has been used for more than two thousand years and is considered a promising resource for potential treatment because a large number of species contain active pharmaceutical ingredients with a wide range of medicinal properties. Pharmacological studies have shown that the aqueous extract of S. baicalensis, which contains mainly flavonoids, exerts various health care and clinical effects, such as anti-inflammatory activity, anti-atherosclerosis activity and the regulation of serum lipid level, and these diseases are closely related to lipid metabolism.13–15 However, the detailed mechanism by which S. baicalensis regulates metabolism to exert its therapeutic efficacy remains to be elucidated. Thus, our study aimed to provide insights into the effects of S. baicalensis on regulating lipid metabolism and clarify its mechanism of action.
Metabolomics, a newly developed discipline, is generally utilized to explore the mechanism of diseases and to evaluate the therapeutic efficacy of drugs by identifying the changes in specific substrates of metabolic pathways and metabolites of products from a comprehensive and holistic perspective.16 Because of the integrity of metabolomics studies, this approach has been extensively utilized to disclose the metabolic mechanism of the whole organism, which reflects physiological and pathological processes.17 In particular, traditional Chinese medicines (TCM) are best characterized by the nature of multiple components and ambiguously revealed mechanisms with clinically confirmed curative effects. Researchers have attempted to elucidate the mechanism of TCM using modern analytical equipment.18 Currently, metabolomics based on chromatography and mass spectrometry coupled with network pharmacology has emerged as an important method for discovering the mechanisms of TCM from a systemic perspective and at the molecular level, accelerating the revelation of the mechanism and medicinal material basis of TCM.19,20 In this study, network pharmacology was initially utilized to identify potential therapeutic compounds and explore the biological mechanism by which S. baicalensis regulates lipid metabolism. In addition, metabolomics based on liquid chromatography-quadrupole time-of-flight-mass spectrometry (LC-QTOF-MS) and gas chromatography-mass spectrometry (GC-MS) were performed to probe metabolic markers and analyze metabolic pathways. Eventually, the molecular mechanism underlying the effects of compounds in S. baicalensis on crucial targets was further validated by performing molecular docking. This study primarily probes the minute mechanism by which S. baicalensis regulates lipid metabolism.
Materials and Methods
Preparation of the S. baicalensis Aqueous Extract and Quality Control
S. baicalensis (200 g) (S. baicalensis Georgi, Lamiaceae; batch number, 1709019024 purchased from Fu Chun Tong Chinese Herbal Pieces Co., Ltd. Anhui, China) was thoroughly decocted in 2000 mL (1:10 v: w) of distilled water for 2 h, and then the dregs were decocted again with 1600 mL distilled water (1:8 v: w) and filtered through gauze. The filtrates were pooled and evaporated by rotary evaporation under vacuum at 60°C and concentrated to 100 mL for further freezing-drying. Eventually, 17.5 g of powder of the aqueous extract was obtained and stored in a desiccator until further drug treatment. The method used for S. baicalensis decoction referred to the technology of the Huang-Lian Jie-Du decoction used in this laboratory to ensure the extraction of effective ingredients.
S. baicalensis was analyzed by HPLC to assure the quality and effectiveness (the details were shown in Supplementary Materials). The results are displayed in Supporting information Figure S1 and Tables S1 and S2.
Experimental Design
C57BL/6 male mice (18–22 g) (batch number: SCKX, 2019–0002) were purchased from the Experimental Animal Center of Qinglongshan (Nanjing China). All mice were housed in a standard environment (12/12 h dark/light cycle, 50±10% relative humidity, 22±2°C) and had free access to standard mouse chow and water. After 10 days of acclimatization, they were randomly divided into two groups in a blinded manner: S. baicalensis group and control group. Animal welfare and experimental procedures were strictly performed in accordance with the Guide for the Care and Use of Laboratory Animals (US National Research Council, 1996) and the related ethics committee of Nanjing University of Chinese Medicine (NO. 202102A006, Item NO. 012071001462) approved the animal experiments. Mice in the S. baicalensis group were intragastrically administered S. baicalensis freeze-dried powder at a dosage of 100 mg/kg body weight during the experimental period. Mice in the control group were intragastrically administered normal saline. The experimental period lasted for 7 days, and the body weights of the mice were recorded daily. On the seventh day of the experiment, 1 h after the last drug administration, all mice were sacrificed by cervical dislocation after orbital blood collection. The cortex region tissues were removed from the brain and stored at −80°C until further metabolomic analysis.
Network Pharmacology Analysis
Identification of the Active Compounds and Target Genes of S. baicalensis
Bioinformatics was utilized to screen and comprehensively identify the potential active druggable ingredients of S. baicalensis. The chemical ingredients of S. baicalensis were obtained from the Traditional Chinese Medicine Systems Pharmacology Analysis Database and Analysis Platform (http://tcmspw.com/tcmsp.php) by importing S. baicalensis as a key word, and compounds were obtained from TCMSP manually supplemented based on a literature search in PubMed Central of the NCBI database (https://www.ncbi.nlm.nih.gov/). The selected compounds were further filtered by oral bioavailability (OB), drug-likeness (DL) and bblood-brainbarrier (BBB) values. These ADME properties are critical for drug discovery and development, and the general filtering criteria were OB≥30%, DL ≥0.18, and Caco-2≥-0.4. Subsequently, the molecular format of the filtered active compounds was converted into an sdf file format, and then these files were uploaded into PharmMapper Server (http://www.lilab-ecust.cn/pharmmapper/), which is a freely accessible web server designed to identify potential candidate target proteins for specific probe small molecules (drugs, natural products, or other newly discovered compounds with binding targets unidentified) using a pharmacophore mapping approach. Eventually, the targets obtained from the aforementioned database search were imported into Venny (http://www.liuxiaoyuyuan.cn/) to be combined, and duplicates were removed. These retrieved target proteins were calibrated to their official name (official symbol).
