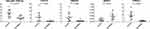Back to Journals » Journal of Inflammation Research » Volume 15
Peripheral Blood Mononuclear Cell Gene Expression in Chronic Obstructive Pulmonary Disease: miRNA and mRNA Regulation
Authors Wang L , Zhao H , Raman I, Yan M, Chen Q, Li QZ
Received 2 November 2021
Accepted for publication 16 March 2022
Published 1 April 2022 Volume 2022:15 Pages 2167—2180
DOI https://doi.org/10.2147/JIR.S337894
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Lijing Wang,1 Hongjun Zhao,2,* Indu Raman,3 Mei Yan,3 Qiong Chen,1 Quan-Zhen Li3,*
1Departments of Geriatrics, Respiratory Medicine, National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, 410008, Hunan, People’s Republic of China; 2Department of Rheumatology and Immunology, Xiangya Hospital, Central South University, Changsha, 410008, Hunan, People’s Republic of China; 3Department of Immunology, University of Texas Southwestern Medical Center, Dallas, TX, 75390, USA
*These authors contributed equally to this work
Correspondence: Quan-Zhen Li, Department of Immunology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390, USA, Tel +1 214-645-6071, Fax +1 214-645-6074, Email [email protected]
Introduction: The mechanisms underlying chronic obstructive pulmonary disease (COPD) remain unclear. Genetic and genomic changes may play a significant role in the pathogenesis of COPD. Identification of differentially expressed genes and miRNAs and their regulatory mechanisms at the whole-genome level will provide a comprehensive understanding of the development of COPD.
Methods: Peripheral blood mononuclear cells (PBMCs) from 12 patients with COPD and 12 normal controls were examined at the miRNA and mRNA expression levels using Affymetrix GeneChip. Microarray data were analyzed with Affymetrix Transcriptome Analysis Console 2.0 and GeneSpring software. Gene interaction pathways of the differentially expressed genes and miRNA-mRNA regulation were analyzed using the Ingenuity Pathway Analysis software. Four differentially expressed genes and one miRNA were further confirmed using RT-qPCR.
Results: One hundred and thirty-three upregulated and 973 downregulated genes were identified in PBMCs of patients with COPD. Pathway analysis on the differentially expressed genes in COPD revealed significant enrichment in IL-8 signaling and iCOS-iCOSL signaling in T helper cells. Seventy-seven upregulated miRNAs and 43 downregulated miRNAs were differentially expressed between PBMCs from patients with COPD and normal controls. Among these 120 differentially expressed miRNAs, 42 miRNAs targeting 28 upregulated genes and 69 miRNAs targeting 498 downregulated genes were identified. The expression of CXCR1, HBEGF, TREM-1, and hsa-miR-148a-3p was more elevated in patients with COPD than in normal controls, whereas NFAT5 was decreased.
Conclusion: miRNAs and mRNAs are differentially expressed in PBMCs of patients with COPD, compared with normal controls. miRNAs regulate the expression of mRNAs, and thus play a role in the pathogenesis of COPD. Investigating these relationships may provide further insight into the mechanisms of COPD.
Keywords: COPD, whole-genome transcription, gene, miRNA, peripheral blood mononuclear cells
Introduction
Chronic obstructive pulmonary disease (COPD) is one of the most common lung diseases in clinical practice. It is a leading cause of morbidity and mortality worldwide, and causes an increasingly substantial socioeconomic burden.1 The pathophysiology of COPD is characterized by progressive airflow limitation. According to the severity of airflow limitation, COPD is classified into four grades—GOLD 1, 2, 3, and 4. Presently, the complex interactions between the environment and genes contribute to the development of COPD. Cigarette smoking is the most important COPD risk factor,1–3 with several studies reporting that smoking can lead to abnormal gene expression in different cells.4,5 Following the genome-wide analysis of lung tissues from patients with COPD, including smokers and non-smokers, certain COPD-related genes were identified.6–8 Additionally, gene expression is related to airflow limitation of COPD.9,10 Worsening airflow limitation is associated with exacerbations of COPD and a significantly increased risk of mortality in patients with GOLD 3 (severe) and GOLD 4 (very severe) COPD.1 However, the gene expression profiles of GOLD 3 and 4 COPD cases are yet to be established.
