Back to Journals » Infection and Drug Resistance » Volume 13
Plasmid-Mediated AmpC β-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates
Authors Aryal SC , Upreti MK, Sah AK , Ansari M , Nepal K, Dhungel B , Adhikari N , Lekhak B, Rijal KR
Received 30 September 2020
Accepted for publication 6 November 2020
Published 24 November 2020 Volume 2020:13 Pages 4249—4261
DOI https://doi.org/10.2147/IDR.S284751
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Subhas Chandra Aryal,1,* Milan Kumar Upreti,1,* Anil Kumar Sah,2 Meharaj Ansari,3 Krishus Nepal,1 Binod Dhungel,4 Nabaraj Adhikari,4 Binod Lekhak,1,4 Komal Raj Rijal4
1Golden Gate International College, Kathmandu, Nepal; 2Annapurna Neurological Institute and Allied Sciences, Kathmandu, Nepal; 3Shi-Gan Int’l College of Science and Technology (SICOST), Kathmandu, Nepal; 4Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu, Nepal
*These authors contributed equally to this work
Correspondence: Komal Raj Rijal
Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu Email [email protected]
Background: Antibiotic resistance mediated by the production of extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases is posing a serious threat in the management of the infections caused by Gram-negative pathogens. The aim of this study was to determine the prevalence of two AmpC β-lactamases genes, blaCITM and blaDHAM, in Gram-negative bacterial isolates.
Materials and Methods: A total of 1151 clinical samples were obtained and processed at the microbiology laboratory of Annapurna Neurological Institute and Allied Science, Kathmandu between June 2017 and January 2018. Gram-negative isolates thus obtained were tested for antimicrobial susceptibility testing (AST) using Kirby–Bauer disk diffusion method. AmpC β-lactamase production was detected by disk approximation method using phenylboronic acid (PBA). Confirmed AmpC β-lactamase producers were further screened for blaCITM and blaDHAM genes by conventional polymerase chain reaction (PCR).
Results: Out of 1151 clinical specimens, 22% (253/1152) had bacterial growth. Of the total isolates, 89.3% (226/253) were Gram-negatives, with E. coli as the most predominant species (n=72) followed by Pseudomonas aeruginosa (n=41). In the AST, 46.9% (106/226) of the Gram-negative isolates were multidrug resistant (MDR). In disk diffusion test, 113 (50%) isolates showed resistance against cefoxitin, among which 91 isolates (83 by disk test and Boronic acid test, 8 by Boronic test only) were confirmed as AmpC β-lactamase-producers. In PCR assay, 90.1% (82/91) and 87.9% (80/91) of the isolates tested positive for production of blaCITM and blaDHAM genes, respectively.
Conclusions: High prevalence of AmpC β-lactamase-producers in our study is an alarming sign. This study recommends the use of modern diagnostic facilities in the clinical settings for early detection and management which can optimize the treatment therapies, curb the growth and spread of the drug-resistant pathogens.
Keywords: ESBLs, AmpC β-lactamase, blaCITM, blaDHAM, MDR, polymerase chain reaction
Background
Drug resistance among some of the Gram-negative bacteria (Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa) has emerged and spread worldwide. Among Enterobacterales family, Escherichia coli and Klebsiella spp. are frequently isolated organisms that have notable drug-resistance properties. β-lactams are the commonly prescribed drugs against these recalcitrant strains, and alone constitute about two-thirds of recent clinical prescriptions.1 This group contains four major chemical classes: penicillins, cephalosporins, carbapenems and monobactams, in which carbapenems are used as the last resort drugs in the empirical therapy against pathogenic strains of Gram-positive, Gram-negative and anaerobic bacteria.2
Resistance to β -lactams is attributable to the production of β-lactamases, those hydrolytic enzymes which are able to inactivate the antibiotics before they reach penicillin binding proteins (PBPs) located at the cytoplasmic membrane.3 Major β-lactamase families include plasmid-mediated extended-spectrum β-lactamases (ESBLs), AmpC, cephalosporinases, and carbapenemase. All classes have been detected globally with few of them concentrated in specific countries.1
The genes encoding AmpC β-lactamase have spread extensively and are widely detected in bacterial plasmids. The first plasmid-encoded AmpC variant was first identified in 1989 from Klebsiella pneumoniae isolated in South Korea. It was named CMY-1 because of its phenotypic trait associated with cephamycinase and was notoriously resistant to cefoxitin.4,5 In a short span of time many families of plasmid-mediated AmpC variants were detected, predominantly from the isolates of K. pneumoniae and E. coli. Bacteria possessing plasmid-mediated AmpC genotypes were attributed on the basis of homology in nucleic acid sequence, forming a larger number of bacterial genera which acted as a source of these plasmids and a number of AmpC families.6 To date, following AmpC families have been reported globally: two families of CMY β-lactamases (CMY-1 and CMY-2) isolated from Aeromonas hydrophila and Citrobacter freundii, respectively; FOX- type and MOX-type enzymes isolated from Aeromonas spp.; the ACC family derived from H. alvei; the LAT family of cephalosporinases isolated from C. freundii; the MIR and ACT families originated from Enterobacter spp.; and the DHA family isolated from Morganella morganii.4,6,7 The majority of plasmid-encoded AmpC genes are found in isolates of E. coli and K. pneumoniae in nosocomial infections whereas resistance among other Gram-negative bacteria such as E. cloacae, C. freundii, S. marcescens, and M. morganii are conferred by chromosome-mediated AmpC β-lactamases, thus enhancing resistance to broad spectrum cephalosporins.8 The organisms producing ESBLs often characterized by co-expression with AmpC β-lactamases are posing a serious threat in the diagnosis and treatment of the pathogens.