Prediction of Targets of S. baicalensis in Lipid Metabolism
Data on the metabolism of lipid-associated targets were collected from six resources, DisGeNET (http://www.disgenet.org/), CTD (http://ctdbase.org/), NCBI Gene (https://www.ncbi.nlm.nih.gov), OMIM (http://omim.org/) and GENECARD (https://www.genecards.org/). The targets related to lipid metabolism were searched with keywords related to lipid metabolism based on the classification of lipid metabolism from KEGG Mapper (https://www.kegg.jp/), and the retrieved results were imported into Excel for merging and removing repeated targets. The predicted targets of S. baicalensis and lipid metabolism-related targets were derived from the intersection as potential targets of S. baicalensis in lipid metabolism.
Enrichment Analysis and Network Construction
Key targets were imported into the Metascape database (http://metascape.org) to obtain information on GO (Gene Ontology)-related biological processes (BP), cellular components (CC), molecular functions (MF) and KEGG pathways, GO enrichment and KEGG pathway enrichment analyses were performed to explore the potential mechanism of S. baicalensis in lipid metabolism at the system level.21 Additionally, the acquired targets were submitted to STRING (https://string-db.org/) and Metascape to obtain a protein–protein interaction network.22 Furthermore, Cytoscape 3.7.1 software (http://www.cytoscape.org/) was utilized to construct a drug molecule-potential target network of S. baicalensis regulating lipid metabolism.
MS-Based Metabolomics
Cortex Region Tissue Collection and Preparation
Mice were sacrificed by cervical dislocation on day 7 after blood collection. The cortex region was removed and stored at −80°C until further metabolomics analysis. Tissues (~20 mg) were homogenized with 200 µL of cold methanol in a homogenizer (multisample tissue lyser-48, Jingxin Technology, Shanghai, China). Then, 600 µL of methyl tert-butyl ether (MTBE) was added and vortexed using a vortexer (multitube vortexer, Targin Technology, Beijing, China) for 1 min, followed by incubation at 4°C for 30 min to fully extract lipid metabolites. After the incubation, 150 µL of deionized water was added to induce phase separation and centrifuged at 10,000×g for 10 min at 4°C to separate the mixture into two phases with a protein interface. The bottom phase (400 µL) was retrieved, and then 800 µL of MTBE were added and the extraction was repeated. The organic solvents were combined, dried under nitrogen, and stored at −80°C until analysis. The nitrogen-dried samples were redissolved in a mixture of acetonitrile (ACN) and isopropyl alcohol (IPA) (1:9 v: v, with 0.1% formic acid and 1 mM ammonium formate) ultrasonically and centrifuged (15,000 rpm for 10 min), and then the supernatants were collected for detection. A mixture of equal quantities was extracted from each lipid extract sample and used as aa quality control(QC) pool sample for quality control during the MS analysis.
Metabolomics Data Acquisition and Processing
An Agilent 5600 Q-TOF LC/MS (Agilent Technologies, Palo Alto, USA) system equipped with a heated electrospray ionization (ESI) probe was used for lipidomics analysis. A 5 µL aliquot of each sample was injected into the system on an ACQUITY UPLC BEH C18 analytical column (2.1×50 mm, 1.7 µm; Waters Crop) at 40°C. A mixture of ACN and deionized water (6:4 v: v, with 10 mM ammonium formate and 0.1% formic acid, mobile phase A) and a mixture of ACN and IPA (1:9 v: v, with ammonium formate and 0.1% formic acid, mobile phase B) were employed as the mobile phases. For better chromatographic separation, a linear gradient with flow rate set to 0.4 mL/min was utilized for 14 min: 32% B at 0 min, 40% B from 0–1 min, 45% B from 1.5–4 min, 50% B from 4–5 min, 60% B from 5–8 min, 70% B from 8–11 min, and 80% B from 11–14 min. MS signal acquisition was performed in both positive and negative scanning modes. Quality controls (QC samples) were prepared with an equal volume of 5 µL and injected after every 5 analytical samples during the experiment to evaluate the stability of the analytical system and monitor the reproducibility of the data.
Mass spectrometry parameters were set as follows: ESI, cation and anion modes; mass scanning range, m/Z 50–1500; air curtain gas pressure, 275.8 kPa; atomization gas pressure, 379.2 kPa; auxiliary gas pressure, 379.2 kPa; ion source temperature, 550°C; spray voltage, −4500 V; and cluster voltage, −100 V.
In an attempt to comprehensively analyze metabolites, particularly neutral lipids, which are notoriously difficult to ionize with ESI sources, we selected GC-MS to analyze neutral lipids as a complement to LC-MS with inadequate abilities to analyze neural lipids for the purpose of obtaining better identification and separation of metabolites. Two steps were performed before sample analysis, namely, profile preprocessing, as described below. The first step was a lipid metabolite extraction protocol including methanol homogenization and an incubation with MTBE and (butylated hydroxytoluene) BHT (0.4 mg/mL) to prevent autoxidation of the extracted lipids, because MTBE shows a better performance in extracting neutral lipids, particularly sterols, including cholesterol and its precursors, than chloroform. After vortexing and phase separation, the organic phase was transferred to new tubes and dried under nitrogen. Sample deviation was involved in the second processing step. For comprehensive deviation, the mixture of N-trimethylsilylimidazole (TSIM) and N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) (1:10, v: v) was optimized as the derivatization reagent, and the reaction period was set to 1 h at 60°C. The vial was shaken at least twice with a vortex mixer for 30 s during the derivatization process to ensure complete sialylation. Eventually, each sample was centrifuged (15,000 rpm for 10 min), and 100 µL of the supernatant was transferred to a GC-MS autosampler for analysis.
For GC-MS data analysis, 1 µL of the derivatized sample was injected (splitless mode), and the inlet temperature was maintained at 250°C. All samples for the metabolomics analysis were analyzed on an Agilent 7890A GC system coupled with an Agilent 5975C MSD quadrupole mass spectrometer using electron impact ionization mode (Agilent, Palo Alto, USA). The injection temperature was 280°C, and helium was used as the carrier gas at a constant flow rate of 1.4 mL/min. An inert 5% phenylmethyl polysiloxane column (HP-5MS 30 m×0.25 mm 0.25 µm from Agilent) was selected to ensure a satisfactory degree of metabolite separation. After injection, the initial column oven temperature was held at 180°C for 1 min, ramped up at a rate of 20°C/min until reaching 280°C, maintained for 7 mins, and eventually raised to 300°C at a rate of 4°C/min. The transfer temperature and ion source temperature were set at 250°C and 230°C, respectively. Eventually, each peak was identified and matched by the NIST14 search program.