Recently, the role of microRNA (miRNA/miR) expression has been widely studied in clinical research. miRNAs are a family of small non-coding RNAs that negatively regulate gene expression post-translation. Considerable evidence has revealed that miRNAs may play important roles in the pathogenesis and development of COPD.11–16 For instance, the expression level of hsa-miR-664a-3p increased in patients with COPD, and its target gene, four and a half LIM domains 1, was downregulated with positive correlation with forced expiratory volume in one second (FEV1)/forced vital capacity (FVC).11
Peripheral blood mononuclear cells (PBMCs) can function as a critical component of immune response. COPD is a systemic disease, and blood is more accessible for genomic and biomarker studies in clinical trials and practice than lung tissue samples. Several studies on gene expression in COPD were completed through PBMC experiments.17–19
This study sought to establish dual profiles for differentially expressed mRNAs and miRNAs in the PBMCs of patients with GOLD 3 and 4 COPD, and study whether miRNA expression may be linked to mRNA expression and thus biological pathways associated with the pathogenesis of COPD.
Materials and Methods
Patients with COPD and Normal Controls
Twelve patients with COPD were recruited from the Xiangya Hospital of Central South University (Changsha, China) between May and September 2016. COPD was diagnosed using the GOLD criteria. Ethics approval was obtained from the Xiangya Hospital, and the experiments were conducted after each patient provided written informed consent. The inclusion criteria were as follows: i) Post-bronchodilator FEV1/FVC <70%; ii) FEV1 <50% predicted; iii) age, 50–75 years; iv) no other co-existing lung disease and no treatment with antibiotics in the past 4 weeks; and v) smokers or ex-smokers of at least 10 pack-years. The exclusion criteria were as follows: i) Primary diagnosis of asthma; ii) chronic inflammatory or infectious disease; iii) cancer; iv) chronic renal failure and chronic liver disease; and v) chronic antibiotic use. The clinical features of patients with COPD included in this study are shown in Table 1. Twelve non-smoking volunteers with normal lung function were concurrently recruited as normal controls. The control group comprised healthy individuals with no history of airway disease. Patients with COPD and controls were matched for age, sex, and ethnicity. All participants underwent spirometry as part of their routine clinical assessment.
 |
Table 1 Clinical Characteristics of COPD Subjects |
RNA Isolation
Peripheral blood samples were obtained from patients with COPD and normal controls. The PBMCs were isolated from heparinized blood samples by density gradient centrifugation using Ficoll-Paque Plus (GE Healthcare, Piscataway, NJ, USA). Total RNA was isolated from PBMCs by standard phenol-chloroform extraction using the TRIzol® reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. The concentration was measured on a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA quality was assessed using a Bioanalyzer Nanochip (Agilent Technologies, Santa Clara, CA, USA), and samples with an RNA integrity number >7 were used for microarray analysis.
Gene Expression Microarray
According to the standard Affymetrix protocol, 100 ng of each RNA sample was processed using an Affymetrix GeneChip Whole Transcript PLUS Reagent kit (Affymetrix, Santa Clara, CA, USA). cRNA (15 μg) was used during the second cycle of the cDNA reaction. ss-cDNA (5.5 μg) was used for fragmentation and labeling. The fragmented cDNA was labeled by terminal deoxynucleotidyl transferase using the Affymetrix proprietary DNA Labeling reagent, which is covalently linked to biotin. Hybridization cocktails containing fragmented and labeled ss-cDNA were injected into the Affymetrix Human Transcriptome Array 2.0 (Affymetrix, Santa Clara, CA, USA). Hybridization was performed at 60 rpm for 16 h at 45°C. Following washing and staining, gene chips were scanned using the Affymetrix GeneChip Command Console software (AGCC; Affymetrix, Santa Clara, CA, USA). After the scan was completed, AGCC was used to save the imaging data and compute the probe intensity data (.cel file).
miRNA Microarray
miRNAs were profiled in all patients using the Affymetrix GeneChip 4.0 Array (Affymetrix, Santa Clara, CA, USA), according to the manufacturer’s instructions. Briefly, total RNA was labeled with FlashTag Biotin HSR by Poly(A) tailing and subsequent FlashTag Biotin HSR ligation, and incubated at 99°C for 5 min and 45°C for 5 min in the Hybridization Master Mix. Arrays were hybridized at 48°C and 60 rpm for 16 h, and washed and stained with phycoerythrin-conjugated streptavidin in an Affymetrix 450 Fluidics Station (Affymetrix, Santa Clara, CA, USA). The arrays were scanned using an Affymetrix GeneChip Scanner 3000 (Affymetrix, Santa Clara, CA, USA) to generate fluorescent images, as described in the Affymetrix Gene Chip protocol. Cell intensity files were generated using AGCC (Affymetrix, Santa Clara, CA, USA). Differentially expressed miRNAs were visualized using hierarchical clustering based on average linkage and Euclidean distance, as implemented in Cluster.