Moreover, AmpC β-lactamase genes, especially MOXM, CITM, DHAM, EBCM, FOXM and ACCM are responsible for the development of broad-spectrum resistance to most of the β-lactams (other than cefepime and carbapenems). In addition, acquisition of these genes in the bacteria can further enhance resistance because inhibitors of class A enzymes (including clavulanic acid, sulbactam, and tazobactam) and p-chloromercuribenzoate are not effective against AmpC β-lactamases, although some of them can be inhibited by tazobactam or sulbactam.6
Although efforts are made to curb the growth and spread of drug-resistant pathogens, the studies on AmpC β-lactamases in resource-limited settings are still inadequate. The most important step in coping with increasing antimicrobial resistance (AMR) is the precise detection of pathogenic (and/or resistant) strains in diagnostic laboratories. Nonetheless, the lab protocols for routine reporting fail to offer the precise result in Nepal because AmpC β-lactamases are difficult to identify by phenotypic tests alone and are often falsely detected as ESBLs in clinical laboratories. Enterobacterales isolates which are positive in screening test of ESBL phenotype but negative on confirmatory assay are usually considered as potential AmpC β-lactamases-producers either conferred by chromosomal depression or plasmid transfer.9
Even the expert clinical microbiologists fail to identify plasmid-mediated AmpC β-lactamase thereby suggesting a need for more precise and specific method for the prompt detection. However, some of these procedures are resource intensive, often requiring reagents that are not easily available.10 For these reasons, AmpC β-lactamases go undetected in clinical specimens. Thus, a more reliable and valid lab protocol consisting multiplex polymerase chain reaction (PCR) has been devised to facilitate the diagnosis of plasmid-encoded AmpC genes which is responsible for AmpC β-lactamase expression in various clinical infections.11
Most of the studies are focused on the prevalence of ESBL enzymes in Gram-negative clinical isolates in Nepal.12–16 There are limited number of studies on AmpC β-lactamases enzymes in Gram-negative bacteria in Nepal. One study conducted in Kathmandu valley reported 27.8% of Enterobacterales isolates as AmpC β-lactamases producers using phenotypic method.17 To our knowledge, there are limited studies related to detection and characterization of AmpC β-lactamases enzymes in Gram-negative bacteria using both phenotypic and genotypic methods in Nepal. It is essential to set up standard phenotypic and genotypic methods on antibiotic susceptibility test to screen the drug-resistant pathogens in hospitals. This study was undertaken to isolate and identify the AmpC β-lactamases genes (blaCITM and blaDHAM) in Gram-negative bacterial isolates using both phenotypic screening and confirmatory methods.
Methods
Study Design
This is a hospital based cross-sectional study carried out from June 2017 to January 2018 at Annapurna Neurological Institute and Allied Sciences (ANIAS). A total of 1151 non-duplicated clinical samples were collected and were processed during the study period. The samples included urine (n=412), blood (n=206), catheter tip (n=163), pus (n=132), stool (n=89), CSF (n=53), wound swab (n=58), and vaginal swab (n=38). The specimens were obtained in a sterile, well-labeled, and leak-proof container; and processed as soon as possible.18 All the visiting patients suspected of having bacterial infections who provided their consent to be involved were included in the study. Study participants with inadequate demographic information were excluded.
Culture of Specimens and Identification of the Isolates
Samples were collected according to standard microbiological guidelines for the collection of urine, stool, pus, blood, CSF and catheter tip. Eligible samples were further inoculated on blood agar (BA) Chocolate agar (CA) and MacConkey agar (MA). In addition, urine samples were inoculated in Chromogenic UTI agar. Isolates were identified using standard microbiological parameters such as morphological appearances of the colonies, staining reactions and biochemical properties.19,20
Antibiotic Susceptibility Testing
All identified Gram-negative isolates further underwent in-vitro antimicrobial susceptibility test using modified Kirby–Bauer disk diffusion method.21 Firstly, inoculums were made by transferring bacterial colonies from nutrient agar to sterile normal saline. The turbidity of the inoculums was set in equivalence to 0.5 McFarland standard as outlined by CLSI guidelines. The carpet culture of the test inoculums was also prepared on Muller-Hinton agar (MHA). Antibiotic disks (HiMedia India Pvt. Ltd, Bengaluru, India) were used in the following proportions: amoxicillin (10 μg), azithromycin (10 μg), amikacin (30 μg), aztreonam (30 μg), cefoxitin (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), piperacillin/tazobactam (100/10 μg), ertapenem (10 μg), meropenem (10 μg), cotrimoxazole (25 μg), and cefepime (30 μg). The inoculated plate was aerobically incubated at 37°C up to 18 hours. After sufficient incubation, the zones of inhibition (ZOI) around the disks were measured and the result was classified as sensitive, intermediate, or resistant.21 Isolates exhibiting resistance to three or more antibiotics of different classes were interpreted as multidrug resistant.22
Screening for AmpC β-Lactamases
The isolates were firstly screened for possible AmpC β-lactamases production according to CLSI guidelines, 2019.23 For screening, ceftazidime or cefotaxime or cefoxitin or ceftriaxone, each of 30 µg were subjected in AST testing and organisms resistant to those antibiotics (showing zone of inhibition of diameter ≤ 18mm) were screened as potential AmpC producers and underwent further confirmatory tests.
Confirmation for AmpC β-Lactamases
Screening positive AmpC β-lactamases producers were confirmed by AmpC disk test and inhibitor-based confirmatory test (Boronic acid test).
