Back to Journals » Infection and Drug Resistance » Volume 15
Polymorphisms in the K13-Propeller Gene in Artemisinin-Resistant Plasmodium in Mice
Authors Zheng S, Liang Y, Wang Z, Liu M, Chen Y, Ai Y, Guo W, Li G, Yuan Y, Xu Z, Wu W, Huang X, Wu Z, Xu Q, Song J, Deng C
Received 26 July 2022
Accepted for publication 26 October 2022
Published 8 November 2022 Volume 2022:15 Pages 6533—6544
DOI https://doi.org/10.2147/IDR.S383127
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shaoqin Zheng1,2 *, Yuan Liang2 *, Zhaojia Wang,1 Min Liu,1 Yingyi Chen,1 Ying Ai,1 Wenfeng Guo,1,3 Guoming Li,1 Yueming Yuan,1,2 Zhiyong Xu,2 Wanting Wu,1 Xinan Huang,1,3 Zhibing Wu,3 Qin Xu,1,3 Jianping Song,1,3 Changsheng Deng1,3
1Artemisinin Research Center, Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China; 2Science and Technology Institute, Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China; 3The First Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianping Song; Changsheng Deng, Artemisinin Research Center, Guangzhou University of Chinese Medicine, Guangzhou, 510405, People’s Republic of China, Tel +86-20-3658 5633 ; +86-20-8746 1723, Email [email protected]; [email protected]
Introduction: Artemisinin-based combination therapies (ACTs) act as first-line antimalarial drugs and play a crucial role in the successful control of falciparum malaria. However, the recent emergence of resistance of Plasmodium falciparum to ACTs in South East Asia is of particular concern. Hence, there is an urgent need to identify the genetic determinants of and understand the molecular mechanisms underpinning such resistance. Artemisinin resistance (AR) is primarily driven by the mutations in the P. falciparum K13 protein, which is widely recognized as the major molecular marker of AR. However, association of K13 mutations with in vivo AR has been ambiguous due to the absence of a tractable model.
Methods: In this study, we have successfully produced artemisinin- and piperaquine-resistant P. berghei K173 following drug administrations. Prolonged parasite clearance and early recrudescence were found following daily exposure to high doses of artemisinin and piperaquine. We have also sequenced the DNA of artemisinin-resistant strains and piperaquine-resistant strains of P. berghei K173 to explore the relationship between PfK13 and AR.
Results: The resistance index of P. berghei K173 reached 12.4 after 30 artemisinin-resistant generations, but AR declined gradually after 30 generations. On the 50th generation, the resistance index of artemisinin-resistant strains was only 5.0 compared with the severe drug resistance of piperaquine-resistant strains (I90=148.8). DNA sequencing of artemisinin-resistant strains showed that there were 9 meaningful mutations at P. berghei K13-propeller domain, but the above mutations did not include common clinical point mutations.
Conclusion: Our data show that artemisinin is less susceptible to severe resistance compared with other antimalarial drugs. In addition, mutation on P. berghei K13 has a multi-drug-resistant phenotype and may be used as a biomarker to monitor its resistance. More studies need to be conducted on the new mutations detected so as to understand their association, if any, with ACT resistance.
Keywords: malaria, Plasmodium berghei, P. berghei K13 gene, artemisinin resistance
Plain Language Summary
Resistance against artemisinin poses a great threat to global malaria control and elimination. So, there is a major need to figure out the mechanism of the resistance to artemisinin. Widely prevalent in the economically less developed regions, it is hard to collect live Plasmodium with resistance to artemisinin. Given this, we cultured artemisinin- and piperaquine-resistant strains in laboratory by the way of drug high-pressure cultivation, and then we analyzed the DNA sequence to detect the possible mechanism of resistance. Through this study, we found that artemisinin-resistant strains evolved more slowly than the piperaquine-resistant strains. More interestingly, the resistance index decreased as the number of breeding generations increased. Moreover, we found that there were 9 meaningful mutations in P. berghei K13 of artemisinin-resistant strains via DNA sequencing, which were different from the clinical mutant points. The points may be related to the multi-drug resistance phenotype of P. berghei K13 variants. Awaiting in-depth study, these meaningful mutations may be used as biomarkers to monitor drug resistance of artemisinin-resistant strains.
