Back to Journals » Infection and Drug Resistance » Volume 11
Phenotypic and genotypic characterizations of extended-spectrum beta-lactamase-producing Escherichia coli in Thailand
Authors Bubpamala J, Khuntayaporn P , Thirapanmethee K , Montakantikul P, Santanirand P , Chomnawang MT
Received 23 August 2018
Accepted for publication 16 September 2018
Published 5 November 2018 Volume 2018:11 Pages 2151—2157
DOI https://doi.org/10.2147/IDR.S174506
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Jiranun Bubpamala,1 Piyatip Khuntayaporn,1 Krit Thirapanmethee,1 Preecha Montakantikul,2 Pitak Santanirand,3 Mullika T Chomnawang1
1Department of Microbiology, Faculty of Pharmacy, Mahidol University, Bangkok, Thailand; 2Department of Pharmacy, Faculty of Pharmacy, Mahidol University, Bangkok, Thailand; 3Department of Pathology, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand
Purpose: Extended-spectrum β-lactamases (ESBLs) have become an issue in community worldwide due to an increase in antibiotic resistance over the past decade. This study was aimed to investigate the phenotypic and genotypic characteristics of ESBL-producing Escherichia coli in Thailand.
Materials and methods: In this study, all clinical isolates collected from tertiary hospitals in Thailand were identified as E. coli by biochemical tests and MALDI-TOF mass spectrometry. ESBL-producing E. coli was preliminary screened with disk diffusion method by cephalosporin disks and confirmed by the method of combination disk diffusion. Antimicrobial susceptibility test was used to determine MIC values of all ESBL-producing E. coli. For genotypic detection, a variety of ESBL genes were determined by PCR. Moreover, multilocus sequence typing (MLST) analysis was performed on internal portions of seven housekeeping genes for the diversity and phylogenetic relatedness of E. coli clonal group.
Results: Of the 285 ESBL-producing E. coli, most were susceptible to carbapenems. These strains showed a high resistance rate to ciprofloxacin (85.26%). The most frequently detected gene was bla CTX-M1 group at about 71.23% followed by bla CTX-M9 group (38.95%). The bla TEM, bla PER, bla GES, bla VEB, and bla SHV genes were identified in 31.93%, 5.96%, 4.56%, 3.51%, and 0.70% of ESBL-producing isolates, respectively. The bla OXA-10 gene was detected in only one strain. ESBL-producing E. coli isolates with high antimicrobial resistance were further investigated. Among those, E. coli sequence type ST38 was mostly found, followed by ST405, ST410, and ST131. It is noteworthy that the bla CTX-M gene was mainly detected in all four ST-type E. coli clones (ST38, ST405, ST410, and ST131).
Conclusion: This study provided a recent evidence of the genetic diversity of ESBL-producing E. coli in Thailand. In addition, the profile related to antimicrobial resistance pattern in this region was also demonstrated.
Keywords: epidemiology, prevalence, antimicrobial resistance, MLST, ESBLs, antibiotic resistance genes
Introduction
The rapid emergence of antibiotic resistance threatens effective prevention and treatment of an increasing range of infections. Some bacteria are naturally resistant to certain antibiotics and others can acquire resistance through mutations in some of their genes when they are exposed to an antibiotic. This acquired resistance can spread to other bacterial species. The main mechanism of antibiotic resistance mostly found is enzyme production such as β-lactamase enzymes. The β-lactamase enzymes produced by some bacteria provide resistance to β-lactam antibiotics by hydrolyzing β-lactam rings.1
Among antimicrobial resistant bacteria, Escherichia coli is one of highly concerned bacteria in the family Enterobacteriaceae. E. coli is a common cause of urinary tract infection and intra-abdominal infection in humans and is the second most common Gram-negative bacteria causing community-acquired bloodstream infection, accounting for 7.3% of all bloodstream infection isolates.2 ESBL-producing E. coli isolates have become an importance in community-onset infections, as well as nosocomial infections. The prevalence of resistance to fluoroquinolones and extended-spectrum cephalosporins in E. coli had highly increased over the past decade rendering severely limited therapeutic options for these infections.3
Extended-spectrum β-lactamases (ESBLs) are extremely broad spectrum β-lactamase enzymes, which can be produced by Gram-negative bacteria. They are mainly found in a family of Enterobacteriaceae. ESBLs are produced by the mutation of the TEM-1, TEM-2, and SHV-1 β-lactamases, which have been discovered since 1980–1990 and first detected in Western Europe.4 To date, more than 350 different natural ESBL variants are known, which have been classified into nine distinct structural and evolutionary families based on amino acid sequence comparisons such as TEM, SHV, CTX-M, PER, VEB, GES, BES, TLA, and OXA.5–7 The main types of ESBL variants include TEM, SHV, CTX-M, and OXA. Interestingly, blaCTX−M has rapidly increased and is now widely found in clinically isolated E. coli across the world.8 ESBLs, especially of the CTX-M type, are strongly associated with specific clonal E. coli strains.9 To date, little is known about the epidemiology of ESBL variants in Thailand. Moreover, it is critical to provide up-to-date resistance pattern which affect the treatment decision in this region. Therefore, this study was aimed to investigate the phenotypic characteristic and variation of genetically related ESBL-producing E. coli in Thailand.
