Back to Journals » OncoTargets and Therapy » Volume 9
PIM1 polymorphism and PIM1 expression as predisposing factors of esophageal squamous cell carcinoma in the Asian population
Authors Wu Y, Lu D, He Z, Jin C
Received 30 December 2015
Accepted for publication 18 March 2016
Published 17 May 2016 Volume 2016:9 Pages 2919—2925
DOI https://doi.org/10.2147/OTT.S103392
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Yuan-Bo Wu,1 Di Lu,1 Zhi-Feng He,1 Chan-Guan Jin2
1Department of Cardiothoracic Surgery, The First Affiliated Hospital of Wenzhou Medical University, 2Department of Hepatobiliary Surgery, The Second Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, People’s Republic of China
Abstract: Our study aimed to identify the association between a PIM1 polymorphism and PIM1 expression levels with clinicopathological features of esophageal squamous cell carcinoma (ESCC). A total of 168 patients with ESCC were recruited as the case group, and 180 healthy individuals were included as the control group. Polymerase chain reaction-direct sequencing was employed to analyze all genotypes containing the PIM1 -1 882 A>T mutation. Immunohistochemistry was used to detect PIM1 expression. The distributions of genotype AA and allele A of PIM1 -1 882 A>T were higher in the case group than in the control group (both P<0.05). AT + TT carriers had a lower risk of ESCC than AA carriers (P<0.05). PIM1 polymorphism was related to the invasion depth, degree of differentiation, and lymphatic metastasis of ESCC (P<0.05). PIM1 expression was associated with lymphatic metastasis of ESCC and PIM1 polymorphism (both P<0.05). PIM1 -1 882 A>T and the overexpression of PIM1 were associated with the clinicopathological features of ESCC, and PIM1 -1 882 A>T may help to reduce the risk of ESCC in the Asian population.
Keywords: PIM1, polymorphism, esophageal squamous cell carcinoma, -1 882 A>T, clinicopathological features, proto-oncogene
Introduction
Esophageal cancer is extremely aggressive and is one of the most fatal cancers worldwide. At present, it ranks eighth in morbidity and sixth in mortality among all cancers in the world.1 Clinically, esophageal squamous cell carcinoma (ESCC), as the predominant histological subtype, accounts for ~90% of the total esophageal cancer patients.2 Interestingly, the incidence of ESCC is higher in males than in females.3 An epidemiological report indicates that the highest rate of ESCC is in Iran, followed by other countries, including the People’s Republic of China, South Africa, and France.4 The etiology of ESCC appears to be involved with factors such as excessive smoke inhalation and overconsumption of pickled vegetables or hot food. Furthermore, a history of reflux esophagitis is significantly associated with an increased risk of ESCC.5 Although the prognosis has improved over the last decade, the 5-year survival rate of ESCC is still only 30%, with tumors recurring within the first year after surgery.6 The postoperative recurrence, invasion, and metastasis of ESCC are influenced by relevant factors such as proto-oncogene activation, tumor-suppressor gene inactivation, and abnormal protein expression.
Pim family kinases, including three closely related mammalian serine or threonine kinases (PIM1, PIM2, and PIM3), are small, constitutively active, and highly evolutionarily conserved serine/threonine-specific kinases.7,8 PIM1 was originally identified from mouse lymphoma samples as a frequently activated gene that was derived from the preferential integration of Moloney leukemia virus into the 3′-untranslated region of the PIM1 gene.9 PIM1 is located on chromosome 6p21, which has been found to play an oncogenic role in the occurrence of T-cell lymphoma in mice. PIM1 promotes oncogenesis primarily through suppression of apoptosis, promotion of cell cycle progression, and promotion of genomic instability.10,11 Overexpression of the PIM1 protein has been implicated in the tumorigenesis of ESCC, gastric cancer, and colon cancer.10,12,13 However, there is little data about the association between PIM1 polymorphism and biological behavior of ESCC. This study aims to investigate the association between PIM1 polymorphisms and clinical pathological features of ESCC through detecting the PIM1 −1 882 A>T mutation and evaluating PIM1 protein expression in ESCC.