Introduction
Since its isolation in the 1970s from the plant Artemisia annua, artemisinin (ART) and artemisinin derivatives have evolved as important components of artemisinin-based combination therapies (ACTs) worldwide.1 ART and artemisinin derivatives are highly effective and act quickly against the blood stages of malaria parasites. The administration of ART and artemisinin derivatives can lead to up to 10,000-fold parasite reductions in the first replication cycle of the Plasmodium.2 Recently, reported reductions in the malaria morbidity and mortality have been partly attributed to the usage of ACTs for malaria treatment and prevention.3 Malaria parasites generally develop resistance to the new antimalarial drugs after 10 to 15 years of usage.4 In 2008–2009, the New England Journal of Medicine published two different articles describing the clinical AR to P. falciparum strains found in Cambodia and Thailand border areas.5,6 These two studies found that there was a markedly decreased sensitivity of P. falciparum to artesunate, parasite clearance time was significantly prolonged, and these changes were not due to pharmacokinetic or patient factors, but rather related to alterations in malarial parasite whole-genome.7
In 2014, Arief et al8 collected and sequenced the whole genome of artemisinin-resistant strains from malaria-endemic areas in Africa and Cambodia. The genetic variation of P. falciparum (Pf3D7_1343700) kelch propeller domain (K13-propeller, amino acid residues 350 to 726) was found to be associated with AR. Phenotypically, AR is defined as a potential delay in the parasite clearance. These AR strains recrudesce more frequently than artemisinin-sensitive parasites after standard 3-day therapeutic courses with ACTs.9 Genetic mutations of K13-propeller domain are associated with delayed parasite clearance in vivo and reduced susceptibility of the ring-stage parasites in vitro in ring-stage survival assays (RSA).6,10 PfK13 regional mutations have been often reported in most areas of South East Asia. The meta-analysis of the WWARN K13 Genotype-phenotype Study Group11 reported that many isolates with the C580Y allele of PfK13 were found at the western border of Thailand, Cambodia, Laos and Vietnam. Moreover, isolates with the Y493H and R539T mutant codons were more commonly found in Cambodia, and those with A578S were more common in Africa. It has been reported that upon being influenced by the drug pressure, life environment, mode of transmission, host body and other factors in the process of its evolution, Plasmodium K13 gene mutates in diverse ways, but the reasons behind this mutation are still unclear. Meanwhile, there are no sufficient evidences indicating that ART might be completely ineffective against artemisinin-resistant strains of P. falciparum, but the reduced sensitivity of P. falciparum to ART could potentially become a huge obstacle in complete eradication of malaria. Hence, it is important to study the mechanism(s) underlying these mutations in detail.
AR in P. falciparum has been associated with PfK13 polymorphisms, and the K13-propeller domain serves as a molecular marker in the surveillance.12 More than 200 nonsynonymous K13 single-nucleotide polymorphisms have been reported previously, including 11 candidate resistance mutations (P441L, G449A, C469F/Y, A481V, R515K, P527H, N537I/D, G538V, V568G, R622I, A675V, associated with delayed parasite clearance) and 10 validated mutations (F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H, P74L, C580Y, reduced in vitro sensitivity).13 Although K13-propeller mutations are highly predictive of resistance, very little is known about the molecular mechanisms that render P. falciparum insensitive to artemisinins. Therefore, it is urgent to further study the mechanisms of resistance of Plasmodium parasites to ART. Meanwhile, ART resistance is well established in Cambodian provinces but which is uncommon and less prevalent. Moreover, P. falciparum cannot be tested in vivo in rodents, and data from rodents do conform to the patterns of many drug resistance that are observed in P. falciparum in nature.14,15 Moreover, P. berghei and P. falciparum K13 structural homology are highly conserved (over 87% sequence identity overall) at DNA sequences. In order to further explore the correlation between K13 gene polymorphism and AR, we obtained artemisinin- and piperaquine-resistant strains in P. berghei through continuous drug high-pressure culture in the laboratory and conducted relevant DNA sequencing. Our results suggested that the mutations at PbK13 can play a causal role in the development of the drug resistance in P. berghei and could serve as the potential target for monitoring and preventing AR in Plasmodium.
Materials and Methods
Ethical Consideration
The experimental protocol for in vivo studies was approved by the Ethics Committee of Science and Technology, Guangzhou University of Chinese Medicine, China (PZ14016). All the procedures related to animals were conducted according to the Regulations of the Experimental Animal Administration, State Committee of Science and Technology of the People’s Republic of China.
Parasite, Animal and Drugs
Parasite strains: We used P. berghei K173, which is a parental, drug-sensitive P. berghei line, developed by the National Institutes of Health (NIH) in 1997. P. berghei K173 was maintained in cryopreserved state in the liquid nitrogen and administered by intraperitoneal injections of infected blood into the mice. P. berghei kelch protein K13 (putative) was downloaded from the European Nucleotide Archive database (coding: CXJ03505).
Among Kunming mice (18–22g) used, half were male and half were female. They were supplied by the Laboratory Animal Center of Guangzhou University of Chinese Medicine or Southern Medical University, Guangdong, China. The license numbers for animal maintenance are SCXK (Yue) 2013–0034 and SCXK (Yue) 2016–0041. The animals were raised in an experimental room at 20–24°C and 70–80% relative humidity and fed on 60 Co irradiated forage and water ad libitum. The mice were infected with parasitized red blood cells (pRBCs) via intraperitoneal (i.p.) injection. Daily monitoring was carried out on infected mice, and infected bloods were transmitted to the receptor mouse.