Gene and miRNA Expression Analysis
Microarray data were analyzed using Affymetrix Transcriptome Analysis Console 2.0 and GeneSpring software (Silicon Genetics, Redwood City, CA, USA) successively to identify statistically significant genes (ANOVA P-value; fold change >1.5 or <–1.5). With regards to GeneSpring, raw data were imported as.cel files and underwent RMA normalization using baseline median values of all samples. In the next step, the raw data were filtered by expression signals of 20–100% to eliminate particularly weak signals on the borderline of background noise. Unpaired t-test was used to identify differentially expressed genes and miRNAs at a significance level of P <0.05 between COPD and control groups. Following false discovery rate and significance analyses, fold change (>1.5 or <–1.5) was used as the secondary criterion for the selection of differentially expressed genes and miRNAs. Using Venn diagram analysis, differentially expressed genes and miRNAs that are common between these two analysis databases (TAC and GeneSpring software) were identified. Hierarchical clustering was performed to show the different mRNA and miRNA expression profiles among samples.
Pathway and Integrative Analysis of mRNA and miRNA Expression
The biological networks, canonical pathways, and functional analyses were generated using Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity). The canonical pathways in IPA summarized the biological function of differentially expressed genes, and the significance values for the canonical pathways were calculated using Fisher’s exact right-tailed test. Significance indicated the probability of an association between differentially expressed genes and the canonical pathway by random chance only, which was referenced in the ratio of the number of genes differentially expressed that mapped to the canonical pathways divided by the total number of molecules in a certain pathway. The lists of the identified candidate genes were uploaded to the IPA site, and IPA core analysis was used to generate a list of activated or inhibited pathways and gene networks with the highest degree of significance. For differentially expressed miRNAs, all predicted targets in human genes overlapping with differentially expressed genes were obtained using the IPA software Target Filter to identify whether the gene was a predicted target of the miRNA. miRNA-mRNA pairs with statistically significant negative correlations were cross-referenced using TargetScan to elucidate the target genes of the miRNAs.
Reverse Transcription-Quantitative (RT-q) PCR Validation of the Differential mRNA and miRNA Expression
RT-qPCR was performed to verify the expression level of some candidate mRNAs and miRNAs, including C-X-C motif chemokine receptor 1 (CXCR1), heparin-binding EGF-like growth factor (HBEGF), nuclear factor of activated T-cells 5 (NFAT5), triggering receptor expressed on myeloid cells 1 (TREM-1), and hsa-miR-148a-3p. These differentially expressed genes and miRNA were validated using TaqMan assays on a 7900HT Fast Real-Time PCR system (Thermo Fisher Scientific, Inc.). The assay was performed using a TaqMan RNA-to-CT 1-Step kit (Thermo Fisher Scientific, Inc.) in a volume of 20 μL, which contained a final concentration of 900 nM sense and antisense primers, 250 nM TaqMan gene probe, 1X TaqMan RT Enzyme Mix, and 1X TaqMan RT-PCR Mix. The cDNA amplification was monitored using 7900HT Fast Real-Time PCR system under the conditions of 48°C for 15 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. All RT-qPCR procedures were performed in three independent experiments conducted in triplicate. Data are presented as 2(-ΔCq) relative to 18s RNA. Unpaired t-test was performed using SPSS version 20.0 (IBM Corp.), and P <0.05 indicated a statistically significant difference.
Results
Participant Characteristics
PBMC samples were obtained from 12 patients with COPD at GOLD stage 3 (30% ≤ FEV1 <50% predicted; n=7) or stage 4 (FEV1 <30% predicted; n=5) and 12 normal controls without airflow obstruction. The mean age of patients with COPD was 59.8±7.1 years and that of the normal controls was 58.6±6.7 years. No statistical difference in age between the two groups was observed. Participants in each group were 11 males and one female. Clinical characteristics of patients with COPD are presented in Table 1.