Metabolomic Data Analysis and Pathway Analysis
The raw LC-MS data were processed using Markerview 1.2.1 software (AB Corp, Milwaukee, WI, USA) to perform t-tests, and then the preprocessed data were imported to SIMCA-P 14.0 (Umetrics AB, Umea, Sweden) software and HMBD (https://hmdb.ca/) for the multivariate statistical analysis to obtain and identify potential differentially altered metabolites. Principal component analysis (PCA) was performed to achieve separation among the experimental cohorts. The quality of the PLS-DA model was evaluated by the permutation test (n=200). Potential lipid biomarkers were extracted from the S-plot and VIP-plots filtered with VIP values >1 and P (corr) absolute values greater than 0.58 mean, and then the filtered metabolites were screened with Peakview1.2.1 to compare the mass spectrometry information. Finally, the peak area and retention time were analyzed using independent-sample t-tests to discriminate significant differences, and all the quantified data are presented as the means ± SEM. S. baicalensis-regulated lipid metabolites were imported into the online Metabolite 4.0 software to explore the potential mechanism by which S. baicalensis regulates lipid metabolism.
Validation of the Binding Capacity Between Active Ingredients and Key Targets by Molecular Docking
Molecular docking has emerged as an important instrument to investigate the interaction between a ligand and receptor macromolecule and to predict their binding patterns and affinity, which has been widely utilized for drug design and discovery. AutoDock 1.5.6 (http://autodock.scripps.edu/) and Vina (http://vina.scripps.edu/) are excellent freely accessible drug discovery platforms with high docking accuracy and speed that integrate predictive physics-based methods with machine learning techniques to accelerate drug discovery.23,24 The sdf file format of active ingredients obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) was prepared and optimized by Chem3D for energy minimization and structure optimization. Based on the node values of the network pharmacology and alterations in metabolomics, squalene monooxygenase (SQLE), and lanosterol synthase (LSS), in total,10 protein targets were selected out as potential acting targets. The PDB files of key protein targets with the best structural resolution coupled with endogenous ligands were downloaded from the RCSB Protein Data Bank (https://www.rcsb.org/). The PDB files of key targets were then processed by Discovery Studio 4.5 to perform structural optimization for further analysis. The parameter settings for the docking site were obtained from a published report and the PyMOL 2.2.0 plugin to obtain grid box parameters. After setting the grid box for the active site of proteins, a genetic algorithm was performed, and docking run options were set to the default parameters; eventually, the dpf file was exported. The docking results were visualized with Discovery Studio Visualizer 4.5 to observe the docking binding pattern.
Validation of Molecular Docking
To confirm the inhibiting effect of baicalin on SQLE, we performed a vitro experiment implemented on Mouse liver microsomes (LM-XS-02M, Purchased from Research institute for Liver disease, Shanghai Ruide). The biochemistry assay of Baicalin activity was performed on Mouse liver microsomes, 7 concentration gradients including 175, 150, 100, 75, 50, 40 and 20 µM baicalin was added and incubated for 1h. The detailed information was expressed on Supplemental Information Figure S5 and Table S6.
Results
Target Prediction and Functional Analysis of the Regulatory Effects of S. baicalensis on Lipid Metabolism
Predicted Targets of S. baicalensis Involved in Lipid Metabolism
Based on the oral bioavailability (OB), drug likeness (DL), BBB and Caco-2 parameters obtained through ADME, thirty-six ingredients were screened as potential druggable active compounds of S. baicalensis (Supplemental Table S3), and the structures of the identified ingredients are shown in Figure 1. The Pharmacy Mapper and TCMSP online databases (https://tcmspw.com/index.php) were utilized to comprehensively explore the possible targets of the active ingredients, and 471 target proteins of S. baicalensis were predicted. The target genes associated with “lipid metabolism” were obtained from the GeneCards (https://www.genecards.org/), OMIM (https://omim.org/) and CTD (http://ctdbase.org/) databases, and they intersected with the 471 active target proteins to yield 89 potential targets of active ingredients involved in lipid metabolism. Eventually, the predicted targets of ingredients and predicted targets of S. baicalensis involved in lipid metabolism were imported into Cytoscape 3.71 (https://cytoscape.org/) to construct the drug-target cross-linking network shown in Figure 2.
 |
Figure 1 The structures of the ingredients of S. baicalensis. |
Based on the corresponding quantity of key targets, the ingredients were endowed with different node sizes, providing ingredients and targets selected for further molecular docking analysis.
Enrichment Analysis and Network Construction
We imported 89 predicted target genes of S. baicalensis involved in lipid metabolism to the Metascape database, which is a comprehensive and powerful online tool integrated with many authoritative databases for gene enrichment analysis, to obtain information on GO-related biological processes, cellular components, molecular functions and KEGG pathways enriched in the predicted targets. After the key predicted targets were imported, the results of the gene enrichment analysis of pathways and processes were obtained and colored by clusters. The KEGG and GO enrichment analysis results are shown in Figure 3.