The frozen tube containing malaria infected blood was removed from the liquid nitrogen and placed in warm water at 40°C until it has completely melted. The pRBCs were injected intraperitoneally into 4~10 mice while maintaining the amount of blood injected into each mouse as constant. Eight days later, parasitemia was assessed in Giemsa-stained thin smears from the tail-vein blood of infected mice. Mice with the highest infection rate were selected as the donor mice. The blood was diluted with physiological saline to contain 1 × 107 pRBCs per 0.2mL of blood. The mice were thereafter infected by intraperitoneal injection of blood containing about 1 × 107 pRBCs.
ART was obtained from Sichuan Tongrentai Pharmaceutical Co., Ltd. (Sichuan, China), Lot No.141101, 160401. Piperaquine (PQ) was obtained from Artepharm Co., Ltd. (Guangdong, China), Lot No. 110804. ART was prepared in a 1:1 mixture of DMSO and Tween 80 (Sigma) and diluted 10-fold in sterile distilled water immediately before administration. PQ was dissolved directly in distilled water. All drugs were prepared fresh before in vivo administration, and drug delivery was carried out by intragastric administration.
Procedures for Exerting Drug-Selection Pressure
The “serial technique” (ST) was used to select resistant line16. Eight Kunming mice were selected as the parent generation and were randomly divided into 2 different groups with 4 mice in each group. The day of P. berghei K173 infection was denoted as D0. The mice were treated by a single dose of ART or PQ on D2 after P. berghei K173 inoculation (The doses used for the drug-selection pressure have been shown in Table 1). From D3 to D7, the parasitemia of each infected mouse was assessed and mice with the highest parasitemia were selected as potential donors of pRBCs for the next generations. Peter’s 4-day suppressive test was performed at every five generations to determine the resistance index. The drug dosage was increased every 5 generations depending on the parasite response.
 |
Table 1 Selection of Artemisinin and Piperaquine Resistance in P. berghei K173 Using the Serial Technique |
Peter’s 4-Day Suppressive Test
Mice were infected intraperitoneally with 1 × 107 pRBCs, and drug was administered at 2h, 24h, 48h, and 72h, respectively, postinfection.17 Parasitemias were assessed by the microscopic examination of Giemsa-stained slides from tail vein blood collected on day 4 post-infection, in order to manually determine the levels of infection.
Assessment of Resistance
Blood was obtained by trimming the tip of the tail and smeared on a microscope slide (thin blood smear) and then fixed with absolute methanol for 10s. After fixation, the slides were dried and stained with 10% Giemsa stain for 15 min, rinsed with running water, and dried at room temperature. The parasite-infected red blood cells were quantified using a light microscopy (CX41RF, OLYMPUS, Corporation, TOKYO, Japan). The blood films which did not visualize protozoa in 50 microscope fields were considered as negative, and the infection rate was found to be 0. The Peters 4-day suppression test was used to measure the resistance index once every five generations.17 Then, the “index of resistance”, I90 (defined as the ratio of the ED90 of the resistant line to that of the sensitive parent line), was calculated. The degree of resistance was grouped into four levels by the values of I90 as previously reported:18–20 (1) I90≦1.0, sensitive, (2) I90=1.01–10.0, slightly resistance, (3) I90=10.01–100.0, moderately resistant and (4) I90>100.0, strongly resistant. The formula for I90 used was as follows: 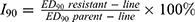 .
.
Blood Sample Collection
In the process of resistance cultivation, after each generation of resistance cultivation, the blood samples were respectively harvested by heparinized capillary tubes from the mice orbitals after the mice were narcotized using diethyl ether. About 700 μL of the blood samples were obtained from each mouse in the anticoagulant collection vessels of Ethylenediamine Tetraacetic Acid (EDTA). The resistance cultivation algebra was then labeled. The plasma was kept at −80°C before use.
DNA Extraction
Genomic DNA of parent strains (sensitive P. berghei K173) and drug-resistant strains (ART- and PQ-resistant strains of the 30th and 50th generation) was extracted from 100 μL of each whole blood sample using DNA blood kit according to the manufacturer’s protocol (Takara Bio Inc. Nojihigashi 7-4-38, Kusatsu, Shiga 525–0058, Japan, Lot No.: AK1901). The extracted DNA was eluted in 60 μL of TE buffer (10 mM Tris–HCl, 0.1 M EDTA, pH 8.0) and stored at −20°C until further use. The quality of genomic DNA was detected using 1.0% agarose gel electrophoresis. The gel was stained with Goldview nucleic acid stain used as an ethidium bromide substitute (Sangon Bio Inc., Shanghai, China).