In boronic acid test, the carpet culture of the test organism was made on MHA plate taking 0.5 McFarland solutions. Two cefoxitin (30µg) disks, one of which was added with phenyl boric acid (400 µg) were placed on carpet culture on MHA plate and incubated and the results were interpreted. If there was an increase of inhibition zone by ≥5 mm when the desired antibiotic disk (ceftazidime or cefotaxime) was assessed in combination with phenylboronic acid than during the assay without the combination (antibiotic disk alone), the isolate was marked as having positive confirmatory test.
In disk approximation test, disks were put over the carpet culture of E. coli ATCC 25922. 30μg ceftazidime disk was placed at the center of the plate followed by 10μg imipenem, 30μg cefoxitin, and 20/10μg amoxicillin/clavulanate disks which were put at a distance of 20mm from the ceftazidime disk. The colonies of test organism were added to a disk and then the plate was inverted and incubated for 24 hours at 37°C aerobically. Any indentation or flattening of the ZOI indicated the production of AmpC β-lactamase by isolates.20,24
Preservation of AmpC β-Lactamase Positive Isolates
Glycerol stock preparation method was used for preservation. Organisms were preserved in Tryptic Soy Broth (TSB) containing 40% glycerol and stored at −20ºC.25
Crude Plasmid DNA Extraction
An isolated colony of the test organism was inoculated in Luria Bertani (LB) broth. The inoculum was incubated aerobically using a water bath shaker at 37°C for 18–24 hours. Thus obtained pure culture was subjected under alkaline-lysis method to extract the plasmid DNA. The extracted plasmid DNA was then suspended in TE buffer and stored at −20°C until further investigation.26
Amplification of the AmpC β-Lactamase Genes (blaCITM and blaDHAM) by PCR
The AmpC β-lactamases genes (blaCITM and blaDHAM) were amplified by PCR using plasmid DNA preparation as template. The primers used for blaCITM gene was CLR5-F (5ʹ TGG CCA GAA CTG ACA GGC AAA −3ʹ) and CLR5-R (5ʹ- TTT CTC CTG AAC GTG GCT GGC −3ʹ) as forward and reverse primer respectively while the primers for blaDHAM gene was forward sequence DHAM for (5ʹ AAC TTT CAC AGG TGT GCT GGG −3ʹ) and reverse sequence DHAM_rev (3ʹ CCG TAC GCA TAC TGG CTT TGC −5ʹ).11 Reaction volume was set as 25µL by adding 12.5µL of 1X master mix (5× HOT FIREPol Blend Master Mix Ready to Load, Solis BioDyne, Estonia), 0.5µL each of the forward and reverse primers and 4µL of DNA template and ddH2O 7.5µL. The optimized PCR amplification of both the genes was 94ºC for 3 mins for initial denaturation; 35 cycles of denaturation at 94ºC for 30 seconds; 25 cycles of annealing at 62ºC for 30 seconds; 35 cycles of extension at 72º for 1 min; and final extension at 72ºC for 7 mins.11,27
Purification of DNA
Plasmid DNA was purified by ethanol precipitation method. In this method, the DNA pellet was rinsed with ice-cold 70% ethanol and left it to dry for 10 minutes which facilitates the alcohol evaporation. The DNA pellet was suspended in a buffer solution containing Tris, EDTA and RNases to eash out the remaining impurities.
Detection of PCR Products by Gel Electrophoresis
The amplified products were visualized by using gel-electrophoresis in 1.5% agarose gel stained with 0.1µL ethidium bromide. After gel preparation, 2µL of 100bp DNA ladder was added to the first well as marker, 2µL of negative control was added to another well, 2µL of positive control was added to another well and 2µL of PCR products were added to the remaining wells. Finally, the gel was visualized under UV trans-illuminator.28
Quality Control
A standard aseptic procedure was adopted for the procedures in this study. All batches of the culture media and chemical reagents were processed with aseptic techniques following CLSI guidelines. In AST, quality control was maintained by using the control strains of E. coli ATCC 25922. During PCR, quality control was assured by the use of Klebsiella isolates carrying both the genes under question, while blank or negative controls were prepared without the application of nucleic acids. All these controls were used in each batch of the PCR assay.
Statistical Analysis
Data were entered and analyzed by using SPSS software version 24.0. The associations were explored using Chi-squared tests at 95% confidence interval (CI) among demographic variables such as gender, and age of the subjects.
Results
Demographic and Clinical Character of Enrolled Patients
Among 1151 patients enrolled, 54.2% (624/1151) were male and 45.8% (527/1151) were female. Of 253 bacterial growth, 47.8% (121/253) were from male and 52.2% (132/253) were from female. Of the total (253) bacterial growth, 26.1% (66/253) were from urine samples, followed by catheter tips (17.4%;44/253), blood (16.2%; 41/253), pus (11.9%; 30/253) and wound swabs (10.7%; 27/253). On age wise distribution of the patients, the highest growth of bacterial pathogens was found in age group 16–45 years (39.5%; 100/253) followed by 46–60 years (30.1%; 76/253), >60 years (24.1%; 61/253), and 0–15 years (6.3%; 16/253) respectively (Table 1).