Materials and methods
Subjects
A total of 168 Chinese patients with ESCC (107 males and 61 females) admitted to the Department of Hepatobiliary Surgery, The Second Affiliated Hospital of Wenzhou Medical University, between September 2012 and September 2014 were enrolled into our study as the case group. Postoperative tumor tissues, adjacent normal tissues, and venous blood (2 mL) were collected from each patient. Patients with ESCC had a mean age of 60.7±12.5 years, with a median age of 61 years. With complete medical records, the ESCC cases with no preoperative chemotherapy or radiotherapy were pathologically confirmed. As defined by the esophageal cancer tumor node metastasis staging manual published by the Union for International Cancer Control/American Joint Committee on Cancer in 2010,14 the case group contained 28 patients in stage I, 85 in stage II, 36 in stage III, and 19 in stage IV; 89 with lymph node metastasis and 79 without; and 63 of high differentiation, 50 moderate, and 55 low differentiation. There was no organ-specific metastasis in these patients with ESCC other than lymphatic metastasis. Additionally, we recruited 180 healthy Chinese individuals (121 males and 59 females) with normal esophageal mucosa as the control group and collected tissue specimens and venous blood (2 mL) from each of them. They were aged 61.4±14.7 years, with a median age of 61 years. There were no significant differences in sex or age between the case and control groups (both P>0.05). All paraffin-embedded tissue specimens were confirmed by a pathology specialist. This study was approved by the Ethics Committee of the Department of Hepatobiliary Surgery, The Second Affiliated Hospital of Wenzhou Medical University. All subjects gave informed written consent.
Single nucleotide polymorphism screening and genotype analysis
Genomic DNA was extracted from the venous blood with the phenol/chloroform method.15 The PIM1 promoter region (−2,376 bp~+10 bp) amplification was conducted with an upstream primer of 5′-GTGGAAAGGCTTTGAATAAA-3′ and a downstream primer of 5′-ATGCAGCCCTCAGTCGTT-3′, designed by Primer 5.0 software (Premier Biosoft, Palo Alto, CA, USA). Direct DNA sequencing (SinoGenoMax Co., Ltd., Beijing, People’s Republic of China) was used to screen the single nucleotide polymorphism. DNAman software (version 5.2; Lynnon Biosoft, San Diego, CA, USA) was used to read the screening results and analyze all genotypes with the −1 882 A>T polymorphism. The total volume of the polymerase chain reaction was 25 μL, containing 100 ng DNA sample and 10 pmol/L primer concentration. The polymerase chain reaction amplification was performed with 5 minutes initial denaturation at 94°C, then 30 cycles of 30 seconds at 94°C, 30 seconds at 57°C, 60 seconds at 72°C, and a final 7 minutes extension at 72°C.
Immunohistochemistry
The streptavidin–peroxidase conjugated method was employed for the detection of PIM1 expression. All samples were fixed in 10% formalin, then embedded with paraffin and cut into sections (4 μm each). Throughout deparaffinization and alcoholic dehydration, the sections were immersed in 3% hydrogen peroxide to block endogenous peroxidase for antigen repairing. Rabbit-anti-human PIM1 polyclonal antibody (Boster Biotech Co., Ltd., Wuhan, People’s Republic of China) was added; the sections were incubated at 37°C for 1 hour and then incubated with a biotinylated secondary antibody. Subsequently, diaminobenzidine was added with a coloration time of 1–2 minutes, and then the sections were washed using phosphate-buffered saline (PBS) solution (3×2 minutes). After 1 minute of counterstaining using hematoxylin, the sections were dehydrated and transparent, and they were then sealed with neutral gum. The primary antibody was replaced by PBS as the negative control.
Immunohistochemistry staining results
The sections were observed under a light microscope, and the double-blind method was employed to count the positive cells. As a baseline, 300 tumor cells in five high-background fields (×200) were counted. The criteria for the quality controls used for the immunohistochemistry were as follows. Using the couple score and the semiquantitative method, the staining intensity (no staining, 0 points; slight staining, 1 point; moderate staining, 2 points; deep staining, 3 points) and the ratio of positive cells (0%–5%, 0 points; 6%–25%, 1 point; 26%–50%, 2 points; 51%–75%, 3 points; over 75%, 4 points) were determined. The final results were the two scores mentioned earlier multiplied together: 0 points, negative (−); 1–4 points, weak positive (+); 5–8 points, positive (++); and 9–12 points, strong positive (+++).
Statistical analysis
Data analysis was performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The distributions of the allele and genotype frequencies were analyzed with the use of the Hardy–Weinberg equilibrium, which was examined using the χ2 test when n≥40 but 1≤ T <5 and checked using Fisher’s exact test when n<40 or T <1. The odds ratio with 95% confidence interval was calculated to estimate the associations between PIM1 polymorphism and the risk of ESCC. Moreover, risk factors, such as the degree of tumor differentiation, invasion depth, and clinical stage, were examined using a rank test. P<0.05 was considered statistically significant.