PCR Amplification and Sequencing of P. berghei K13-Propeller
Kelch13 fragments of P. berghei sensitive and resistant strains were amplified and were identified by agarose gel electrophoresis. PCR amplification products were sent to Shenzhen BGI Technology Co., Ltd. (Guangdong, China) for sequencing of P. berghei K13 fragments. The sequencing was completed by using three reactions. DNASTAR and DNAMAN9.0 were used to compare the above sequences with ENA CXJ03505. The primers were designed for P. berghei K13, 2126752 to 2128968 base pairs on chromosome 13 (GenBank: LT160033.1, locus_tag=“PBK173_000418100”). PCR primer:21 Upstream primer sequence: 5’-AGTCAAACAGTATCTCTAACT-3’, downstream primer sequence: 5’-ACGGAATGTCCAAATCTTG-3’. The PCR amplification reaction of components was thereafter combined in a master mix composed of TAKARA Master Mixture (5U/μL), 25μL; PCR Forward Primer (100μM), 2μL; PCR Reverse Primer (100μM), 2μL; Template DNA (20~50ng/μL), 2μL and RNAse Free dH2O used to reach a total reaction volume of 50μL. The cycling program was set at 1 cycle of predenaturation at 95°C for 5 min, denaturation at 94°C for 1 min, annealing process at 51°C for 30s, and extension at 72°C for 1.5 min, followed by 2~4 cycle for 30 times, and finally repair extension at 62°C for 10 min.
Quantification and Statistical Analyses
All the data were statistically analyzed using SPSS (IBM SPSS Statistics 20.0). Statistical analysis was performed using one-way analysis of variance (ANOVA). A significance level of P<0.05 was required for all tests and considered as statistically significant.
Results
P. berghei Did Not Develop Severe Resistance to ART
Artemisinin- or piperaquine-resistance P. berghei K173 strain was obtained by a serial passaging of the sensitive malaria parasites under incremental ART or PQ dosages for 50 generations in our laboratory. The resistance index of PQ was determined to be 148.8 (I90>100, Table 1) in our laboratory, which increased rapidly and indicated severity of infection. Interestingly, the resistance index of ART increased from the 5th generation to 30th, which indicated a moderate resistance in the 30th generation (I90=12.4, 10≤I90<100, Table 1). However, in subsequent 31–50 generations, although the dosage of artemisinin was increased by up to 53.95% (Table 1), the resistance index of ART decreased significantly to less than 10, showing mild resistance (I90=5.0, I90<10, Table 1). From 21 to 50 generations of resistant culture, the difference between artemisinin- and piperaquine-resistant strain I90 was statistically significant (P<0.05, Figure 1). In conclusion, P. berghei K173 strain did not effectively develop severe resistance to artemisinin under continuous drug pressure, which may be related to the special mechanism of artemisinin. Artemisinin has been used clinically for several decades. Although resistance has been reported (prolonged protozoa clearance time), there has not been a large-scale spread of severe resistance, thereby proving that potential application of artemisinin is more advantageous and irreplaceable than other antimalarial drugs.
Variation at the K13 Locus of P. falciparum or P. berghei Can Be Used as a Possible Biomarker to Predict the Resistance Development
According to the primers, a total of 2179 bp fragments from position 20 to position 2198 were amplified. After sequencing, the splicing sequences were compared in NCBI-BLAST (Table 2), which confirmed that the measured sequence was the K173 KELCH13 fragment of P. berghei. We next downloaded the K173 KELCH13 gene sequence CXJ03505 from the European Nucleotide Archive (ENA). DNAMAN9.0 was used to compare the gene sequences with K173 sensitive strains, ART-resistant strains of the 30th generation (A30), ART-resistant strains of the 50th generation (A50), PQ-resistant strains of the 30th generation (P30) and PQ-resistant strains of the 50th generation (P50), and the changed locations were marked blue (Supplementary Figure 1). It could be seen that the nucleotide sequence of P. berghei resistant strains (A30, A50, P30 and P50) has many identical changes compared with CXJ03505 (Table 3), while the similarity between the sensitive strains (P. berghei K173) and CXJ03505 was up to 99.95%.
 |
Table 2 Comparison of Each Sequence with K13 Gene Sequence of P. berghei |
 |
Table 3 Potential Changes of P. berghei K13 Caused by Resistance Breeding |
It was found that compared with the sensitive strains, resistant strains had 9 missense mutations, such as A128T, A189G, A311G, G361A, A368G, G398A, A464G, A531T and C1643T. The corresponding amino acid changes were Y43F, I63M, N104S, A121T, N123S, S133N, N155S, E177D and S548L, and there were 29 silent mutations. However, PfK13 mutation sites, such as C458Y, R539T, Y493H, M476I, which have been closely related to artemisinin-resistant strains of P. falciparum in South East Asia, were not detected. It was observed that the variation of P. berghei K13 genes under the drug pressure was diverse due to the influence of environment, transmission route, host and other factors. The P. berghei K13 variation detected in this study needs to be further verified.