In summary, the analysis of enriched KEGG pathways indicated that 32 pathways and 16 key targets were involved in sphingolipid metabolism, 26 target genes were enriched in metabolic pathways, 9 target genes were involved in steroid biosynthesis, 8 target genes were involved in the PPAR signaling pathway and 7 target genes were involved in glycerolipid metabolism. This result showed that S. baicalensis mainly disturbs the sphingolipid metabolism and cholesterol biosynthesis pathways. The GO enrichment analysis information is illustrated from 3 perspectives. Target genes mainly participate in lipid biosynthesis processes, including cholesterol metabolism and biosynthesis, sphingolipid biosynthesis and lipid catabolic processes, and these genes are mostly located and exert their functions inthe vacuolar lumen, lipid droplets and endoplasmic reticulum lumen. In addition, the biosynthesis process and two major enriched cellular components and molecular functions were imported into OmicStudio tools at (https://www.omicstudio.cn/tool) for bioinformatics analysis and to obtain an UpSet graph (Figure 4). This result offered the relevance and intersection of pathways and GO enrichment analysis.
 |
Figure 4 The UpSet graph coupled with elements of overlapping enriched GO terms. (A) GO biological process and (B) two major GO processes of cellular components and molecular functions. |
According to the GO and KEGG enrichment analyses, S. baicalensis exerts its regulatory effects on lipid metabolism mainly by modulating cholesterol biosynthesis, which has an important role in steroid metabolism and sphingolipid metabolism. Furthermore, with the purpose of tapping into the core regulatory genes, the predicted target proteins of S. baicalensis were imported into Metascape (http://metascape.org) STRING (https://string-db.org/) to build the PPI (Figure 5). The PPI result shows that the clustered proteins groups mainly involved in metabolism of lipids, lipid biosynthetic process and cholesterol biosynthesis process. The analysis of protein interaction networks is one of the key challenges in the study of biology, which connects genotypes to phenotypes, and the disruption of PPIs often leads to diseases.
Results of Cortex Region Tissue Metabolomics
Analysis of Metabolic Profiles
We performed PCA to reflect the clustering degree of the QC samples, verify the QC samples, and analyze the reproducibility and repeatability of the analytical equipment. The PCA results indicated that the QC samples were closely clustered, indicating excellent reproducibility and stability (Supplemental Figures 2 and 3). Furthermore, the model of the PCA class was built to discriminate between the analyzed groups, and the control cohorts and S. baicalensis cohorts were clearly separated. The OPLS-DA model was performed to focus categorical information on principal components and to determine metabolic variations.
R2 and Q2 (positive mode is shown in Supplemental Figure S2, and negative mode is shown in Supplemental Figure S3) indicate the fitness and predictive accuracy of the model. Subsequently, 200 permutations and a threshold VIP value prediction were performed, and a VIP value greater than 1 and a P value less than 0.05 were used to select significantly differentially expressed lipids. Eventually, the ion mass information was imported into HMDB (the Human Metabolome Database) and further processed with Peakview1.1.2 to obtain identified metabolites. Among the identified lipids, 14 metabolites were significantly different between the control group and the S. baicalensis group including sphingomyelin, galabiosylceramide (d18:1/9Z-18:1), lactosylceramide (LacCer) (d18:1/12:0), glucosylceramide (GlcCer) (d18:1/24:0), sulfatide, cholesterol ester (CE) (16:1(9Z)), phosphatidylcholine, phosphatidylethanolamine and Triacylglycerol (TG) (16:0/16:0).
Metabolic Pathway Analysis
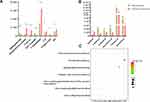
Fourteen differentially altered endogenous metabolites were identified between the S. baicalensis group and the control group, and detailed information on altered metabolites and corresponding trends is presented in Supplemental Table S4. These metabolites are primarily associated with cholesterol biosynthesis and sphingolipids, fatty acids and steroid hormones.25 A metabolic pathway analysis was performed by introducing these differentially altered metabolites into MetaboAnalyst 4.0 (http://www.metaboanalyst.ca/) to further explore the potential role of S. baicalensis and the possible mechanism by which it regulates lipid metabolism. The pathway analysis based on different biomarkers revealed that treatment with S. baicalensis resulted in significant enrichment of the cholesterol de novo biosynthesis pathway and sphingolipid metabolism when filtered with the threshold value of P≤ 0.05. Eventually, the histogram of significantly alternated lipid metabolites after treatment with S. baicalensis and bubble diagram enriched pathways was constructed (Figure 6). The metabolomics results initially supported the regulative effect of S. baicalensis on lipid metabolism and displayed the main metabolic pathways, which S. baicalensis was involved. Therefore, significant differences in endogenous metabolites between S. baicalensis and control group were proposed.
Network Pharmacology and Metabolomics Integration Analysis
We assayed the correlation between metabolites and possible upstream targets, integrated the results of metabolomics and network pharmacological analyses, and comprehensively explored the underlying mechanisms to further understand the potential therapeutic and effective substrates of S. baicalensis in lipid metabolism. In particular, we mentioned the relationship between metabolites of key pathways and their associated proteins, such as enzymes identified from the human metabolome database. Finally, a network containing the relationships among metabolites, metabolic pathways, enzymes and target genes was established (Figure 7). This constructed graph combined the results of network pharmacology and results of metabolomics, which provides general view of pathways S. baicalensis is involved.
Molecular Docking
Notably, the force of interaction between drugs and targets is fundamental to biological effects. Numerous interaction patterns play an important role in the recognition and binding of proteins and drugs. However, because of the difference in binding energy and effective radius, the stability and binding strength are different. In this context, three main existing interaction patterns were introduced, as described. Hydrogen bonds with an average binding energy of 5 kJ/mol, which are weak electrostatic attraction between electropositive hydrogen atoms and electronegative heteroatoms, play an essential role in drug–target interactions.26 In addition, relatively weak attractions mediated by van der Waals forces widely exist and decay with an increasing interaction radius. Hydrophobic interactions are formed between the hydrocarbyl moieties of drug molecules and hydrophobic groups of targets, extruding water molecules that were originally configured on the surface of proteins and drugs. This interaction pattern is very important in maintaining the stability and formation of complexes.