Identification of Differentially Expressed Genes in COPD
The whole-genome transcription profiles, including 67,528 genes (or transcripts), were compared between patients with COPD and normal controls. The differential transcriptome profiles in COPD were analyzed using TAC and Gene Spring software. A total of 2412 upregulated and 2622 downregulated genes were initially identified in patients with COPD using the TAC software (Supplemental Table 1A–C; Table S1A–C). Additionally, 161 upregulated and 1223 downregulated genes were found to be aberrantly expressed in patients with COPD using the GeneSpring software (Supplemental Table 2A–C; Table S2A–C). Integrated comparison by Venn diagram analysis identified 1106 common genes between the two mRNA profiles (Supplemental Table 3A–C; Table S3A–C). As shown in the heat map, the 1106 genes clearly differentiated the COPD cases from the normal controls (Figure 1). Among the 1106 differentially expressed genes, 133 genes were upregulated, 113 (85%) of which were coding genes, and 973 genes were downregulated, 638 (66%) of which were coding genes.
Differentially Expressed miRNAs in COPD
The miRNA data were analyzed using the same protocol as in mRNA data analysis. Using the TAC software, 148 miRNAs, which were differentially expressed in COPD, were identified in human PBMCs. Among these 148 miRNAs, 104 miRNAs were upregulated, and 44 miRNAs were downregulated (Supplemental Table 4A–C; Table S4A–S4C). A total of 689 upregulated and 215 downregulated miRNAs were identified using Gene Spring software (Supplemental Table 5A–C; Table S5A–C). Venn diagram analysis revealed 120 common miRNAs, as illustrated in Figure 2. The differentially expressed miRNAs in COPD included 77 upregulated and 43 downregulated miRNAs (Supplemental Table 6A–C; Table S6A–C).
Canonical Pathways of Differential Gene Expression by the IPA Software
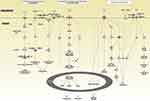
Biological network and pathway analyses were performed using the IPA software to identify canonical biological pathways of the differentially expressed genes. The canonical pathways by which the 133 upregulated genes were mostly enriched are shown in Figure 3. The top five pathways involving the upregulated genes included “granulocyte adhesion and diapedesis”, “role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis”, “interleukin (IL)-8 signaling”, “glucocorticoid receptor signaling and role of osteoblasts”, and “osteoclasts and chondrocytes in rheumatoid arthritis.” Figure 4 illustrates the canonical pathways by which the 973 downregulated genes were mostly enriched. The top five pathways involving the downregulated genes are “natural killer cell signaling”, “crosstalk between dendritic cells and natural killer cells”, “inducible co-stimulator (iCOS)-iCOS ligand (iCOSL) signaling in T helper cells”, “glucocorticoid receptor signaling”, and “role of BRCA1 in DNA damage response”.
The predicted pathway activation or inhibition was assessed with the z-score and ratio. Among the canonical pathways modulated by aberrantly expressed genes in COPD, the IL-8 signaling pathway showed the highest significance, represented by the upregulated genes in COPD (Figure 5). Five genes in the IL-8 signaling pathway, including C-X-C motif chemokine receptor 1(CXCR1), C-X-C motif chemokine receptor 2 (CXCR2), heparin binding EGF-like growth factor (HBEGF), insulin receptor substrate 2 (IRS2), and LIM domain kinase 2 (LIMK2), were overexpressed in patients with COPD, as compared with the normal control group (Figure 6). This finding suggests that the activation of the IL-8 signaling pathway may play a significant role in the pathogenesis of COPD. Similarly, downregulated genes were highly enriched in iCOS-iCOSL signaling in the T helper cell pathway in COPD. As shown in Figure 7, the expression levels of AKT serine/threonine kinase 3 (AKT3), ATM serine/threonine kinase (ATM), calcium/calmodulin-dependent protein kinase IV (CAMK4), CD40 ligand (CD40LG), lymphocyte cytosolic protein 2 (LCP2), nuclear factor of activated T-cells 5 (NFAT5), nuclear factor of activated T cells 2 (NFATC2), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha (PIK3C2A), and T cell receptor-associated transmembrane adaptor 1 (TRAT1) were reduced in patients with COPD. The downregulation of these genes in iCOS-iCOSL signaling in the T helper cell pathway indicated a dysregulated T cell response in COPD.