Mutations at P. berghei K13 occurred in both artemisinin-resistant and piperaquine-resistant strains of P. berghei. In A30 and A50 strains, although the I90 resistance decreased from moderate to mild, there were no observed differences between them. We conclude that the detection of P. berghei K13 variants can be used as the biomarkers to predict the development of malaria resistance but could not be employed to quantify the severity of resistance.
Resistant Strains in P. berghei Display Multidrug Resistance Phenotypes
We complied both artemisinin- and piperaquine-resistance P. berghei, and the P. berghei K13 fragment of the two drug-resistant strains showed almost identical mutations. This finding clearly indicated that Plasmodium resistant strains have a multidrug resistance phenotype, making Plasmodium resistant to either artemisinin or artemisinin combination. Moreover, sequencing results also revealed that the 1643th nucleotide change is C (cytosine) into T (thymine), which can cause the 548th in the amino acid sequence generated by the S (ser), the change of L (leucine) (Supplementary Figure 2), the specific variation may be in P. berghei K13 protein six propellers area, or directly related to artemisinin resistance. The multidrug resistance phenotype of Plasmodium directly leads to the inevitable resistance of all the antimalarial drugs after long-term chronic use. It has been established that different drugs have different antimalarial mechanisms, so the time and severity of the resistance are also different. Artemisinin is activated mainly by the degradation products of hemoglobin in Plasmodium, thus producing Reactive Oxygen Species (ROS) to kill Plasmodium. Although there is evidence that PfK13 can significantly attenuate the sensitivity of Plasmodium to artemisinin by functionally reducing its endocytosis of hemoglobin22 (Figure 2), hemoglobin is still the main raw material required for the maintenance of growth and development of Plasmodium, and artemisinin can act through modulating multiple binding location or pathways. Activated artemisinins can effectively form adducts with a variety of biological macromolecules, including haem, translationally controlled tumor protein (TCTP), P. falciparum phosphatidylinositol-3-kinase (PfPI3K), inhibiting a calcium ATPase (PfATP6) and other high-molecular-weight proteins. Moreover, Bing Zhou and his team23,24 have reported that artemisinin can also cause mitochondrial depolarization, and mitochondria can serve as potential targets of both artemisinin and activators of artemisinin. This may be an important reason why malaria parasites have not yet been developed widespread and severe resistance to artemisinin. The problem is the opportunity as the multi-resistant phenotype of Plasmodium can act as both the cause of multi-resistant Plasmodium and the potential target for the future treatment of Plasmodium resistance.
Discussion
Our study showed that compared with PQ, ART was less prone to severe drug resistance. We found that P. berghei K13 gene mutation could effectively predict the development of drug resistance in Plasmodium. Furthermore, we also found that P. berghei K13 gene mutation could predict the emergence of resistance to Plasmodium. During the breeding of P. berghei 1–50 generations in the laboratory, we noticed that ART did not show severe resistance, and its I90 was maintained at around 5 after 30 generations. The subsequent DNA sequencing of the parent strains (P. berghei K173) and drug-resistant strains (A30, A50, P30 and P50) almost displayed the same mutations, while the parent strains did not exhibit the same mutations. Overall, combined with the existing literature, our study provided biological support for the hypothesis that P. berghei K13 site mutation can lead to Plasmodium resistance.
We successfully cultivated the P. berghei K173ʹs resistant strains against ART and PQ in our laboratory. Artemisinin-resistant strains hardly showed severe resistance and their resistance was unstable. One study found that artemisinin-resistant P. falciparum isolates displayed a loss of health, the drug-resistant strains could not undergo the transition from ring forms to trophozoites as well as schizonts, and fewer offspring were found in red blood cells during development cycle.25 In case of an insufficient supply of exogenous amino acids, the loss of adaptability of the resistant strains will increase significantly. The drug-resistant parasites are immature, so the anti-artemisinin phenotype is often unstable. Repeated Plasmodium resistance was first reported by Elfawal et al in their study on artemisinin-resistant strains of P. yoelii.26 Their study also found that ART was less prone to severe drug resistance.24 Moreover, when drug dosage was increased, the antimalarial effect of ART could be temporarily restored.27 The above findings indicated that in areas where artemisinin-resistant strains were endemic, increasing the amount of artemisinin in ACTs or prolonging the treatment course of ACTs can be applied as an important strategy to improve the clinical efficacy of ACTs but not as a long-term solution.