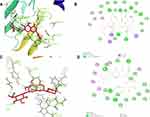
Autodock1.5.6 was utilized to elucidate the interactions between active components and identify different lipid-related protein targets. The docking results are shown in Supplemental Table S5. A lower docking energy represents a stronger affinity between proteins and ligands. Moreover, the docking pattern was visualized using Discovery Studio 4.5 to predict the binding pattern. Based on the results of the target-ingredient interaction network, 9 targets and 10 ingredients were selected to perform molecular docking and the targets of crystal structure file were obtained from PDB bank. The docking results are presented in a heat map (Figure 8). The docking energy and predicted Ki (inhibition constant) were considered. Each target corresponded to the optimal ingredients with the lowest energy and highest Ki and LSS, and SQLE was identified as having the best Ki and binding energy and used to predict its interaction mechanism and binding pattern with baicalin (shown in Figure 9). In the dissection of the binding pattern predicted in Discovery Studio presented in the 3D and 2D graphs, 6 hydrogen bonds between LSS and baicalin and 4 hydrogen bonds between SQLE coupled with FAD and baicalin were observed. In the complex of OSC and baicalin, TYR 98 (2.09 Å), GLY380 (2.12 Å), CYS456 (3.08 Å), TYR 503 (1.82 Å), and TYR 704 (2.47 Å) interact with the oxhydryl group of baicalin, forming 5 hydrogen bonds; moreover, TRP192, TRP230, PHE521 and ILE524 constitute the hydrophobic pocket that is beneficial to the stabilization of the complex. Additionally, the alkyl group of baicalin is stacked between LEU324, PHE509, PRO505, VAL506 and LEU509, and the alkoxy and oxhydryl groups are hydrogen-bonded to TY195 (2.71 Å), PRO415 (1.87 Å), LEU416 (1.95 Å), and the oxygen atom (2.9 Å) of FAD. Furthermore, the optimal conformation of baicalin was closely clustered with endogenous ligands of LSS and SQLE, indicating that baicalin accurately lies in the active pocket of the target (Supplemental Figure 4). Based on the results described above, we conclude that baicalin presented a favorable binding pattern and may serve as a potential inhibitor of LSS and SQLE.
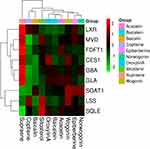 |
Figure 8 Heat map of docking energies. |
Vitro Experiment for SQLE Inhibition
Considering the SQLE protein is principally located on endoplasmic reticulum, we chose Mouse liver microsomes to perform vitro inhibiting experiment of baicalin. In the dissection of the result of seven different concentrations of baicalin treatment on Mouse liver microsomes, similar but concentration-dependent inhibiting effects was observed (shown in Supplemental Information Figure S5 and Table S6). Squalene, the substrate of SQLE, was gradually increased with the increase of baicalin, which represented the inhibiting effect of baicalin was concentration-dependent. The results of Mouse liver microsomes and metabolomics both evidenced the molecular docking results.
Discussion
The effects of S. baicalensis on lipid metabolism have been widely investigated and reported, and clinical evidence has also probed its regulatory role in lipid metabolism, which involves in glycolipid metabolism, triglyceride signaling pathway and cholesterol metabolism.27–30 In many recently published studies, S. baicalensis and its main bioactive and effective ingredients have yielded many antiobesity, antihyperlipidemic and anti-hepatic steatosis activities, and some of the underlying mechanisms that closely associated with lipid metabolism have been revealed.31–33 As demonstrated in vitro and vivo experiments, S. baicalensis and baicalin exhibited anti-obesity and regulative effective on blood cholesterol and triglyceride level through regulating metabolism, in an experiment combined with chemoproteomics revealed that baicalin can directly activate carnitine palmitoyltransferase 1 (CPT1) and accelerate the oxidation of lipids.33–35 In the present study, the mechanism by which S. baicalensis regulates lipid metabolism was elucidated by identifying differentially altered lipid metabolites and performing a network pharmacology prediction. The metabolomic and network pharmacology results indicate that the effect of S. baicalensis on lipid metabolism involved the regulation of sphingolipid and cholesterol de novo biosynthesis and glycerophospholipid metabolism. Additionally, 16 targets and 12 ingredients were identified based on the constructed network. The targets participating in cholesterol biosynthesis, including SQLE, LSS, FDFT1, and SOAT, are potential targets, and baicalin, baicalein, coptisine, epiberberine, wogonin, and sitosterol may be potential active ingredients of S. baicalensis that modulate cholesterol biosynthesis. Moreover, as disclosed in this context, ARSA, KDSR, GALC, GBA, GLA, NEU, UGCG, UGT8, DEGS1, SPHK1, B4GALT6, GAL3ST1, CERS2, SGPP1, ACER1, SGMS1, AKT2, and MAPK1 were potential targets in sphingolipid metabolism. In conclusion, multiple ingredients of S. baicalensis exert their regulatory effects on lipid metabolism by targeting multiple metabolic pathways. Ultimately, our results indicated that S. baicalensis mainly acts on cholesterol biosynthesis and sphingolipid metabolism, which may be the potential mechanism by which S. baicalensis exerts its therapeutic effects.
Cholesterol has multiple essential functions in the human body, particularly in the CNS, which contains 25% of the total human cholesterol distributed in neurons, glial cells and myelin mainly in the form of free cholesterol.36 Cholesterol not only plays an important role in the constitution of the cell membrane but also exerts a signal transduction function. The cholesterol turnover rate in neurons and glial cells may reach an estimate of 20% per day.37 In addition, based on accumulating genetic, biochemical and clinical evidence, the alteration of cholesterol metabolism may be the cause of the predisposition to many neurodegenerative diseases.9,37 In addition to cholesterol, many precursors in distal cholesterol biosynthesis, including and starting with squalene and 24-OH cholesterol, have recently emerged as promising drug targets in several diseases.38,39 For example, squalene accumulation has been reported to be associated with lung and colorectal cancer.40,41 Moreover, numerous studies have confirmed the relationship between high cholesterol levels and Alzheimer’s disease, and cholesterol drugs, including statins and efavirenz, have yielded pronounced curative effects.42 In the present study, after intervention with S. baicalensis, metabolomics results revealed that cholesterol biosynthesis was significantly inhibited. The metabolomics results, which were consistent with the results of treatment with NB598, as an inhibitor of SQLE, showed that squalene was prominently increased while cholesterol and beta-sitosterol were decreased.43 In the dissection of the trends of altered metabolites using metabolomics and their upstream and downstream relationships, we speculated that SQLE, namely, squalene monooxygenase, may be the crucial target of S. baicalensis. In combination with the evidence obtained from molecular docking and network pharmacology, SQLE, which serves as one of the rate-limiting enzymes in cholesterol biosynthesis, may be a target. In the dissection of results from network pharmacology and molecular docking, baicalin with the best binding energy and node degree was screened as a potential inhibitor of SQLE. Intriguingly, SQLE has been shown to be associated not only with cholesterol biosynthesis but also with cancer and tumors.44 Considering the clinical therapeutic effects of S. baicalensis and baicalin and evidence based on our research, we are confident in our conclusion that SQLE is an important target of S. baicalensis.45 Collectively, based on the results of network pharmacology, metabolomics and molecular docking, baicalin, the principal component of S. baicalensis, is a potential effective medicinal element that regulates cholesterol biosynthesis by interacting with SQLE. Accordingly, further in-depth studies should be conducted to verify and elucidate the detailed mechanism underlying the effect of baicalin on SQLE.