Results
Distributions of genotype and allele of PIM1 polymorphism and the risk of ESCC
The distribution of the allele and genotype of PIM1 −1 882 A>T in the case and control groups agreed with the Hardy–Weinberg equilibrium (both P>0.05). The frequencies of allele −1 882 A in the case and control groups were significantly different (P<0.05) at 92.9% and 87.8%, respectively. The frequencies of the AA, TT, and AT genotypes were, respectively, 86.3%, 0.6%, and 13.1% in the case group and 77.2%, 1.7%, and 21.1% in the control group, which also demonstrated a significant difference between these two groups (all P<0.05). We also found that AT + TT carriers had a lower risk of ESCC than AA carriers (odds ratio =0.538, 95% confidence interval =0.307–0.943, P<0.05) (Table 1).
  | Table 1 Genotype and allele frequency and risks of PIM1 polymorphism in the case and control groups |
PIM1 polymorphism and clinicopathological features of ESCC
The distribution of the PIM1 polymorphism changed remarkably, which influenced the invasion depth, degree of differentiation, and lymphatic metastasis of ESCC (all P<0.05). However, there was no significant difference in the frequency of the PIM1 polymorphism with regard to sex, age, clinical stage, or tumor size according to the rank test (all P>0.05) (Table 2).
  | Table 2 PIM1 polymorphism and clinicopathological features of esophageal squamous cell carcinoma |
PIM1 protein expression in ESCC tissues and adjacent normal tissues
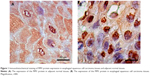
The PIM1 protein was primarily expressed in the cell cytoplasm and membrane. Staining for the PIM1 protein in ESCC tissues was uniformly brown-yellow or sepia, with a positive rate of 74.4% (125/168). However, a small amount of PIM1 protein expression was observed in the adjacent normal tissues, which were colorless or light yellow, and the positive rate was 23.2% (39/168), which was significantly lower than that in the corresponding ESCC tissue (P<0.05) (Table 3; Figure 1).
PIM1 protein expression and clinicopathological features of ESCC
PIM1 protein expression was associated with lymphatic metastasis of ESCC (P<0.05). However, there was no significant difference in PIM1 protein expression with regard to sex, age, invasion depth, degree of differentiation, clinical stage, or tumor size according to the rank test (all P>0.05) (Table 4).
  | Table 4 PIM1 protein expression and clinical pathologic features of esophageal squamous cell carcinoma |
Association between PIM1 polymorphism and PIM1 protein expression
As shown in Table 5, the rate of PIM1 protein expression was significantly different (P<0.05) among the AA (77.9%), AT (54.6%), and TT (0) PIM1 genotypes. The AA genotype exhibited the highest level of positive protein expression, suggesting that the PIM1 polymorphism is associated with PIM1 protein expression.
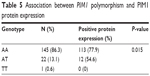  | Table 5 Association between PIM1 polymorphism and PIM1 protein expression |
Discussion
ESCC is the most prevalent pathological type of esophageal cancer in both Western and Asian countries.16 Recently, progress has been made in diagnosis and therapeutic options for ESCC, particularly in exploring its relationship with gene polymorphisms.17,18 PIM1 is a proto-oncogene that encodes a serine/threonine kinase with multiple cellular functions involving the regulation of cell apoptosis and progression, as well as transcription through various signaling pathways, including the STAT3 → PIM1 → NFkappaB stress response pathway and signaling pathways downstream of Akt.19–21 Accumulating evidence has demonstrated that PIM1 plays an important role in tumor progression, including colon cancer, laryngeal cancer, lymphocytic leukemia, and head and neck squamous cell carcinomas.13,22–24 In our study, we found that PIM1 polymorphism and alteration of PIM1 expression participated in the development of ESCC and were closely related to the clinicopathological features of ESCC, revealing that PIM1 could be useful for predicting biological behavior in patients with ESCC. Kim et al25 found that the T-C-T-C haplotypes of PIM1 (-1,196 T>C, IVS4 +55 T>C, IVS4 +1,416 T>A, and +3,684 C>A) were associated with an increased risk of lung cancer. Akagi et al26 found that the E135K mutation of PIM1 may be associated with acute myeloid leukemia. PIM1 −1 882 A>T is a novel single nucleotide polymorphism that has not been reported. However, we found that individuals with the AA genotype exhibited the highest positive protein expression, and there was just one patient carrying the TT genotype among the 168 patients with ESCC, suggesting that the PIM1 polymorphism was associated with PIM1 protein expression. A possible reason is that PIM1 polymorphism can reduce PIM1 protein expression levels. Our study first explored the PIM1 −1 882 A>T polymorphism and found that the AA genotype and A allele might contribute to the development and progression of ESCC, while the −1 882 A>T mutation (TT genotype and T allele) might decrease the risk of ESCC.