 |
Table 1 Demographic and Clinical Character of Patients Attending at Annapurna Neurological Institute and Allied Sciences |
Distribution of Bacterial Isolates in Clinical Specimens
Among a total of 1151 clinical samples, 22% (253/1151) showed growth on culture media. Of 253 bacterial isolates, most (89.3%; 226/253) were Gram-negative bacteria. Among the bacterial growth, E. coli (28.5%; 72/253) was the predominant organism followed by Pseudomonas aeruginosa (16.2%; 41/253), Acinetobacter baumannii (15.8%; 40/253), Gram-positive bacteria (10.7%; 27/253), Klebsiella pneumoniae (9.9%; 25/253), and Klebsiella oxytoca (4.7%; 12/253) (Figure 1)
 |
Figure 1 Distribution of bacterial genera in culture-positive clinical specimens (n=253). |
Antibiotic Susceptibility Pattern of Isolated Gram-Negative Bacteria
Out of 226 Gram-negative bacterial isolates, 66.8% (151/226) were found resistant to cefoxitin, followed by ceftazidime (58.4%; 132/226), ciprofloxacin (53.5%; 121/226), and cefepime (51.8%; 117/226) whereas isolates were most susceptible to meropenem (69%; 156/226), ertapenem (68.6%;155/226), imipenem (66.8%; 151/226), amikacin (61.1%;138/226), piperacillin/tazobactam (56.6%;128/226) and azithromycin (53.1%; 120/226) (Table 2).
 |
Table 2 Antibiotic Susceptibility Pattern of Gram-Negative Bacterial Isolates (n=226) |
Multidrug Resistance (MDR) Pattern in the Isolates
Of 226 isolates, 46.9% (106/226) of the isolates were MDR. Highest percentage of MDR was found among E. coli (31.1%; 33/106) followed by P. aeruginosa (20.8%; 22/106), A. baumannii (18.9%; 20/106), K. pneumoniae (9.4%; 10/106) and K. oxytoca (4.7%; 5/106) respectively (Figure 2). In individual species, the highest percentage of multidrug resistance was found among C. freundii (100%; 8/8) followed by P. aeruginosa (53.6% 22/41), C. koseri (50%; 2/4), A. baumannii (50%; 20/40) and E. coli (45.8% 33/72), respectively (Figure 2).
 |
Figure 2 Distribution of MDR bacteria and overall MDR % and MDR within each species. |
AmpC Detection by Various Tests
Out of 226 Gram-negative isolates, 50% (113/226) showed resistance against cefoxitin. AmpC β-lactamase producing isolates were confirmed by two different confirmatory tests, Disk tests and Boronic acid tests. Of the ninety-one isolates, 91.2% (83/91) showed positive result in both tests and 8.8% (8/91) isolates showed positive only in boronic acid test confirmed by at least one test and were considered as AmpC β-lactamase producers.
Prevalence of blaCITM and blaDHAM Genes in AmpC β-Lactamase Producing Gram-Negative Isolates
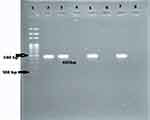
In PCR assay, 90.1% (82/91) and 87.91% (80/91) of the isolates were tested positive for blaCITM and blaDHAM. Respectively, the amplified blaCITM and blaDHAM genes with their amplicon size 465bp and 405bp were detected (Figures 3 and 4).
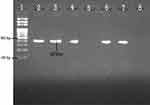 |
Figure 4 Agarose gel electrophoresis (1.5%) used for separation of PCR products. Lane 2, positive control; Lane 3, 4, 6 and 7 are Dham positive; Lane 5, Dham negative; and Lane 8, negative control. |
Distribution of AmpC β-Lactamase, blaCITM and blaDHAM Genes Among Gram-Negative Isolates and Their Relation to Gender, Age, Clinical Specimens and Clinical Isolates
Out of 91 AmpC producers, 50.6% (46/91) of the isolates were from female and 49.4% (45/91) were from male patients. Similarly, equal percentages (50%; 41/82) of blaCITM genes were obtained from specimens of both genders while 51.3% (41/80) and 48.7% (39/80) of blaDHAM genes were detected in the specimens obtained from male and female patients respectively. There was no any significant association of patient’s gender with the production of AmpC enzymes and AmpC genes (Table 3).
 |
Table 3 Distribution of AmpC β-Lactamase, CITM and DHAM Genes Among Gram-Negative Bacterial Isolates and Their Relation to Gender, Age and Clinical Specimens |
Highest number of AmpC β-lactamase producers (40.7%; 37/91) and prevalence of isolates producing blaCITM (40.2%; 33/82) and blaDHAM (41.2%; 33/80) were obtained from the age group (46–60) years followed by (16–45) years (30.8%; 28/91), (29.3%;24/82) and 31.3%; 25/80) respectively. There was a significant association between AmpC β-lactamase enzyme production and age group (p=0.01) while others factors including the acquisition of blaCITM and blaDHAM genes had no significant association with patient’s age (Table 3).
Among clinical specimens, the highest number of AmpC β-lactamase-producing isolates (20.9%; 19/91) were detected from blood and catheter tips, followed by urine (18.7%; 17/91), pus (13.3%; 12/91) and wound swabs (9.9%; 9/91). Similarly, highest number of blaCITM and blaDHAM producing isolates were detected from catheter tips (21.9%; 18/82); (22.5%; 18/80), followed by blood (19.5%; 16/82); (21.3%; 17/80) urine (17.0%; 14/82); (17.5%; 14/80) and pus (13.4%; 11/82); (8.8%; 7/80), respectively. There was no significant association between clinical specimens, AmpC β-lactamase production, and genes: blaCITM and blaDHAM (Table 3).
Distribution of AmpC β-Lactamases and Acquisition of blaCITM and blaDHAM Genes in Various Gram-Negative Bacteria
The highest prevalence of AmpC β-lactamase enzymes was found in E. coli (28.6%; 26/91), followed by P. aeruginosa (26.4%; 24/91), A. baumannii (13.2%; 12/91) and K. pneumoniae (10.9%; 10/91). Highest prevalence of blaCITM and blaDHAM genes were detected in E. coli (30.6%; 25/82) (31.3%; 25/80) followed by P. aeruginosa (25.7%; 21/82), (22.5%; 18/80) A. baumannii (12.2%; 10/82), (12.4%; 10/80) and K. pneumoniae (10.9%; 9/82), (12.4%; 10/80), respectively (Table 3).