Sphingolipids are a component of all membranes but are particularly abundant in the myelin sheath.46 The hydrolysis of sphingolipids, which are important bioactive metabolites, has been reported to be involved in the regulation of many important signal transduction processes.47 Previous research studies have revealed that alterations in sphingolipid metabolism are associated with a high risk of neurodegenerative diseases.48 A recent study by Vladimir Rudajev reported that sphingomyelin triggers oligomerization of Aβ40, which is one of the major biochemical hallmarks of Alzheimer’s disease.49 In addition, previous lipidomics studies performed on the brains of wild-type mice at different ages revealed that the content of glucosylceramide, glucosylsphingosine, and GM1a was increased with aging.50 In the present study, SM (d18:1/18:0) and glucosylceramide were significantly decreased after treatment with S. baicalensis compared with the normal cohort, and supporting evidence was also obtained for the serum levels of these lipids after treatment with Huang-Lian-Jie-Du-Tang.51 As reported in many studies, the increase in the SM content is closely associated with inflammation and cell apoptosis.52,53 From the perspective of cell function, SM is mainly located in the endoplasmic reticulum and regulated by sphingomyelin phosphodiesterase (SMPD1), which mainly converts sphingomyelin to ceramide to maintain the relative balance of SM.54 Virtual screening using Instadock was conducted to identify bioactive ingredients and explore the mechanism by which S. baicalensis regulates SMPD1.55 Wogonin, norwogonin and coptisine showed good binding activity with SMPD1 in our study, which may contribute to the variation observed in lipidomics.
Conclusions
In the present study, based on bioinformatics encompassing network pharmacology, GO and KEGG enrichment analyses and molecular docking, we screened 10 candidate ingredients and two lipid-associated pathways: cholesterol biosynthesis and sphingolipid metabolism. The integrated and comprehensive strategy may circumvent the drawback of conventional research on traditional Chinese medicine, enabling the biological mechanisms to be more accurately and precisely determined. The results of lipidomics and network pharmacology analyses revealed that SQLE, LSS, SOAT, CPT1A and SMPD1 were predicted as lipid-related targets of S. baicalensis in lipid metabolism. Intriguingly, SQLE and LSS may be targets with which baicalin directly interacts to exert its clinical therapeutic effect on lowering cholesterol. Collectively, this study preliminarily elucidated the pharmacological mechanism of S. baicalensis in regulating lipid metabolism through network pharmacology and lipidomics, which may contribute to further drug discovery and clinical application of S. baicalensis in diseases associated with lipid metabolism dyshomeostasis.
Abbreviations
PPARs, peroxisome proliferator-activated receptor; SQLE, squalene monooxygenase; LSS, lanosterol synthase; BBB, bblood-brainbarrier; TCM, traditional Chinese medicine; OB, oral bioavailability; DL, drug-likeness; HPLC, high-performance liquid chromatography; GO, Gene Ontology; BP, biological processes; CC, cellular components; MF, molecular functions; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, pprotein–proteininteraction; PCA, principal component analysis; PLSDA, partial lleast-squaresdiscrimination analysis; LacCer, lactosylceramide; GlcCer, glucosylceramide; CE, cholesterol ester; TG, triacylglycerol; SM, sphingomyelin; SOAT, sterol O-acyltransferase; CPT1A, carnitine O-palmitoyltransferase 1, liver isoform; SMPD1, sphingomyelin phosphodiesterase; TYR, tyrosinase; GLY, glycine; CYS, cysteine; PHE, phenylalanine; ILE, isoleucine; VAL, valine; LEU, leucine; PRO, proline.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (Project Nos. 81873027 and 81573635), the Qing-Lan Project of Jiangsu Province, the Open Project Program of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica (No. JKLPSE201820), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Project of the Innovation Research Team of Nanjing University of Chinese Medicine, and the Project Funded by the Six Talent Project in Jiangsu Province.
Disclosure
The authors reported no conflicts of interest for this work and declare that the study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Balla T. Cell biology: lipid code for membrane recycling. Nature. 2016;529(7586):292–293. doi:10.1038/nature16868
2. Fielding CJ, Fielding PE. Membrane cholesterol and the regulation of signal transduction. Biochem Soc Trans. 2004;32(1):65–69. doi:10.1042/bst0320065
3. Chen JH, Hsieh CJ, Huang YL, et al. Genetic polymorphisms of lipid metabolism gene SAR1 homolog B and the risk of Alzheimer’s disease and vascular dementia. J Formos Med Assoc. 2016;115(1):38–44. doi:10.1016/j.jfma.2015.01.008
4. Hussain G, Anwar H, Rasul A, et al. Lipids as biomarkers of brain disorders. Crit Rev Food Sci. 2020;60(3):351–374. doi:10.1080/10408398.2018.1529653
5. Bales KR. Brain lipid metabolism, apolipoprotein E and the pathophysiology of Alzheimer’s disease. Neuropharmacology. 2010;59(4–5):295–302. doi:10.1016/j.neuropharm.2010.01.005
6. Cheng TJ, Chuu JJ, Chang CY, Tsai WC, Chen KJ, Guo HR. Atherosclerosis induced by arsenic in drinking water in rats through altering lipid metabolism. Toxicol Appl Pharmacol. 2011;256(2):146–153. doi:10.1016/j.taap.2011.08.001
7. Zarrouk A, Debbabi M, Bezine M, et al. Lipid biomarkers in Alzheimer’s disease. Curr Alzheimer Res. 2018;15(4):303–312. doi:10.2174/1567205014666170505101426
8. Pena-Bautista C, Alvarez L, Durand T, et al. Clinical utility of plasma lipid peroxidation biomarkers in Alzheimer’s disease differential diagnosis. Antioxidants (Basel). 2020;9(8).