miRNA-mRNA Interactions and Target Gene Prediction
To elucidate the regulatory role of miRNA on mRNA expression in COPD, the IPA Target Filter was applied to the miRNA and mRNA list based on the source and confidence level of the miRNA-mRNA target relationships. Of the 120 differentially expressed miRNAs, 42 putative miRNAs targeting 28 upregulated genes and 69 putative miRNAs targeting 498 downregulated genes were identified. Among them, 18 high-confidence miRNA-upregulated mRNA pairs and 561 high-confidence miRNA-downregulated mRNA pairs in human PBMCs were identified (Supplemental Table 7A and B; Table S7A and B). Differentially expressed genes involved in the IL-8 signaling and iCOS-iCOSL signaling in T helper cell pathways were also regulated by certain miRNAs (see Tables 2 and 3). Our analysis showed that the expression of hsa-miR-4736 significantly decreased by 2.3-fold, targeting four genes in the IL-8 signaling pathway (CXCR1, CXCR2, HBEGF, and LIMK2). In addition, the expression of hsa-miR-148a-3p increased by 2.54-fold, which was linked to the downregulation of four genes involved in iCOS-iCOSL signaling in the T helper cell pathway (LCP2, NFAT5, PIK3CA, and PIK3C2A). The inverse relationship between the expression level of miRNAs and mRNAs indicated the regulatory role of miRNAs on aberrant gene expression in COPD.
 |
Table 2 Differentially Expressed miRNAs Targeting Upregulated Genes Involved in the IL-8 Signaling Pathway |
 |
Table 3 Differentially Expressed miRNAs Targeting Downregulated Genes Involved in iCOS-iCOSL Signaling T Helper Cells Pathway |
RT-qPCR Validation of Differentially Expressed Genes and miRNAs
The expression of CXCR1, HBEGF, NFAT5, and TREM-1 was further confirmed using RT-qPCR based on their differential expression in COPD and/or their significant enrichment in the identified pathways. Additionally, hsa-miR-148a-3p was validated by the markedly increased expression in COPD and the finding that it had target genes. As illustrated in Figure 8, the expressions of hsa-miR-148a-3p, CXCR1, HBEGF, NFAT5, and TREM-1 in patients with COPD by RT-qPCR were different (P <0.05) from those in the normal controls, which was consistent with the microarray data.
Discussion
This study performed a comprehensive analysis of miRNA and mRNA expression in PBMCs obtained from patients with GOLD 3 and 4 COPD, and compared it to that of normal controls. Although multiple studies have been conducted on gene expression in COPD, to the best of our knowledge, no studies on miRNA-mRNA regulation in PBMCs from patients with GOLD 3 and 4 COPD have been conducted.
Several genes (133 upregulated and 973 downregulated genes) were found to be differentially expressed in COPD, compared with the controls. Further pathway analysis was performed to elucidate the potential pathways regulated by the aberrantly expressed genes in COPD. As expected, the upregulated genes in COPD were significantly enriched in inflammatory reaction and inflammation regulatory pathways, such as “granulocyte adhesion and diapedesis”, “role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis”, and “IL-8 signaling”. Particularly, the ‘granulocyte adhesion and diapedesis’ and “IL-8 signaling” pathways were found to be strongly associated with COPD. The downregulated genes in COPD were enriched in immune regulatory pathways, such as “iCOS-iCOSL signaling in T helper cells”, “natural killer cell signaling”, and “crosstalk between dendritic cells and natural killer cells”.
Several genes that participate in the IL-8 signaling pathway were differentially expressed in the current data. CXCR1, CXCR2, HBEGF, IRS2, and LIMK2 were upregulated. CXCR1 and HBEGF expression was validated by RT-qPCR. IL-8 is a member of the C-X-C family of chemokines that play a significant role in inflammation and angiogenesis. The cell surface receptors for IL-8, which are coupled to G proteins, include CXCR1 and CXCR2. CXCR1 is selectively activated by IL-8 only. IL-8 induces NF-κB through a TNF receptor-associated factor 6-dependent pathway, leading to inflammation and angiogenesis. Activation by IL-8 can trigger inflammation in cells, leading to chemotaxis, respiratory burst, granule release, and increased cell adhesion. IL-8 has been the subject of several prior investigations in COPD. Studies have shown that IL-8 levels are significantly elevated in the blood of patients with COPD, and that IL-8 levels directly correlate with mortality, exacerbation rate, and BODE scores, and inversely correlate with FEV1 and diffusing capacity of the lung for carbon monoxide.20–22 In summary, several factors in COPD cause an increase in IL-8 expression, which in turn further induces an inflammatory response, leading to persistent chronic inflammation.