Discussion
The increasing incidence of antimicrobial resistance remains as one of the major public health problems in developing countries like Nepal29 which has led to the prolonged hospital stay, increased treatment costs, constraining of therapeutic options and increased morbidity and mortality.29,30 Production of β-lactamases is the main defensive mechanism against β-lactam antibiotics. This study was carried out to explore the presence of Amber Class C β-lactamases (AmpC) in Gram-negative organisms derived from clinical samples obtained at ANIAS, Kathmandu, Nepal. This study further assessed the prevalence of AmpC β-lactamase genes (blaCITM and blaDHAM) by PCR assay. We found high prevalence of these enzymes and genes, although the prevalence of such enzymes varied from one geographical region to another, within and between the countries.
Out of 1151 clinical samples, only 253 (21.9%) showed significant growth of organisms. Of the total (253) bacterial growth, more than ninety percent were Gram-negative bacteria, E. coli was the most predominant isolates among them. Our findings are consistent with previous studies reported from Everest Hospital, Baneshwor,14 Alka Hospital, Jawlakhel,31 National Public Health Laboratory, Teku,32 Universal College of Medical Sciences, Bhairahawa,33 B.P Koirala Institute of Health Sciences, Dharan,34 Nobel Medical College, Biratnagar,35 New Delhi, India,36 Al-Najaf City, Iraq,37 Shashemene Referral Hospital, Ethiopia,38 and Mexico City, Mexico.39 Slightly higher rate of Gram-negative bacterial infections were observed among female patients and could be due to higher prevalence of urinary tract infections among females. This is consistent with previous studies from Model Hospital, Kathmandu,17 and Andra-Pradesh, India.40 Consistent with this study, higher rate of pus infections on post-operative cases was reported in females (63.33%) than in males (43.75%) from one study conducted at Uttar Pradesh state in India.41
In this study, Gram-negative bacterial isolates exhibited higher resistance to the following antibiotics: cefoxitin, ceftazidime, ciprofloxacin and cefepime, followed by cotrimoxazole whereas meropenem, imipenem, piperacillin-tazobactam and azithromycin were reported as the most sensitive antibiotics. Comparable pattern was observed in some of the previous findings from Chitwan Medical College, Chitwan,42 Padma Hospital, Pokhara.43 Cefoxitin and ceftazidime with low susceptibility rates are the first line drugs which are easily hydrolyzed by the bacterial enzymes and are less useful in treating the infections caused by Gram-negative pathogens. Similar to our study, imipenem/meropenem was found as the most sensitive drugs in the previous studies from Model Hospital, Kathmandu,17 Madrid Spain,44 and Wisconsin, USA.45
Antimicrobial Resistance (AMR) with increasing spread of MDR is a global concern, and its impacts are higher among the low- and middle-income countries where the burden of infectious diseases are high.30 The treatment of MDR microbes are expensive and difficult and are further compounded by the high prevalence of nosocomial infections.46 In this study, almost half of the isolated Gram-negative isolates (46.9%; 106/226) were found to be MDR. Higher prevalence of MDR bacteria has been reported from some other studies conducted in Kathmandu46–49 and other districts of Nepal.15,50,51 Production of ESBL, metallo β-lactamase (MBL) or AmpC β-lactamase could be the reason behind the reduced susceptibility towards newer generation of antibiotics.52 Moreover, multidrug resistance occurs because of the aggregation and expression of different genes on resistant (R) plasmids or genes that encode multidrug efflux pumps.53 In addition, the genes responsible for the expression of β-lactamase enzymes are constantly associated with the non-β-lactam antibiotics like aminoglycosides and fluoroquinolones.
In this study, AmpC production was found higher among male patients (41.67%) than females (38.98%). The findings of this study are consistent with some other studies from Kano, North-West Nigeria.54 However, our findings differed with studies reported from Benin, Nigeria.55 Higher prevalence of UTI with multidrug resistance among females has been reported by many studies, this may be due to higher prevalence of AmpC producing pathogens among women as they are more prone to UTI than men.
The use of cefoxitin-resistance in diagnostic laboratories serves as a reliable screening agent/marker to detect AmpC production. Moreover, this marker provides a good negative predictive value.
In spite of their broad scope, some of the studies have stressed the use of cefoxitin as a poor screening agent for AmpC production. This could be due to the existence of alternative mechanisms other than AmpC (one of them is porin channel mutation) which may lead to false positive interpretation (as cefoxitin resistance).52 Our study revealed that one third of the Gram-negative isolates (>40%) were responsible for AmpC production. The findings of this study contrasted with the study from Model Hospital, Kathmandu.17 The difference may be due to the phenotypic detection method used in this study which used cefoxitin resistivity test and AmpC disk test method using cefoxitin (30µg). The confirmatory test used was phenylboronic acid test using cefoxitin 30µg/400µg phenylboronic acid. Boronic acid derivatives were proved as reversible inhibitors of AmpC β-lactamase enzymes.56
In this study, AmpC production was observed in 91 (80.53%) isolates out of 113. The AmpC production rate in our study is slightly higher than other studies reported from Model Hospital, Kathmandu,17 Chitwan Medical College,57 Bharatpur, Chitwan,58 and India.59
In order to compensate the limitations due to one of the method, integration of both techniques in routine diagnosis can increase sensitivity and specificity of the tests in the clinical settings.