The P. berghei K13 gene mutation of the artemisinin-resistant strain cultivated in our laboratory was different from the PfK13 mutation point of the artemisinin-resistant strain of P. falciparum detected in South East Asia. This may be due to the difference of the life cycle of the two malaria parasites and the difference of the host bodies. In the existing literature, PfK13 mutation of artemisinin-resistant strains in South East Asia mostly occurred in C580Y, E25Q, Y493H, R539T;28 while in Africa,29 PfK13 mutation primarily occurred in A578S, T549C, and G553A. One study in Uganda30 monitored P. falciparum resistance in 3 different local villages and found that the K13 mutations were closely associated with artemisinin resistance of Plasmodium in the target areas were not common and did not increase over time. Another study conducted in Equatorial Guinea (Ngonamanga and Miyobo)31 sequenced the K13-propeller domain of P. falciparum in two different villages in 2005 and 2013, respectively, and found that the mutations in the K13 region in 2005 and 2013 were not consistent. It can be observed that the mutation of PfK13 was inherently polymorphic. Many studies have confirmed that the artemisinin resistance of the malaria parasite could be directly related to the mutation of the K13 gene, thus regular detection of PfK13 gene sequence mutations in malaria-endemic areas can potentially serve as an effective means to predict the occurrence of malaria parasite resistance, and adjust malaria treatment plans in a timely manner. Although there are no reports of large-scale transmission of resistant strains of P. falciparum in Asia and Africa recently, there are evidences to suggest that Anopheles gambiae can be used as the main vector for the transmission of the resistant strains of Plasmodium in Asia and Africa.32 Therefore, monitoring the drug susceptibility of the malaria parasite strains and controlling the population movement in malaria-endemic areas could be two important means to prevent and control the spread of malaria-resistant strains on a global scale.
Mutations in PfK13 can induce it to express a multi-drug resistance phenotype. The experiment of Paloque L at al.33 showed that PfK13 mutation can cause cross-resistance between artemisinin and endoperoxide-based antimalarials. The highly homologous variation on artemisinin- and piperaquine-resistant strain P. berghei K13 in this experiment also illustrated this point. Mbengue A et al34 proved that in the resistant clinical strains, increased PfPI3K expression was related to the C580Y mutation of P. falciparum Kelch13. It is currently known that PfK13 mutations can increase phosphatidylinositol 3-phosphate (PI3P) tubules/vesicles, and elevated PI3P can functionally induce the resistant parasite unfolded protein response (UPR) of P. falciparum, thereby neutralizing the Plasmodium protein affected by the toxicity of artemisinin and thereby reduce the sensitivity of resistant strains to artemisinin.35 In addition, PfK13 mutation can also induce Plasmodium to reduce the endocytosis of the hemoglobin,27,36 thereby reducing the activation of artemisinin. It should be noted that in the previous studies on Plasmodium resistance genes, some other locus mutations have also been confirmed that can lead to resistance. Pholwat S et al37 used TaqMan array cards to test the samples from 18 different laboratory parasite strains and 87 clinically resistant strains. It was found that except for the samples from Malawi, most of the clinical samples showed mutations in pfcrt. All the samples showed pfdhfr and pfdhps mutations. The number and types of pfmdr1 and PfK13 mutations varied based on the country of origin, and pfmdr means multi-drug resistance. The Nag S team38 performed high-throughput sequencing using Illumina NGS (Miseq) platform on the samples of 457 patients from the Bandim and Belém Health Care Centers in Guinea-Bissau. The results showed that various polymorphisms can be found in pfcrt, pfmdr1, pfdhfr, pfdhps, and PfK13. Plasmodium resistance has been found to be often accompanied by multi-site mutations. Therefore, multi-resistant phenotype can also arise because of the synergistic effects of multi-site mutations.
The study may be useful to explore the drug resistance mechanism of P. falciparum, but which also has several limitations. Firstly, in vivo malaria models have their own constraints. We used mouse tail vein blood with high malaria infection rate to dilute and vaccinate the healthy mice. This method is different from the clinical transmission of malaria caused through the bite of female Anopheles mosquitoes. Therefore, mice malaria models cannot completely include all the distinct characteristics of human malaria, which might be one of the reasons why the variation of resistant strains found in this study was significantly different from the clinical ones. Secondly, the sample size for DNA sequencing in this study was limited. We have reported 9 different missense mutations in the resistant strains of Plasmodium, which need to be verified by further experiments.
In the future, we will cultivate Pf3D7 and P. berghei K173 strains in vitro, with successional drug-selection pressure forming AR strains, followed by sequencing. At the same time, gene editing technology will use to knock out specific mutation sites in the various susceptible strains, and thus it can be verified that whether this strategy can be effectively used to prolong the clearance time and early relapse of Plasmodium.
Conclusions
In response to the artemisinin challenge, the slow clearance phenotype of P. falciparum parasite populations can at least involve one non-synonymous change in the P. falciparum-specific single mutations in the propeller region of the PfK13 gene. In order to further define the other genomic changes in the parasite populations that can support PfK13 mutants to potentially overcome the fitness deficits, genomic studies needed to validate whether these different propeller mutations can compromise the parasite fitness.