Similarly, iCOS-iCOSL signaling in the T helper cell pathway is another molecular pathway in which downregulated genes were found to be enriched in COPD in the present study. In total, 10 downregulated genes were involved in the regulation of iCOS-iCOSL signaling in the T helper cell pathway. Among them, NFAT5 was an important member of the pathway, and its expression was validated using RT-qPCR. During an immune response, T cells are optimally activated to appropriately migrate into areas of inflamed tissues. iCOS-mediated signaling plays important physiological roles in the regulation of T helper type 1 (Th1) cells in the endothelium and control of the selective entry of Th1 cells into inflamed peripheral tissues. The potency of iCOS is enhanced following its ligation to iCOSL. Despite its inflammatory etiology, COPD is considered an immune-deficient state, since the abundant activated T cells in the airways of patients with COPD do not eradicate bacterial infections. Although Th1 cells are crucial for immune responses to bacterial infections,23 they display impaired immune response in COPD, such as the production of interferon-γ and phagocytosis.23–26 Impaired Th1 function is implicated in the susceptibility of patients with COPD to respiratory infections, which are common causes of acute exacerbation of COPD (AECOPD).26 The present data revealed aberrant gene expression in iCOS- iCOSL signaling in the T helper cell pathway in COPD, suggesting a potential mechanism of impaired T cell response in COPD. This implied that patients with GOLD 3 and 4 COPD are more susceptible to infections and acute exacerbations, which may be due to impaired immune function.
TREM-1 is an activating receptor on neutrophil, monocyte, and macrophage subsets. Its expression is upregulated by microbial products.27 TREM-1 is considered to amplify both infectious and non-infectious inflammation28 and to elicit the release of TNF-α, IL-8 myeloperoxidase, and nitric oxide by innate immune cells.29 According to previous reports, levels of soluble TREM-1 in the serum of patients with stable COPD and AECOPD significantly increase.27,30 Furthermore, serum levels of sTREM-1 were shown to be significantly negatively correlated with lung function impairment.30 In the present study, TREM-1 was highly expressed in the PBMCs of patients with COPD. Therefore, we hypothesized that TREM-1 plays a significant role in the pathogenesis of COPD. However, the mechanism remains unclear and requires further research.
Following miRNA expression analysis at the whole-genome level, a group of miRNAs that were differentially expressed in COPD subjects, compared with normal controls, was identified. Due to the distinct characteristics of participants and the different methods used, it was possible for results to be different. The present study compared the obtained list of differentially expressed miRNAs with those previously reported in COPD. The miRNAs that were differentially expressed in this study and previous studies, included hsa-miR-19a-3p,31 hsa-miR-29b-3p,32,33 hsa-miR-183-5p,31 hsa-miR-223,31,34 hsa-miR-142-5p,31 hsa-miR-451,35 hsa-miR-664,35 hsa-miR-148a,35 and hsa-miR-99b.35 Following integrated analysis of gene transcription and miRNA expression, 42 miRNAs targeting 28 upregulated genes and 69 miRNAs targeting 498 downregulated genes were identified, suggesting that miRNAs may have a regulatory effect on most of the differentially expressed genes in COPD. This study validated the significant upregulation of hsa-miR-148a-3p in PBMCs from patients with COPD, but its functional role in COPD has not been studied. Downregulated genes, including LCP2, NFAT5, PIK3CA, and PIK3C2A, were predicted to be targeted genes of hsa-miR-148a-3p, and were involved in iCOS-iCOSL signaling in the T helper cell pathway. Therefore, hsa-miR-148a-3p and its targeted genes may play a significant role in COPD; however, the exact mechanisms require further study. Profiling mRNA and miRNA expression in the same set of COPD and control samples enabled us to investigate the complexity of gene dysregulation in the context of this disease. Investigating these relationships may provide further insight into the mechanisms of COPD.
Conclusion
In conclusion, miRNAs and mRNAs are differentially expressed in PBMCs of patients with COPD, compared with normal controls. miRNAs regulate the expression of mRNAs, and, therefore, play a role in the pathogenesis of COPD. This study had limitations. First, it included a relatively small sample size, and the participants were almost exclusively male (91.7%). Second, the study was restricted to participants at relatively high-risk; 58.3% of patients with COPD were at GOLD stage 3 and 41.7% were at stage 4. Further studies are required to confirm the role of certain differentially expressed genes and miRNAs and the underlying regulatory mechanism.