Because we measured the PIM1 expression levels in both the ESCC tumor tissues and the adjacent normal tissues, we discovered that the PIM1 protein is mainly expressed in the cytoplasm and cell membrane, and its expression in the ESCC tumor tissues was significantly higher when compared with the adjacent normal tissues. The PIM protein kinases are overexpressed in multiple human malignancies, including prostate cancer, lymphoma, leukemia, and pancreatic ductal adenocarcinoma and breast tumors.27–31 Additionally, the histologic grade, clinicopathological staging, and lymphatic metastasis are the most crucial prognostic factors for determining clinical outcomes in ESCC and, thus, clinical and pathological parameters, such as age, sex, histologic grade, clinical staging, tumor size, and lymph node metastasis, were used to investigate the expression of PIM1 in the progression of ESCC. In our study, the levels of positive PIM1 protein staining in ESCC tissues were significantly higher than those in the adjacent normal group and were influenced by the lymphatic metastasis of ESCC, which might indicate that the overexpression of PIM1 is correlated with the progression of ESCC. Liu et al12 reported the overexpression of PIM1 in patients with ESCC, suggesting that PIM1 may act as a molecular marker for predicting the prognosis of patients with ESCC.
Interestingly, our study also found that the invasion depth, degree of differentiation, and lymphatic metastasis were also associated with the distribution of the PIM1 −1 882 A>T genotype. It is possible that the PIM1 −1 882 A>T polymorphism affects the combination of the promoter region and signal factor, leading to an increase in PIMI expression and further inhibiting the malignant transformation of cells.
Our research explored the PIM1 polymorphism and PIM1 expression in the progression of ESCC and was the first to study the possible effect of the novel −1 882 A>T single nucleotide polymorphism on the development of ESCC. Collectively, our study demonstrated that the PIM1 −1 882 A>T polymorphism may help to reduce the risk of ESCC, and PIM1 expression may also serve as an index to detect biological behavior in the Asian population. Given the complexity of the role of gene functions in ESCC, further studies on PIM1 polymorphisms with a larger sample size or greater ethnic diversity are needed to help evaluate the utility of PIM1 as a therapeutic target for the treatment of ESCC.
Acknowledgment
We would like to acknowledge the helpful comments received from our reviewers.
Disclosure
The authors report no conflicts of interest in this work.
References
Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19(34):5598–5606. | ||
He X, Zhang Z, Li M, et al. Expression and role of oncogenic mirna-224 in esophageal squamous cell carcinoma. BMC Cancer. 2015;15:575. | ||
Bodelon C, Anderson GL, Rossing MA, Chlebowski RT, Ochs-Balcom HM, Vaughan TL. Hormonal factors and risks of esophageal squamous cell carcinoma and adenocarcinoma in postmenopausal women. Cancer Prev Res (Phila). 2011;4(6):840–850. | ||
Jessri M, Rashidkhani B, Hajizadeh B, Jessri M, Gotay C. Macronutrients, vitamins and minerals intake and risk of esophageal squamous cell carcinoma: a case-control study in Iran. Nutr J. 2011;10:137. | ||
Peng XE, Chen HF, Hu ZJ, Shi XS. Independent and combined effects of environmental factors and cyp2c19 polymorphisms on the risk of esophageal squamous cell carcinoma in Fujian Province of China. BMC Med Genet. 2015;16:15. | ||
Liu NB, Zhang JH, Liu YF, et al. High DEPTOR expression correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Onco Targets Ther. 2015;8:3449–3455. | ||
Magnuson NS, Wang Z, Ding G, Reeves R. Why target pim1 for cancer diagnosis and treatment? Future Oncol. 2010;6(9):1461–1478. | ||
Santio NM, Vahakoski RL, Rainio EM, et al. Pim-selective inhibitor DHPCC-9 reveals pim kinases as potent stimulators of cancer cell migration and invasion. Mol Cancer. 2010;9:279. | ||
Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–150. | ||