A total of 73 Gram-negative isolates showed the presence of both blaCITM and blaDHAM genes. These findings are consistent with a previous study reported from Ilam, Iran.27 In the similar type of study reported from India, it showed that blaCITM genes were more prevalent in E. coli.60 In our study, the prevalence of AmpC associated genes (blaCITM and blaDHAM) was investigated by phenotypic and genotypic methods. This study provides a broad analysis of the Gram-negative bacteria associated with AmpC β- lactamase production and their antibiotic susceptibility patterns among clinical isolates. The findings of this study are important in informing the resistance patterns of various organisms towards cephalosporins. Surveillance of the multidrug resistance with β-lactamase including ESBL, MBL, and AmpC production are needed for a routine clinical practice to optimize the treatment and prevent further resistance among β-lactams antibiotics.13,61,62
Strengths and Limitations
This is the first study exploring AmpC β-lactamase genes (blaCITM and blaDHAM) using both phenotypic and molecular test among patients attending a tertiary health care center of Nepal. The findings of this study can inform the antimicrobial policy for tertiary care centers including preparing the management of hospital infections, treatment protocol and diagnostic procedure. There are a few limitations of this study that includes investigation of limited AmpC β-lactamases, short duration of the study, and being conducted at a single tertiary care center. Future studies can build on it to conduct a longitudinal study at multiple tertiary care centers with exploration of all β- lactamases such as ESBL, MBL, KPC and the major genes responsible for resistance. Nonetheless, as a first study of its types triangulating the phenotypic and molecular methods, this study will be a valuable reference for future studies on AmpC β-lactamases and prevalence of blaCITM and blaDHAM genes in other settings/hospitals of Nepal.
Conclusion
Our findings show that reduced susceptibility to cefoxitin in Gram-negative isolates is linked to the presence of plasmid-mediated AmpC genes. As there is a lack of single definitive method, it is suggested to utilize several phenotypic detection methods simultaneously for the precise detection of AmpC β-lactamase producing bacteria. In this study, PCR detected almost 90% of AmpC β-lactamase genes (blaCITM and blaDHAM) among phenotypically confirmed isolates. High prevalence of resistant genes and MDR among Gram-negative isolates is an alarming sign, calling for urgent intervention measures to curb the growth and spread of these isolates.
Abbreviations
AST, antimicrobial susceptibility test; CAC, ceftazidime and clavulanic acid; CAZ, ceftazidime; CLSI, Clinical Laboratory Standard Institute; CTX, cefotaxime; CX, cCefoxitin; ESBL, extended-spectrum β-lactamase; EUCAST, European committee on antimicrobial susceptibility testing; GNB, Gram-negative bacteria; ICU, intensive care unit, kDa, Kilo-Dalton; LPS, lipopolysaccharide; MDR, multidrug resistance; MHA, Mueller Hinton agar; NA, nutrient agar; MIC, minimum inhibitory concentration; PBA, phenylboronic acid; PCR, polymerase chain reaction; UTI, urinary tract infection; WHO, World Health Organization; XDR, extensive/xeno-drug resistance.
Ethical Approval
This study was approved by the Ethical Review Board (ERB) of Nepal Health Research Council (NHRC), Kathmandu, Nepal (Reg. No. 369/2017). Written informed consent was obtained from each study subject. Parents/guardians were interviewed in case of children. The research was in compliance with the Helsinki Declaration. This study is free from selection bias.
Acknowledgments
We would like to express our sincere gratitude and admiration to all the staffs and faculties of Golden Gate International College, Kathmandu, and Annapurna Neurological Institutes and Allied Sciences, Kathmandu for their support and guidance to complete this study. Above all, we are grateful to the patients for their involvement in this study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was financially supported by Golden Gate International College, Old Baneshwor, Kathmandu, Nepal.
Disclosure
All the authors hereby declare that they have no competing interests.
References
1. Bush K, Bradford PA. β-lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247. doi:10.1101/cshperspect.a025247
2. Perez F, El Chakhtoura NG, Papp-Wallace KM, Wilson BM, Bonomo RA. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin Pharmacother. 2016;17:761–781.
3. Falagas ME, Karageorgopoulos DE. Extended-spectrum β-lactamase-producing organisms. J Hosp Infect. 2009;73:345–354. doi:10.1016/j.jhin.2009.02.021
4. Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi:10.1128/AAC.46.1.1-11.2002
5. Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other beta-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi:10.1128/AAC.40.8.1926
6. Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi:10.1128/CMR.00036-08
7. Alvarez M, Tran JH, Chow N, Jacoby GA. Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–537. doi:10.1128/AAC.48.2.533-537.2004
8. Choi SH, Lee JE, Park SJ, et al. Emergence of antibiotic resistance during therapy for infections caused by Enterobacteriaceae producing AmpC beta-lactamase: implications for antibiotic use. Antimicrob Agents Chemother. 2008;52:995–1000. doi:10.1128/AAC.01083-07
9. Yu WL, Cheng KC, Chi CJ, Chen HE, Chuang YC, Wu LT. Characterisation and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacter cloacae isolated from a district teaching hospital in Taiwan. Clin Microbiol Infect. 2006;12:579–582.