Ethics Approval and Consent to Participate
The study was approved by the Xiangya Hospital of Central South University Institutional Review Board Committees. Patients provided written informed consent before participation.
Consent for Publication
All patient data were de-identified.
Acknowledgments
The authors thank all staff of the Microarray Core Lab of the Department of Immunology, University of Texas Southwestern Medical Center, for their selfless help.
Funding
This research was supported by the Freedom Explore Program of Central South University (grant no. 2012QNZT128). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare no competing conflicts of interest for this work.
References
1. Global strategy for prevention, diagnosis and management of COPD: 2022 Report. Available from: https://goldcopd.org/2022-gold-reports-2/.
2. Postma DS, Bush A, van den Berge M. Risk factors and early origins of chronic obstructive pulmonary disease. Lancet. 2015;385(9971):899–909. doi:10.1016/S0140-6736(14)60446-3
3. Berndt A, Leme AS, Shapiro SD. Emerging genetics of COPD. EMBO Mol Med. 2012;4:1144–1155. doi:10.1002/emmm.201100627
4. Sundar IK, Rahman I. Gene expression profiling of epigenetic chromatin modification enzymes and histone marks by cigarette smoke: implications for COPD and lung cancer. Am J Physiol Lung Cell Mol Physiol. 2016;311(6):L1245–L1258. doi:10.1152/ajplung.00253
5. Aguiar JA, Tamminga A, Lobb B, et al. The impact of cigarette smoke exposure, COPD, or asthma status on ABC transporter gene expression in human airway epithelial cells. Sci Rep. 2019;9(1):153. doi:10.1038/s41598-018-36248-9
6. de Vries M, Faiz A, Woldhuis RR, et al. Lung tissue gene-expression signature for the ageing lung in COPD. Thorax. 2017. doi:10.1136/thoraxjnl-2017-210074
7. Morrow JD, Zhou X, Lao T, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep. 2017;7:44232. doi:10.1038/srep44232
8. Morrow JD, Cho MH, Platig J, et al. Ensemble genomic analysis in human lung tissue identifies novel genes for chronic obstructive pulmonary disease. Hum Genomics. 2018;12(1):1. doi:10.1186/s40246-018-0132-z
9. Bhattacharya S, Srisuma S, Demeo DL, et al. Molecular biomarkers for quantitative and discrete COPD phenotypes. Am J Respir Cell Mol Biol. 2009;40:359–367. doi:10.1165/rcmb.2008-0114OC
10. Savarimuthu Francis SM, Larsen JE, Pavey SJ, et al. Genes and gene ontologies common to airflow obstruction and emphysema in the lungs of patients with COPD. PLoS One. 2011;6(3):e17442. doi:10.1371/journal.pone.0017442
11. Zhong S, Chen C, Liu N, et al. Overexpression of hsa-miR-664a-3p is associated with cigarette smoke-induced chronic obstructive pulmonary disease via targeting FHL1. Int J Chron Obstruct Pulmon Dis. 2019;14:2319–2329. doi:10.2147/COPD.S224763
12. Tang K, Zhao J, Xie J, et al. Decreased miR-29b expression is associated with airway inflammation in chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. 2019;316(4):621–629. doi:10.1152/ajplung.00436.2018
13. Xue H, Li MX. MicroRNA-150 protects against cigarette smoke- induced lung inflammation and airway epithelial cell apoptosis through repressing p53: microRNA-150 in CS-induced lung inflammation. Hum Exp Toxicol. 2018;37(9):920–928. doi:10.1177/0960327117741749
14. Shen W, Liu J, Zhao G, et al. Repression of toll-like receptor-4 by microRNA-149-3p is associated with smoking-related COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:705–715. doi:10.2147/COPD.S128031
15. Conickx G, Mestdagh P, Avila Cobos F, et al. MicroRNA profiling reveals a role for microRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2017;195(1):43–56. doi:10.1164/rccm.201506-1182OC
16. Xu H, Sun Q, Lu L, et al. MicroRNA-218 acts by repressing TNFR1-mediated activation of NF-κB, which is involved in MUC5AC hyper-production and inflammation in smoking-induced bronchiolitis of COPD. Toxicol Lett. 2017;280:171–180. doi:10.1016/j.toxlet.2017.08.079