Our study demonstrated that 6 missense mutations and 29 silent mutations of P. berghei K13 propeller mutants could be considered molecular markers associated with prolonged parasite clearance. With its complexity of the phenotype, mutations in the propeller region of the PfK13 gene were served as valuable markers for the surveillance of artemisinin responsiveness diminishing in the parasite populations. At the same time, the P. berghei K13 fragment of artemisinin- and piperaquine-resistant strains showed almost similar mutations in the study. This finding is sufficient to indicate that Plasmodium drug-resistant strains have a multidrug resistance phenotype, thereby attributing Plasmodium resistance to either artemisinin or artemisinin combination. Meanwhile, these P. berghei K13 mutant parasites from artemisinin-resistance P. berghei rodent model of malaria also provide evidence of their important role in mediating artemisinin resistance in vivo, which further supports observations related to in vitro artemisinin resistance. These obtained findings should be considered as a preliminary study and might provide a new insight into the drug resistance mechanisms against ARTs.
Abbreviations
ACTs, Artemisinin-based combination therapies; AR, Artemisinin resistance; RSA, Ring stage survival assays; NIH, National Institutes of Health; pRBCs, parasitized red blood cells; ART, Artemisinin; PQ, Piperaquine; DMSO, Dimethyl sulfoxide; ST, Serial technique; ED90, 90% effective dose; EDTA, Ethylenediamine Tetraacetic Acid; A30, Artemisinin-resistant strains of the 30th generation; A50, Artemisinin-resistant strains of the 50th generation; P30, Piperaquine-resistant strains of the 30th generation; P50, Piperaquine-resistant strains of the 50th generation; ROS, Reactive Oxygen Species; TCTP, Translationally controlled tumor protein; PfPI3K, Plasmodium falciparum phosphatidylinositol-3-kinase; PfATP6, Plasmodium falciparum calcium ATPase 6; PfK13, P. falciparum Kelch 13; PbK13, P. berghei Kelch 13; UPR, Unfolded protein response; ANOVA, One-way analysis of variance.
Data Sharing Statement
Data in tables used to support the findings of this study are included within the article. The sequences have been deposited in Supplementary Figures 1 and 2.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from Natural Science Foundation of China [Grant Number 82074301,81403295 and 81873218] to CD and SJ, and Project of Traditional Chinese Medicine Bureau of Guangdong [Grant Number 20201084] to SZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. World Health Organization. Guidelines for the Treatment of Malaria. World Health Organization; 2010.
2. White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–334. doi:10.1126/science.1155165
3. World Health Organization. World Malaria Report 2020. Geneva: World Health Organization; 2020.
4. Ariey F, Witkowski B, Amaratunga C, et al. molecular marker of artemisinin -resistant Plasmodium falciparum malaria. Nature. 2014;2:e12876.
5. Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin resistant malaria in western Cambodia. N Eng J Med. 2008;359:2619–2620. doi:10.1056/NEJMc0805011
6. Arjen M. Dondorp, François Nosten, Poravuth Yi, Debashish Das, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Eng J Med. 2009;361:455–467. doi:10.1056/NEJMoa0808859
7. Anderson TJ, Nair S, Nkhoma S, et al. High Heritability of Malaria Parasite Clearance Rate Indicates a Genetic Basis for Artemisinin Resistance in Western Cambodia. Brief Report. 2010;201(1):1326–1330.
8. Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505(7481):50–55. doi:10.1038/nature12876
9. Wang J, Xu C, Liao FL, et al. Solution to “Artemisinin Resistance”. N Engl J Med. 2019;380(22):2087–2089. doi:10.1056/NEJMp1901233
10. Witkowski B, Amaratunga C, Khim N, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13(12):1043–1049. doi:10.1016/S1473-3099(13)70252-4
11. WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments a WWARN individual patient data meta-analysis. BMC Med. 2019;17(1):548. doi:10.1186/s12916-018-1207-3
12. Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55.
13. World Health Organisation. Global Malaria Programme. Status Report on Artemisinin Resistance and Artemisinin-Based Combination Therapy Efficacy. Geneva: The Organisation; 2018.
14. Peters W. The chemotherapy of rodent malaria. LVII. Drug combinations to impede the selection of drug resistance, part 1: which model is appropriate? Ann Trop Med Parasitol. 1999;93(6):569–587. doi:10.1080/00034983.1999.11813461
15. Wykes MN, Michael F. Infectious disease: malaria What have we learnt from mouse models for the study of malaria? Eur J Immunol. 2009;39:1991–2058. doi:10.1002/eji.200939552
16. Kiboi DM, Irungu BN, Langat B, et al. Plasmodium berghei ANKA: selection of resistance to piperaquine and lumefantrine in a mouse model. Exp Parasitol. 2009;122:196–202. doi:10.1016/j.exppara.2009.03.010
17. Peters W. Chemotherapy and Drug Resistance in Malaria. New York, NY: Academic Press; 1970.
18. Merkli B, Richie R, Peters W. The inhibitory effect of a drug combination on the development of mefloquine resistance in Plasmodium berghei. Ann trop Med Parasit. 1980;74:1–9. doi:10.1080/00034983.1980.11687304
19. Peters W. Drug resistance in Plasmodium berghei, Vincke and Lips. 1948.1. Chloroquine resistance. Exp Parasit. 1965;17:80–89. doi:10.1016/0014-4894(65)90012-3
20. Peters W. Drug resistance in Plasmodium berghei, Vincke and Lips. 1948.1. Chloroquine resistance. Exp Parasit. 1965;17:80–89.