10. Jacoby GA, Walsh KE, Walker VJ. Identification of extended-spectrum, AmpC, and carbapenem- hydrolyzing beta-lactamases in Escherichia coli and Klebsiella pneumoniae by disk tests. J Clin Microbiol. 2006;44:1971–1976. doi:10.1128/JCM.00062-06
11. Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi:10.1128/JCM.40.6.2153-2162.2002
12. Kayastha K, Dhungel B, Karki S, et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella species in pediatric patients visiting International Friendship Children’s Hospital, Kathmandu, Nepal. Infect Dis (Auckl). 2020;13:1178633720909798. doi:10.1177/1178633720909798
13. Thapa Shrestha U, Shrestha S, Adhikari N, et al. Plasmid profiling and occurrence of beta-lactamase enzymes in multidrug-resistant uropathogenic Escherichia coli in Kathmandu, Nepal. Infect Drug Resist. 2020;13:1905–1917. doi:10.2147/IDR.S250591
14. Guragin N, Pradhan A, Dhungel B, Banjara MR, Rijal KR, Ghimire P. Extended spectrum B-lactamase producing Gram negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. TUJM. 2019;6(1):26–31.
15. Raut S, Adhikari B. ESBL and their identification in peripheral laboratories of Nepal. Nepal Med Coll J. 2015;17(3–4):176–181.
16. Koirala A, Agrahari G, Dahal N, Ghimire P, Rijal KR. ESBL and MBL mediated resistance in clinical isolates of nonfermentating Gram negative bacilli (NFGNB) in Nepal. J Microb Antimicrob Agents. 2017;3(1):18–24.
17. Baral P, Neupane S, Shrestha B, Ghimire KR, Marasini BP, Lekhak B. Clinical and microbiological observational study on AmpC-beta -lactamase producing Enterobacteriaceae in a hospital of Nepal. Braz J Infect Dis. 2013;17(2):256–259. doi:10.1016/j.bjid.2012.09.012
18. Isenberg HD. Clinical Microbiology Procedures Handbook.
19. Forbes BA, Sahm DF, Weissfelt AS. Study Guide for Bailey and Scott’s Diagnostic Microbiology. USA, Mosby: Mosby Publication; 2007.
20. Cheesbrough M. District Laboratory Practice in Tropical Countries. United Kingdom: CUPC; 2006.
21. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
22. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi:10.1111/j.1469-0691.2011.03570.x
23. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
24. Tan TY, Ng SY, Teo L, Koh Y, Teok CH. Detection of plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J Clin Pathol. 2008;61:642–644. doi:10.1136/jcp.2007.053470
25. Tille P. Bailey and Scott’s Diagnostic Microbiology.
26. Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual.
27. Maleki A, Khosravi A, Ghafourian S, et al. High prevalence of AmpC beta-lactamases in clinical isolates of Escherichia coli in Ilam, Iran. Osong Public Health Res Perspect. 2015;6:201–204. doi:10.1016/j.phrp.2015.02.001
28. Gurung S, Kafle S, Dhungel B, et al. Detection of OXA-48 gene in carbapenem -resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Infect Drug Resist. 2020;13:2311–2321. doi:10.2147/IDR.S259967
29. Pokharel S, Adhikari B. Antimicrobial resistance and over the counter use of drugs in Nepal. J Glob Health. 2020;10:010360. doi:10.7189/jogh.10.010360
30. Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Glob Health. 2019;4:e002104. doi:10.1136/bmjgh-2019-002104
31. Shakya P, Shrestha D, Maharjan E, Sharma VK, Paudyal R. ESBL production among E. coli and Klebsiella spp. causing urinary tract infection: a hospital based study. Open Microbiol J. 2017;11:23–30. doi:10.2174/1874285801711010023
32. Upadhyaya G, Bhattarai A, Rijal KR, Ghimire P, Upadhyaya B. Urinary tract infections in kidney transplant patients of Kathmandu Valley. Int J Microbiol Res Rev. 2013;3(1):1–6.
33. Raut S, Rijal KR, Khatiwada S, et al. Trend and characteristics of Acinetobacter baumannii infections in patients attending Universal College of Medical Sciences, Bhairahawa, Western Nepal: a longitudinal study of 2018. Infect Drug Resist. 2020;13:1631–1641. doi:10.2147/IDR.S257851
34. Shrestha LB, Baral R, Poudel P, Khanal B. Clinical, etiological and antimicrobial susceptibility profile of pediatric urinary tract infections in a tertiary care hospital of Nepal. BMC Pediatr. 2019;19:36. doi:10.1186/s12887-019-1410-1
35. Thakur P, Ghimire P, Rijal KR, Singh GK. Antimicrobial resistance pattern of Escherichia coli isolated from urine samples in patients visiting tertiary health care centre in eastern Nepal. Sunsari Tech Coll J. 2012;1(1):22–26. doi:10.3126/stcj.v1i1.8657
36. Kaur N, Sharma S, Malhotra S, Madan P, Hans C. Urinary tract infection: aetiology and antimicrobial resistance pattern in infants from a tertiary care hospital in northern India. J Clin Diagn Res. 2014;8:1–3.
37. Majeed HT, Aljanaby AAJ. Antibiotic susceptibility patterns and prevalence of some extended spectrum beta-lactamases genes in Gram-Negative bacteria isolated from patients infected with urinary tract infections in Al-Najaf City, Iraq. Avicenna J Med Biotechnol. 2019;11:192–201.
38. Seifu WD, Gebissa AD. Prevalence and antibiotic susceptibility of Uropathogens from cases of urinary tract infections (UTI) in Shashemene referral hospital, Ethiopia. BMC Infect Dis. 2018;18:30. doi:10.1186/s12879-017-2911-x
39. Ponce-de-Leon A, Rodriguez-Noriega E, Morfin-Otero R, et al. Antimicrobial susceptibility of Gram-negative bacilli isolated from intra-abdominal and urinary-tract infections in Mexico from 2009 to 2015: results from the study for monitoring antimicrobial resistance trends (SMART). PLoS One. 2018;13:e0198621. doi:10.1371/journal.pone.0198621
40. John MS, Meenakshi K, Lakshmi PM, Reddy PS. Prevalence and distribution of bacterial pathogens causing urinary tract infections in humans: a study from tertiary care hospital in AP, India. Int J of Curr Microbio and App Sci. 2015;4:251–257.