9. van der Kant R, Langness VF, Herrera CM, et al. Cholesterol metabolism is a druggable axis that independently regulates tau and amyloid-beta in iPSC-derived Alzheimer’s disease neurons. Cell Stem Cell. 2019;24(3):363–375 e369. doi:10.1016/j.stem.2018.12.013
10. Fonteh AN, Ormseth C, Chiang J, Cipolla M, Arakaki X, Harrington MG. Sphingolipid metabolism correlates with cerebrospinal fluid beta amyloid levels in Alzheimer’s disease. PLoS One. 2015;10(5):e0125597. doi:10.1371/journal.pone.0125597
11. McHale-Owen H, Bate C. Cholesterol ester hydrolase inhibitors reduce the production of synaptotoxic amyloid-beta oligomers. Biochim Biophys Acta Mol Basis Dis. 2018;1864(3):649–659. doi:10.1016/j.bbadis.2017.12.017
12. Nakamura N, Pence LM, Cao Z, Beger RD. Distinct lipid signatures are identified in the plasma of rats with chronic inflammation induced by estradiol benzoate and sex hormones. Metabolomics. 2020;16(9):95. doi:10.1007/s11306-020-01715-w
13. Ming J, Zhuoneng L, Guangxun Z. Protective role of flavonoid baicalin from S. baicalensis in periodontal disease pathogenesis: a literature review. Complement Ther Med. 2018;38:11–18. doi:10.1016/j.ctim.2018.03.010
14. Na HY, Lee BC. S. baicalensis alleviates insulin resistance in diet-induced obese mice by modulating inflammation. Int J Mol Sci. 2019;20(3):727. doi:10.3390/ijms20030727
15. Zhao T, Tang H, Xie L, et al. S. baicalensis Georgi. (Lamiaceae): a review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J Pharm Pharmacol. 2019;71(9):1353–1369. doi:10.1111/jphp.13129
16. Pang H, Jia W, Hu Z. Emerging applications of metabolomics in clinical pharmacology. Clin Pharmacol Ther. 2019;106(3):544–556. doi:10.1002/cpt.1538
17. Chen H, Zhang F, Zhang J, Zhang X, Guo Y, Yao Q. A holistic view of berberine inhibiting intestinal carcinogenesis in conventional mice based on microbiome-metabolomics analysis. Front Immunol. 2020;11:588079. doi:10.3389/fimmu.2020.588079
18. Efferth T, Xu AL, Lee DYW. Combining the wisdoms of traditional medicine with cutting-edge science and technology at the forefront of medical sciences. Phytomedicine. 2019;64:153078. doi:10.1016/j.phymed.2019.153078
19. Zhang W, Chen Y, Jiang H, et al. Integrated strategy for accurately screening biomarkers based on metabolomics coupled with network pharmacology. Talanta. 2020;211:120710. doi:10.1016/j.talanta.2020.120710
20. Arya H, Coumar MS. Virtual screening of traditional Chinese medicine (TCM) database: identification of fragment-like lead molecules for filariasis target asparaginyl-tRNA synthetase. J Mol Model. 2014;20(6):2266. doi:10.1007/s00894-014-2266-9
21. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
22. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
23. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi:10.1002/jcc.21256
24. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
25. Xia JG, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi:10.1093/nar/gkp356
26. Wood PA, Pidcock E, Allen FH. Interaction geometries and energies of hydrogen bonds to C[double bond]O and C[double bond]S acceptors: a comparative study. Acta Crystallogr B. 2008;64(4):491–496. doi:10.1107/S0108768108015437
27. You CL, Su PQ, Zhou XX. [Study on effect and mechanism of S. baicalensis stem-leaf total flavonoid in regulating lipid metabolism]. Zhongguo Zhong Yao Za Zhi. 2008;33(9):1064–1066. Chinese.