Material and methods
Bacterial isolates
ESBL-producing E. coli isolates were collected from tertiary care hospitals located in various regions in Thailand from 2014 to 2015. Tertiary care hospitals were defined over 500-bed hospitals that usually provided a full complement of services. These hospitals were regional hospitals which were generally referral for patients with serious conditions. A total of 285 E. coli isolates were obtained from clinical specimens, including urine, sputum, pus, blood, and feces, which were a part of routine hospital procedures. Confirmation and identification of E. coli strains were performed by biochemical tests and MALDI-TOF mass spectrometry. This study was approved by Mahidol University Institutional Review Board (MU-IRB) [Approval No. MU-IRB 2014/019.0705].
Determination of ESBL-producing E. coli isolates
ESBL-producing E. coli was screened with disk diffusion method by cephalosporin disks as recommended by the CLSI guideline.10 To confirm ESBL production in E. coli, the method of combination disk diffusion technique was performed. Briefly, disks of ceftazidime (30 µg), ceftazidime/clavulanate (30/10 µg), cefotaxime (30 µg), and cefotaxime/clavulanate (30/10 µg) were placed on the Mueller–Hinton agar plate (Difco) with 30 mm distance from the center of each and were incubated at 37°C for 18 hours. The test result is considered as positive if the inhibition zone diameter is 5 mm or larger with clavulanate than without. The strain of Klebsiella pneumoniae ATCC700603 (carrying blaSHV-18 gene) was used for the positive control and E. coli ATCC25922 was used as the negative control in this study.
Antimicrobial susceptibility testing
The determination of MIC values was performed by broth microdilution method with nine antimicrobial agents, namely, ciprofloxacin, prulifloxacin, ceftazidime, fosfomycin, imipenem, meropenem, doripenem, biapenem, and piperacillin/tazobactam. E. coli isolates were grown in Mueller Hinton broth (MH broth), then E. coli isolates were diluted with MH broth to 0.5 McFarland (cell approximately 108 CFU/mL), and diluted with normal saline to adjust the cells to 106 CFU/mL before adding into 96-well plates containing antibiotics in triplicates. The working stock concentrations of antibiotics were based on the CLSI guideline.10 Finally, the plate was kept at 37°C for 18 hours. The results were evaluated by the MIC values from the minimum concentration of the drugs that gave no visible growth.
Characterization of ESBL genotypes
The standard PCR was performed to screen for the presence of ESBL genes: blaTEM, blaSHV, blaCTX-M, blaOXA, blaGES, blaVEB, and blaPER using specific primers described in Table 1. PCR reactions contained 1× buffer, 1.5 mM of MgCl2, 400 µM of dNTPs, 0.5 µM of forward and reverse primers each, 1 U Taq polymerase, and the concentration of DNA template depending on specific primers. For PCR cycling condition, the denaturation step was achieved at 96°C for 30 seconds, followed by the annealing step, the temperature of which depended upon the specific primers, and the final step of extension at 72°C for 30 seconds. All steps were repetitively performed for 30 cycles. Then, the PCR products were analyzed by agarose gel electrophoresis.