41. Bisht HS, Bisht T, Kumar D. The prevalence of post-operative infection among the male and female patients in a Tertiary Care Hospital of Western UP India. ISJR. 2014;3(5):381–383.
42. Gautam R, Chapagain M, Acharya A, et al. Antimicrobial susceptibility patterns of Escherichia coli from various clinical sources. JCMC. 2013;3(1):14–17. doi:10.3126/jcmc.v3i1.8459
43. Tamang K, Shrestha P, Koirala A, Khadka J, Gautam N, Rijal KR. Prevalence of bacterial uropathogens among diabetic patients attending padma Nursing Hospital of Western Nepal. HiJOST. 2017;1:15–19.
44. Oteo J, Delgado-Iribarren A, Vega D, et al. Emergence of imipenem resistance in clinical Escherichia coli during therapy. Int J Antimicrob Agents. 2008;32(6):534–537. doi:10.1016/j.ijantimicag.2008.06.012
45. Kehl SC, Dowzicky MJ. Global assessment of antimicrobial susceptibility among Gram-negative organisms collected from pediatric patients between 2004 and 2012: results from the tigecycline evaluation and surveillance trial. J Clin Microbiol. 2015;53:1286–1293. doi:10.1128/JCM.03184-14
46. Mishra SK, Awal BK, Kattel HP, et al. Drug resistant bacteria are growing menace in a University Hospital in Nepal. Amer J Epidem Infec Dis. 2014;2:19–23.
47. Neupane S, Pant ND, Khatiwada S, Chaudhary R, Banjara MR. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrob Resist Infect Control. 2016;5:5.
48. Nepal K, Pant ND, Neupane B, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16:62. doi:10.1186/s12941-017-0236-7
49. Parajuli NP, Maharjan P, Parajuli H, et al. High rates of multidrug resistance among uropathogenic Escherichia coli in children and analyses of ESBL producers from Nepal. Antimicrob Resist Infect Control. 2017;6:9. doi:10.1186/s13756-016-0168-6
50. Upadhyay A, Parajuli P. Extended spectrum β-lactamases (ESBL)-producing Klebsiella species isolated at National Medical College and Teaching Hospital Nepal. Asian J Pharma and Clin Res. 2013;1:161–164.
51. Shrestha S, Amatya R, Dutta R. Prevalence of extended spectrum beta lactamase (ESBL) production in gram negative isolates from pyogenic infection in tertiary care hospital of eastern Nepal. Nepal Med Coll J. 2011;13:186–189.
52. Kaur DC, Puri JS, Kulkarni SS, Jayawant AN. Prevalenc of Ampc beta-lactamases in clinical isolates of E. coli from a tertiary care rural hospital. Int J Pharm and Pharmaceut Sci. 2015;7:165–168.
53. Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–146. doi:10.1146/annurev.biochem.78.082907.145923
54. Yusuf I, Haruna M, Yahaya H. Prevalence and antibiotic susceptibility of AmpC and ESBLs producing clinical isolates at a tertiary health care center in Kano, north-west Nigeria. Afr J Clin and Experi Microbiol. 2013;14:109–119.
55. Ogefere HO, Osikobia JG, Omoregie R. Prevalence of AmpC β-lactamase among Gram-negative bacteria recovered from clinical specimens in Benin City, Nigeria. Tropic J Pharmaceut Res. 2016;15:1947–1953. doi:10.4314/tjpr.v15i9.20
56. Tondi D, Calo S, Shoichet BK, Costi MP. Structural study of phenyl boronic acid derivatives as AmpC beta-lactamase inhibitors. Bioorg Med Chem Lett. 2010;20:3416–3419. doi:10.1016/j.bmcl.2010.04.007
57. Khan GM, Thappa RK, Adhikari DS, et al. Cancer prevalence trend in central region of Nepal. J Chitwan Med Colleg. 2013;3:22–25. doi:10.3126/jcmc.v3i1.8461
58. Ansari S, Nepal HP, Gautam R, et al. Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care center of Nepal. Antimicrob Resist Infect Control. 2015;4:15. doi:10.1186/s13756-015-0059-2
59. Hemalatha V, Padma M, Sekar U, Vinodh TM, Arunkumar AS. Detection of Amp C beta lactamases production in Escherichia coli & Klebsiella by an inhibitor based method. Indian J Med Res. 2007;126:220–223.
60. Chakraborty AP, Shenoy S, Saralaya V. Characterization of plasmid mediated AmpC producing Escherichia coli clinical isolates from a tertiary care hospital in South India. Indian J Pathol Microbiol. 2014;57:255–258. doi:10.4103/0377-4929.134700
61. Chaudhary U, Agarwal S, Raghuraman K. Identification of extended spectrum beta lactamases, AmpC and carbapenemase production among isolates of Escherichia coli in North Indian tertiary care centre. Avicenna J Med. 2018;8:46–50. doi:10.4103/ajm.AJM_156_17
62. Dhungana K, Awal BK, Dhungel B, Sharma S, Banjara MR, Rijal KR. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo betalactamae (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. Acta Scientific Microbiol. 2019;2(12):60–69. doi:10.31080/ASMI.2019.02.0432
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.