17. Qu X, Dang X, Wang W, et al. Long noncoding RNAs and mRNA regulation in peripheral blood mononuclear cells of patients with chronic obstructive pulmonary disease. Mediators Inflamm. 2018;2018:7501851. doi:10.1155/2018/7501851
18. Pniewska E, Sokolowska M, Kuprys-Lipinska I, et al. Exacerbating factors induce different gene expression profiles in peripheral blood mononuclear cells from asthmatics, patients with chronic obstructive pulmonary disease and healthy subjects. Int Arch Allergy Immunol. 2014;165(4):229–243. doi:10.1159/000370067
19. Bahr TM, Hughes GJ, Armstrong M, et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2013;49(49):316–323. doi:10.1165/rcmb.2012-0230OC
20. Celli BR, Locantore N, Yates J, et al. Inflammatory biomarkers improve clinical prediction of mortality in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1065–1072. doi:10.1164/rccm.201110-1792OC
21. Pinto-Plata V, Toso J, Lee K, et al. Profiling serum biomarkers in patients with COPD: associations with clinical parameters. Thorax. 2007;62(7):595–601. doi:10.1136/thx.2006.064428
22. Agusti A, Edwards LD, Rennard SI, et al. Persistent systemic inflammation is associated with poor clinical outcomes in COPD: a novel phenotype. PLoS One. 2012;7(5):e37483. doi:10.1371/journal.pone.0037483
23. Knobloch J, Chikosi SJ, Yanik S, et al. A systemic defect in toll-like receptor 4 signaling increases lipopolysaccharide-induced suppression of IL-2-dependent T-cell proliferation in COPD. Am J Physiol Lung Cell Mol Physiol. 2016;310(1):L24–L39. doi:10.1152/ajplung.00367.2014
24. Bengoechea JA, Ito K. Chronic obstructive pulmonary disease Th1 cells display impaired response to endotoxin. Am J Respir Crit Care Med. 2011;183(2):148–150. doi:10.1164/rccm.201008-1275ED
25. Knobloch J, Schild K, Jungck D, et al. The T-helper cell type 1 immune response to gram- negative bacterial infections is impaired in COPD. Am J Respir Crit Care Med. 2011;183(2):204–214. doi:10.1164/rccm.201002-0199OC
26. Tan DBA, Teo TH, Setiawan AM, et al. Increased CTLA-4+ T cells may contribute to impaired T helper type 1 immune responses in patients with chronic obstructive pulmonary disease. Immunology. 2017;151(2):219–226. doi:10.1111/imm.12725
27. Rohde G, Radsak MP, Borg I, et al. Levels of soluble triggering receptor expressed on myeloid cells 1 in infectious exacerbations of chronic obstructive pulmonary disease. Respiration. 2012;83(2):133–139. doi:10.1159/000328413
28. Tammaro A, Derive M, Gibot S, et al. TREM-1 and its potential ligands in non-infectious diseases: from biology to clinical perspectives. Pharmacol Ther. 2017;177:81–95. doi:10.1016/j.pharmthera.2017.02.043
29. Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164(10):4991–4995. doi:10.4049/jimmunol.164.10.4991
30. Radsak MP, Taube C, Haselmayer P, et al. Soluble triggering receptor expressed on myeloid cells 1 is released in patients with stable chronic obstructive pulmonary disease. Clin Dev Immunol. 2007;2007:52040. doi:10.1155/2007/52040
31. Wang R, Xu J, Liu H, et al. Peripheral leukocytes as novel biomarkers for COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:1101–1112. doi:10.2147/COPD.S130416;.
32. Molina-Pinelo S, Pastor MD, Suarez R, et al. MicroRNA clusters: dysregulation in lung adenocarcinoma and COPD. Eur Respir J. 2014;43(6):1740–1749. doi:10.1183/09031936.00091513
33. Soeda S, Ohyashiki JH, Ohtsuki K, et al. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int J Mol Med. 2013;31(3):533–539. doi:10.3892/ijmm.2013.1251
34. Hua L, Zheng W, Xia H, et al. Integration of multi-microarray datasets to identify chronic obstructive pulmonary disease-related miRNAs. Biomed Mater Eng. 2015;26(Suppl 1):S1903–S1915. doi:10.3233/BME-151493
35. Ezzie ME, Crawford M, Cho JH, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. 2012;67(2):122–131. doi:10.1136/thoraxjnl-2011-200089
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.