  | Table 1 Specific primers for ESBL genes detection Abbreviation: ESBL, extended-spectrum β-lactamase. |
Molecular typing by MLST
Forty-eight strains of ESBL-producing E. coli were selected and determined sequence types by MLST. The criteria for E. coli strain selection were based on ESBL gene pattern. PCR method was used to amplify internal portions of seven housekeeping genes of E. coli (adk, fumC, gyrB, icd, mdh, purA, and recA) with specific primers. Seven-locus MLST was performed using published criteria and primers.11 Amplification products were submitted to the commercial sequencing service (Macrogen Sequencing, Korea). DNA sequences of each genes were identified using the website: https://pubmlst.org/bigsdb?db=pubmlst_mlst_seqdef&page=sequenceQuery. The allelic profile for each isolate was determined using the BioNumerics MLST Plug-in in accordance with the Achtman scheme available at http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search.
Results
Phenotypic detection of ESBL-producing E. coli
All 285 E. coli collected from tertiary care hospitals located in various regions in Thailand were determined by combination disk diffusion. Among these positive ESBL-producing E. coli, 55 isolates were from the northeast region, 61 isolates from the north region, 45 isolates from the east region, 60 from the central region, and 64 isolates from the south region of Thailand. Antibiotic resistance patterns of isolated ESBL-producing E. coli were further examined according to the CLSI guideline. The results demonstrated that all of ESBL-producing E. coli were sensitive to carbapenems with 100% susceptibility to meropenem and doripenem followed by biapenem (99.65%) and imipenem (98.86%). High susceptibility was also observed with piperacillin and tazobactam (86.32%). Nevertheless, most of them showed a high resistance rate to ciprofloxacin at 85.26% (243/285 isolates), ceftazidime at 80% (228/285 isolates), prulifloxacin at 79.30% (226/285 isolates), and fosfomycin at 10.88% (31/285 isolates) (Figure 1). It was of interest that MIC90 of meropenem and doripenem were about ≤0.125 µg/mL followed by those of biapenem (MIC90=0.25 µg/mL) and doripenem (MIC90=0.5 µg/mL), respectively (Table 2).
  | Figure 1 Antimicrobial susceptibility test of antimicrobial agents against ESBL-producing E. coli performed by broth dilution method. Abbreviation: ESBL, extended-spectrum β-lactamase. |
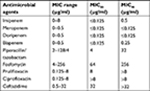  | Table 2 Antimicrobial susceptibility of nine antimicrobial agents against ESBL-producing E. coli Abbreviation: ESBL, extended-spectrum β-lactamase. |
Molecular detection of ESBL gene variants
The presence of ESBL genes including blaTEM, blaSHV, blaCTX-M (CTX-M1 and CTX-M9 group), blaOXA (OXA-2 and OXA-10 group), blaGES, blaVEB, and blaPER genes in all clinical isolates were investigated by PCR method (Figure 2). The results showed that blaCTX-M1 group genes were predominantly presented in Thailand at about 71.23% (203/285 isolates) followed by 38.95% (111/285 isolates) blaCTX-M9 group, 31.93% (91/285 isolates) blaTEM, 5.96% (17/285 isolates) blaPER, 4.56% (13/285 isolates) blaGES, 3.51% (10/285 isolates) blaVEB, 0.70% (2/285 isolates) blaSHV, and the least common 0.35% (1/285 isolate) blaOXA. It was noteworthy that the blaOXA gene was not detected in the chromosome but found on the plasmid. Interestingly, 52.63% (150/285 strains) of ESBL-producing E. coli carried only one ESBL gene. Among these, 33.33% (95/285 strains) of these contained two ESBL genes and 12.63% (36/285 strains) carried three ESBL genes. In addition, the coexistence of all three genes mostly was blaCTX-M1 group, blaCTX-M9 group, and blaTEM.