21. Ndung’u L, Langat B, Magiri E, et al. Amodiaquine resistance in Plasmodium berghei is associated with PbCRT His95Pro mutation, loss of chloroquine, artemisinin and primaquine sensitivity, and high transcript levels of key transporters. Wellcome Open Res. 2017;2:44. doi:10.12688/wellcomeopenres.11768.1
22. Birnbaum J, Scharf S, Schmidt S, et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367(6473):51–59. doi:10.1126/science.aax4735
23. Wang J. Artemisinin Directly Targets Malarial Mitochondria through Its Specific Mitochondrial Activation. PLoS One. 2010;5:12.
24. Yeast Model LW. Uncovers Dual Roles of Mitochondria in the Action of Artemisinin. PLo S Genet. 2005;1:e36. doi:10.1371/journal.pgen.0010036
25. Bunditvorapoom D, Kochakarn T, Kotanan N, et al. Fitness Loss under Amino Acid Starvation in Artemisinin-Resistant Plasmodium falciparum Isolates from Cambodia. Sci Rep. 2018;8(1):12622. doi:10.1038/s41598-018-30593-5
26. Elfawal MA, Towler MJ, Reich NG, Weathers PJ, Rich SM. Dried whole-plant Artemisia annua slows evolution of malaria drug resistance and overcomes resistance to artemisinin. Proc Natl Acad Sci U S A. 2015;112(3):821–826. doi:10.1073/pnas.1413127112
27. De Lucia S, Tsamesidis I, Pau MC, Kesely KR, Pantaleo A, Turrini F. Induction of high tolerance to artemisinin by sub-lethal administration: a new in vitro model of P. falciparum. PLoS One. 2018;13(1):e0191084. doi:10.1371/journal.pone.0191084
28. Cerqueira GC, Cheeseman IH, Schaffner SF, et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18(1):78. doi:10.1186/s13059-017-1204-4
29. Nguetse CN, Adegnika AA, Agbenyega T, et al. Molecular markers of anti-malarial drug resistance in Central, West and East African children with severe malaria. Malar J. 2017;16(1):217. doi:10.1186/s12936-017-1868-y
30. Tumwebaze P, Tukwasibwe S, Taylor A, et al. Changing Antimalarial Drug Resistance Patterns Identified by Surveillance at Three Sites in Uganda. J Infect Dis. 2017;215(4):631–635. doi:10.1093/infdis/jiw614
31. Guerra M, Neres R, Salgueiro P, et al. Plasmodium falciparum Genetic Diversity in Continental Equatorial Guinea before and after Introduction of Artemisinin-Based Combination Therapy. Antimicrob Agents Chemother. 2016;61(1):e02556–15. doi:10.1128/AAC.02556-15
32. St Laurent B, Miller B, Burton TA, et al. Artemisinin-resistant Plasmodium falciparum clinical isolates can infect diverse mosquito vectors of Southeast Asia and Africa. Nat Commun. 2015;6:8614. doi:10.1038/ncomms9614
33. Paloque L, Witkowski B, Lelièvre J, et al. Endoperoxide-based compounds: cross-resistance with artemisinins and selection of a Plasmodium falciparum lineage with a K13 non-synonymous polymorphism. J Antimicrob Chemother. 2018;73(2):395–403. doi:10.1093/jac/dkx412
34. Mbengue A, Bhattacharjee S, Pandharkar T, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520(7549):683–687. doi:10.1038/nature14412
35. Bhattacharjee S, Coppens I, Mbengue A, et al. Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood. 2018;131(11):1234–1247. doi:10.1182/blood-2017-11-814665
36. Birnbaum J, Scharf S, Schmidt S, et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367(6473):51–59.
37. Pholwat S, Liu J, Stroup S, et al. The Malaria TaqMan Array Card Includes 87 Assays for Plasmodium falciparum Drug Resistance, Identification of Species, and Genotyping in a Single Reaction. Antimicrob Agents Chemother. 2017;61(5):e00110–17. doi:10.1128/AAC.00110-17
38. Nag S, Dalgaard MD, Kofoed PE, et al. High throughput resistance profiling of Plasmodium falciparum infections based on custom dual indexing and Illumina next generation sequencing -technology. Sci Rep. 2017;7(1):2398. doi:10.1038/s41598-017-02724-x
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.