28. Cui X, Qian DW, Jiang S, Shang EX, Zhu ZH, Duan JA. Scutellariae radix and coptidis rhizoma improve glucose and lipid metabolism in T2DM rats via regulation of the metabolic profiling and MAPK/PI3K/Akt signaling pathway. Int J Mol Sci. 2018;19(11):3634. doi:10.3390/ijms19113634
29. Xiao S, Liu C, Chen M, et al. Scutellariae radix and coptidis rhizoma ameliorate glycolipid metabolism of type 2 diabetic rats by modulating gut microbiota and its metabolites. Appl Microbiol Biotechnol. 2020;104(1):303–317. doi:10.1007/s00253-019-10174-w
30. Seo MJ, Choi HS, Jeon HJ, Woo MS, Lee BY. Baicalein inhibits lipid accumulation by regulating early adipogenesis and m-TOR signaling. Food Chem Toxicol. 2014;67:57–64. doi:10.1016/j.fct.2014.02.009
31. Hang Y, Qin X, Ren T, Cao J. Baicalin reduces blood lipids and inflammation in patients with coronary artery disease and rheumatoid arthritis: a randomized, double-blind, placebo-controlled trial. Lipids Health Dis. 2018;17(1):146. doi:10.1186/s12944-018-0797-2
32. He XW, Yu D, Li WL, et al. Anti-atherosclerotic potential of baicalin mediated by promoting cholesterol efflux from macrophages via the PPARgamma-LXRalpha-ABCA1/ABCG1 pathway. Biomed Pharmacother. 2016;83:257–264. doi:10.1016/j.biopha.2016.06.046
33. Dai J, Liang K, Zhao S, et al. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc Natl Acad Sci U S A. 2018;115(26):E5896–E5905. doi:10.1073/pnas.1801745115
34. Song KH, Lee SH, Kim BY, Park AY, Kim JY. Extracts of S. baicalensis reduced body weight and blood triglyceride in db/db Mice. Phytother Res. 2013;27(2):244–250. doi:10.1002/ptr.4691
35. Wang ZY, Jiang ZM, Xiao PT, Jiang YQ, Liu WJ, Liu EH. The mechanisms of baicalin ameliorate obesity and hyperlipidemia through a network pharmacology approach. Eur J Pharmacol. 2020;878:878. doi:10.1016/j.ejphar.2020.173103
36. Rosenheim MC. The cholesterol of the brain. III. Note on the cholesterol contents of human and animal brain. Biochem J. 1914;8(1):82–83. doi:10.1042/bj0080082
37. Jin U, Park SJ, Park SM. Cholesterol metabolism in the brain and its association with Parkinson’s disease. Exp Neurobiol. 2019;28(5):554–567. doi:10.5607/en.2019.28.5.554
38. Locatelli S, Lutjohann D, Schmidt HH, Otto C, Beisiegel U, von Bergmann K. Reduction of plasma 24S-hydroxycholesterol (cerebrosterol) levels using high-dosage simvastatin in patients with hypercholesterolemia: evidence that simvastatin affects cholesterol metabolism in the human brain. Arch Neurol. 2002;59(2):213–216. doi:10.1001/archneur.59.2.213
39. Lukiw WJ. Cholesterol and 24S-hydroxycholesterol trafficking in Alzheimer’s disease. Expert Rev Neurother. 2006;6(5):683–693. doi:10.1586/14737175.6.5.683
40. Sui Z, Zhou J, Cheng Z, Lu P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumour Biol. 2015;36(8):6173–6179. doi:10.1007/s13277-015-3301-x
41. Ge H, Zhao Y, Shi X, et al. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal-regulated kinase signaling. Thorac Cancer. 2019;10(3):428–436. doi:10.1111/1759-7714.12944
42. Rahman SO, Hussain S, Alzahrani A, Akhtar M, Najmi AK. Effect of statins on amyloidosis in the rodent models of Alzheimer’s disease: evidence from the preclinical meta-analysis. Brain Res. 2020;1749:147115. doi:10.1016/j.brainres.2020.147115
43. Xu F, Rychnovsky SD, Belani JD, Hobbs HH, Cohen JC, Rawson RB. Dual roles for cholesterol in mammalian cells. Proc Natl Acad Sci U S A. 2005;102(41):14551–14556. doi:10.1073/pnas.0503590102
44. Mahoney CE, Pirman D, Chubukov V, et al. A chemical biology screen identifies a vulnerability of neuroendocrine cancer cells to SQLE inhibition. Nat Commun. 2019;10(1):96. doi:10.1038/s41467-018-07959-4
45. Cheng CS, Chen J, Tan HY, Wang N, Chen Z, Feng Y. S. baicalensis and cancer treatment: recent progress and perspectives in biomedical and clinical studies. Am J Chin Med. 2018;46(1):25–54. doi:10.1142/S0192415X18500027
46. Giussani P, Prinetti A, Tringali C. The role of Sphingolipids in myelination and myelin stability and their involvement in childhood and adult demyelinating disorders. J Neurochem. 2020;156:403–414. doi:10.1111/jnc.15133
47. Avila-Garcia R, Valdes J, Jauregui-Wade JM, Ayala-Sumuano JT, Cerbon-Solorzano J. The metabolic pathway of sphingolipids biosynthesis and signaling in Entamoeba histolytica. Biochem Biophys Res Commun. 2020;522(3):574–579. doi:10.1016/j.bbrc.2019.11.116
48. Amaro M, Sachl R, Aydogan G, Mikhalyov II, Vacha R, Hof M. GM1 ganglioside inhibits beta-amyloid oligomerization induced by sphingomyelin. Angew Chem Int Ed Engl. 2016;55(32):9411–9415. doi:10.1002/anie.201603178
49. Rudajev V, Novotny J. The role of lipid environment in ganglioside GM1-induced amyloid beta aggregation. Membranes (Basel). 2020;10(9). doi:10.3390/membranes10090226
50. Parveen F, Bender D, Law SH, Mishra VK, Chen CC, Ke LY. Role of ceramidases in sphingolipid metabolism and human diseases. Cells. 2019;8(12):1573. doi:10.3390/cells8121573
51. Sun LM, Zhu BJ, Cao HT, et al. Explore the effects of Huang-Lian-Jie-Du-Tang on Alzheimer’s disease by UPLC-QTOF/MS-based plasma metabolomics study. J Pharm Biomed Anal. 2018;151:75–83. doi:10.1016/j.jpba.2017.12.053
52. Andrieu-Abadie N, Levade T. Sphingomyelin hydrolysis during apoptosis. Biochim Biophys Acta. 2002;1585(2–3):126–134. doi:10.1016/S1388-1981(02)00332-3
53. Kim MH, Ahn HK, Lee EJ, et al. Hepatic inflammatory cytokine production can be regulated by modulating sphingomyelinase and ceramide synthase 6. Int J Mol Med. 2017;39(2):453–462. doi:10.3892/ijmm.2016.2835
54. Oninla VO, Breiden B, Babalola JO, Sandhoff K. Acid sphingomyelinase activity is regulated by membrane lipids and facilitates cholesterol transfer by NPC2. J Lipid Res. 2014;55(12):2606–2619. doi:10.1194/jlr.M054528
55. Mohammad T, Mathur Y, Hassan MI. InstaDock: a single-click graphical user interface for molecular docking-based virtual high-throughput screening. Brief Bioinform. 2020. doi:10.1093/bib/bbaa279
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.