Yan B, Yau EX, Samanta S, et al. Clinical and therapeutic relevance of PIM1 kinase in gastric cancer. Gastric Cancer. 2012;15(2):188–197. | ||
Li S, Xi Y, Zhang H, et al. A pivotal role for Pim-1 kinase in esophageal squamous cell carcinoma involving cell apoptosis induced by reducing akt phosphorylation. Oncol Rep. 2010;24(4):997–1004. | ||
Liu HT, Wang N, Wang X, Li SL. Overexpression of Pim-1 is associated with poor prognosis in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2010;102(6):683–688. | ||
Peng YH, Li JJ, Xie FW, et al. Expression of pim-1 in tumors, tumor stroma and tumor-adjacent mucosa co-determines the prognosis of colon cancer patients. PLoS One. 2013;8(10):e76693. | ||
Rice TW. Esophageal cancer staging. Korean J Thorac Cardiovasc Surg. 2015;48(3):157–163. | ||
Di Pietro F, Ortenzi F, Tilio M, Concetti F, Napolioni V. Genomic DNA extraction from whole blood stored from 15- to 30-years at -20 degrees c by rapid phenol-chloroform protocol: A useful tool for genetic epidemiology studies. Mol Cell Probes. 2011;25(1):44–48. | ||
Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: Orient to Occident. Effects of chronology, geography and ethnicity. J Gastroenterol Hepatol. 2009;24(5):729–735. | ||
Zhang L, Zhu Z, Wu H, Wang K. Association between SNP309 and del1518 polymorphism in mdm2 homologue and esophageal squamous cell carcinoma risk in Chinese population of Shandong Province. Ann Clin Lab Sci. 2015;45(4):433–437. | ||
Xu X, Chen G, Wu L, Li L. Association of genetic polymorphisms in PTEN and additional gene-gene interaction with risk of esophageal squamous cell carcinoma in Chinese Han population. Dis Esophagus. Epub 2015 Nov 6. | ||
Zemskova M, Sahakian E, Bashkirova S, Lilly M. The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. J Biol Chem. 2008;283(30):20635–20644. | ||
Muraski JA, Rota M, Misao Y, et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat Med. 2007;13(12):1467–1475. | ||
Willert M, Augstein A, Poitz DM, Schmeisser A, Strasser RH, Braun-Dullaeus RC. Transcriptional regulation of Pim-1 kinase in vascular smooth muscle cells and its role for proliferation. Basic Res Cardiol. 2010;105(2):267–277. | ||
Choi JY, Cho SI, Do NY, Kang CY, Lim SC. Clinical significance of the expression of galectin-3 and Pim-1 in laryngeal squamous cell carcinoma. J Otolaryngol Head Neck Surg. 2010;39(1):28–34. | ||
Decker S, Finter J, Forde AJ, et al. PIM kinases are essential for chronic lymphocytic leukemia cell survival (PIM2/3) and CXCR4-mediated microenvironmental interactions (PIM1). Mol Cancer Ther. 2014;13(5):1231–1245. | ||
Gorogh T, Quabius ES, Meyer P, Hoffmann M. Characterisation of seven newly established head and neck squamous cell carcinoma cell lines. Eur Arch Otorhinolaryngol. 2015;272(5):1251–1258. | ||
Kim DS, Sung JS, Shin ES, et al. Association of single nucleotide polymorphisms in PIM-1 gene with the risk of Korean lung cancer. Cancer Res Treat. 2008;40(4):190–196. | ||
Akagi T, Shih LY, Ogawa S, et al. Single nucleotide polymorphism genomic arrays analysis of t(8;21) acute myeloid leukemia cells. Haematologica. 2009;94(9):1301–1306. | ||
Holder SL, Abdulkadir SA. PIM1 kinase as a target in prostate cancer: roles in tumorigenesis, castration resistance, and docetaxel resistance. Curr Cancer Drug Targets. 2014;14(2):105–114. | ||
Baron BW, Anastasi J, Hyjek EM, Baron JM. Expression of PIM1 protein in chronic lymphocytic leukemia/small lymphocytic lymphoma. Leuk Lymphoma. 2014;55(11):2658–2659. | ||
Li J, Hu XF, Loveland BE, Xing PX. Pim-1 expression and monoclonal antibody targeting in human leukemia cell lines. Exp Hematol. 2009;37(11):1284–1294. | ||
Reiser-Erkan C, Erkan M, Pan Z, et al. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2008;7(9):1352–1359. | ||
Malinen M, Jaaskelainen T, Pelkonen M, et al. Proto-oncogene PIM-1 is a novel estrogen receptor target associating with high grade breast tumors. Mol Cell Endocrinol. 2013;365(2):270–276. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.