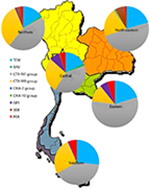  | Figure 2 Epidemiology of genotypic variants of ESBL-producing E. coli in all regions of Thailand. Abbreviation: ESBL, extended-spectrum β-lactamase. |
Forty-seven strains of ESBL-producing E. coli with high antimicrobial resistance were selected to determine 23 STs using the Achtman MLST scheme. The majority of ST-type high antimicrobial resistant strains was ST38 (10/47 isolates), followed by ST405 (6/47 isolates), ST410 (5/47 isolates), and ST131 (4/47 isolates) (Table 3). Among E. coli ST38 isolates, eight strains carried blaPER genes, four strains contained blaGES genes, and two strains possessed blaTEM genes. Interestingly, all of these strains carried blaCTX-M genes and were sensitive to carbapenems, prulifloxacin, and ciprofloxacin. However, 8 out of 10 E. coli ST38 isolates were resistant to ceftazidime and two strains were intermediate sensitive. Most E. coli ST38 strains were sensitive to piperacillin/tazobactam. All of E. coli ST405 contained blaCTX-M genes, while blaVEB could be detected in only two strains and only one strain contained blaTEM gene. E. coli ST405 were sensitive to carbapenems and fosfomycin but were resistant to ceftazidime, prulifloxacin, and ciprofloxacin. All E. coli ST410 carried blaCTX-M and two of them carried blaTEM. Interestingly, only one strain possessed blaOXA and blaVEB genes. This ST type was sensitive to carbapenems and fosfomycin but was resistant to prulifloxacin, ciprofloxacin, ceftazidime, and piperacillin/tazobactam. For E. coli ST131, all of them contained blaCTX-M genes, one strain carried blaTEM, and one strain carried blaVEB. All of these were sensitive to meropenem, doripenem, and fosfomycin. They were resistant to ciprofloxacin, prulifloxacin, and ceftazidime. Two strains of E. coli ST131 were resistant to imipenem and one strain showed intermediate sensitivity to biapenem.
  | Table 3 STs of ESBL-producing E. coli Abbreviations: ESBL, extended-spectrum β-lactamase; STs, Sequence Types. |
Discussion
ESBLs are bacterial enzymes that mediate resistance to the third-generation cephalosporins and monobactams. The spreading and outbreaks of ESBLs are often found in the family of Enterobacteriaceae especially E. coli.13 Our findings were based on the evaluation of E. coli strains isolated from clinical specimens obtained from tertiary care hospitals in Thailand. This study clearly showed high resistance rates of ESBL-producing E. coli to ciprofloxacin (85.26%), ceftazidime (80%), and prulifloxacin (79.30%), which raised serious concern and became a challenge for clinicians. In 2013, researchers reported high resistance to cefotaxime, ceftriaxone, and ceftazidime, and only some isolates were resistant to ciprofloxacin while most strains were susceptible to carbapenems in Thailand.14 In Korea, the study of Kang et al determined antimicrobial susceptibility of ESBL-producing E. coli against imipenem and meropenem and the resistance rates were only 1.5% (1/68) and 0% (0/50), respectively.15 The previous study in Thailand showed that CTX-M family had the highest prevalence of ESBL-related bla genes (87.3%) similar to our findings in which CTX-M type was still predominant.12 This study reported the presence of 31.93% blaTEM genes, while in 2011, blaTEM genes were found at 42% and the previous study in 2007 demonstrated 67% blaTEM genes. This indicated that the spreading of blaTEM ESBL-producing E. coli strains were decreased.16,17 In addition, only few blaSHV carrying strains were detected by the study of Kiratisin et al in 2007 and our study found only two strains of blaSHV ESBL-producing strains.16 In India, the prevalence of blaTEM genes was reported at about 48.7% (38/78 strains), followed by blaCTX-M at 7.6% (6/78 strains) and blaSHV at 5.1% (4/78 strains) which were different from our findings in Thailand.7,18 This study revealed that the majority of ST type with high antimicrobial resistant rate was ST38 which showed high sensitivity to piperacillin/tazobactam. According to the MLST results described by previous study, ST38 E. coli isolates were identified in China (CTX-M-14 producer),19 Japan (CTX-M-9 and CTX-M-14 producers),20 and France (OXA-48 producer).21 Additionally, Rodríguez et al investigated MLST in blaCTX-M ESBL-producing E. coli from Germany, Netherlands, and UK. The study showed high prevalence of ST38.22 Moreover, Alghoribi et al found CTX-M-positive ST38, ST131, and ST405 in Saudi Arabia.23
Conclusion
This study demonstrated that the CTX-M type remained the most common spreading in Thailand. Although carbapenems remain drugs of choice to treat ESBL-producing bacterial infections, we reported that ESBL-producing E. coli have increased the severity of resistance to antibiotics. It was clearly shown that ESBL outbreaks have been a problem worldwide and we hope to raise the awareness on proper antibiotics use to control spreading of these strains in both hospitals and community.
Acknowledgments
The authors wish to thank all staffs in the Department of Microbiology for technical support and suggestion on this work. The authors also thank all staffs at the hospital sites for their help and collaboration.
Disclosure
The authors report no conflicts of interest in this work.
References
Munita J, Arias C. Mechanisms of antibiotic resistance. In Kudva I, Cornick N, Plummer P, Zhang Q, Nicholson T, Bannantine J, Bellaire B (ed), Virulence Mechanisms of Bacterial Pathogens. 5th ed. Washington: ASM Press, DC; 2016: 481–511. | ||
Musicha P, Feasey NA, Cain AK, et al. Genomic landscape of extended-spectrum β-lactamase resistance in Escherichia coli from an urban African setting. J Antimicrob Chemother. 2017;72(6):1602–1609. | ||
Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol. 2018;61:185–188. | ||
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. | ||
Sharma J, Sharma M, Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332–336. | ||
Shahid M, Singh A, Sobia F, et al. bla(CTX-M), bla(TEM), and bla(SHV) in Enterobacteriaceae from North-Indian tertiary hospital: high occurrence of combination genes. Asian Pac J Trop Med. 2011;4(2):101–105. | ||
Bajpai T, Pandey M, Varma M, Bhatambare GS. Prevalence of TEM, SHV, and CTX-M Beta-Lactamase genes in the urinary isolates of a tertiary care hospital. Avicenna J Med. 2017;7(1):12–16. | ||
Miao Z, Li S, Wang L, Song W, Zhou Y. Antimicrobial resistance and molecular epidemiology of esbl-producing Escherichia Coli isolated from outpatients in town hospitals of shandong province, China. Front Microbiol. 2017;8:63. | ||
D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303(6–7):305–317. | ||
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24, Clinical and Laboratory Standards Institute (CLSI), Wayne, PA, USA. 2014;34(1). | ||
Wirth T, Falush D, Lan R, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. | ||
Ryoo NH, Kim EC, Hong SG, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum beta-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005;56(4):698–702. | ||
Cantón R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466–475. | ||
Sharma M, Pathak S, Srivastava P. Prevalence and antibiogram of Extended Spectrum β-Lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. J Clin Diagn Res. 2013;7(10):2173–2177. | ||
Kang CI, Song JH, Chung DR, et al; Korean Network for Study of Infectious Diseases (KONSID). Risk factors and treatment outcomes of community-onset bacteraemia caused by extended-spectrum beta-lactamase-producing Escherichia coli. Int J Antimicrob Agents. 2010;36(3):284–287. | ||
Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis. 2007;58(3):349–355. | ||
Kiratisin P, Henprasert A. Resistance phenotype-genotype correlation and molecular epidemiology of Citrobacter, Enterobacter, Proteus, Providencia, Salmonella and Serratia that carry extended-spectrum β-lactamases with or without plasmid-mediated AmpC β-lactamase genes in Thailand. Trans R Soc Trop Med Hyg. 2011;105(1):46–51. | ||
Kiratisin P, Apisarnthanarak A, Laesripa C, Saifon P. Molecular characterization and epidemiology of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008;52(8):2818–2824. | ||
Peng C, Zong Z. Sequence type 38 Escherichia coli carrying bla(CTX-M-14). J Med Microbiol. 2011;60(Pt 5):694–695. | ||
Suzuki S, Shibata N, Yamane K, Wachino J, Ito K, Arakawa Y. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother. 2009;63(1):72–79. | ||
Poirel L, Bernabeu S, Fortineau N, Podglajen I, Lawrence C, Nordmann P. Emergence of OXA-48-producing Escherichia coli clone ST38 in France. Antimicrob Agents Chemother. 2011;55(10):4937–4938. | ||
Rodríguez I, Thomas K, Van Essen A, et al; SAFEFOODERA-ESBL Consortium. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents. 2014;43(6):553–557. | ||
Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother. 2015;70(10):2757–2762. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
