Back to Journals » OncoTargets and Therapy » Volume 13
Peripheral Blood Leukocyte N6-methyladenosine is a Noninvasive Biomarker for Non-small-cell Lung Carcinoma
Authors Pei Y , Lou X, Li K, Xu X, Guo Y, Xu D, Yang Z, Xu D, Cui W, Zhang D
Received 11 June 2020
Accepted for publication 15 October 2020
Published 19 November 2020 Volume 2020:13 Pages 11913—11921
DOI https://doi.org/10.2147/OTT.S267344
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yuqing Pei,1 Xiaoying Lou,1 Kexin Li,1 Xiaotian Xu,1 Ye Guo,2 Danfei Xu,1 Zhenxi Yang,1 Dongsheng Xu,3 Wei Cui,1 Donghong Zhang4
1State Key Laboratory of Molecular Oncology, Department of Clinical Laboratory, National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, People’s Republic of China; 2Department of Laboratory Medicine, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100730, People’s Republic of China; 3Hematopathology Program, CBL Path, Inc, Rye Brook, NY 10753, USA; 4Center for Molecular and Translational Medicine, Georgia State University, Research Science Center, Atlanta, GA 30303, USA
Correspondence: Wei Cui; Donghong Zhang Tel +86-87788448
Email [email protected]; [email protected]
Background: N6-methyladenosine (m6A) triggers a new layer of epi-transcription. However, the potential noninvasive screening and diagnostic value of peripheral blood m6A for cancer are still unknown. Here, we intend to investigate whether leukocyte m6A can be a novel biomarker for non-small-cell lung cancer (NSCLC).
Materials and Methods: Peripheral blood was collected from 119 NSCLC patients and 74 age-matched healthy controls. Total RNA was isolated from leukocytes for m6A measurement, and clinical information of participants was reviewed. The sensitivity, specificity, and area under the curve (AUC) of m6A for cancer diagnosis were evaluated by the receiver-operating characteristic (ROC) curve analysis. Flow cytometry and the Human Protein Atlas (HPA) database were used to characterize m6A in leukocyte differentials. Pearson’s correlation was applied to indicate the relationship between m6A level and hematology variables. qPCR and bioinformatic analysis were used to identity the expression of m6A regulators in leukocyte.
Results: Leukocyte m6A was significantly elevated in 119 NSCLC patients compared with 74 healthy controls (P< 0.001). We did not find significant association between m6A and age or gender. Elevated m6A level in NSCLC was associated with tumor stage (P< 0.05) and tumor differentiation (P< 0.05), and was significantly reduced after surgery (P< 0.01). ROC curve analysis revealed that leukocyte m6A could significantly discriminate patients with lung adenocarcinoma (LUAD) (AUC=0.736, P< 0.001) and lung squamous cell carcinoma (LUSC) (AUC=0.963, P< 0.001) from healthy individuals. m6A displayed superior sensitivity (100%) and specificity (85.7%) for LUSC than squamous cell carcinoma (SCC) antigen and cytokeratin fragment 211 (Cyfra211). Flow cytometry analysis showed m6A modification was mainly localized on T cells and monocytes among leukocyte differentials. Leukocyte m6A was positively correlated with the number of lymphocytes and negatively correlated with monocytes in NSCLC but not in healthy controls. qPCR and bioinformatic analysis showed that elevated leukocyte m6A in NSCLC was caused by upregulated methyltransferase complex and downregulated FTO and ALKBH5.
Conclusion: Leukocyte m6A represents a potential noninvasive biomarker for NSCLC screening, monitoring and diagnosis.
Keywords: leukocyte, N6-methyladenosine, lung cancer, biomarker
Introduction
Recent studies have demonstrated that epigenetic alterations to genomic DNA, histone modifications and microRNA (miRNA) play an important role in cancer initiation and progression by controlling gene expression and chromatin structure.1,2 Global DNA hypomethylation in peripheral blood cells is also associated with or increases the risk of various cancers, including breast,3 lung,4 prostate,5 ovary,6 endometrial,7 colon and rectum.8 Although epigenetic alterations in peripheral blood cells may not exactly represent the epigenetic changes in the primary tumors, global epigenetic alterations can reflect individual genomic instability and serve as an integrated exposure signature of multiple known and unknown carcinogenic factors.9,10 Therefore, epigenetic biomarkers in peripheral blood cells are becoming a new tool to predict the risk and prognosis of various cancers.11
Reversible N6-methyladenosine (m6A), the most abundant RNA modification in eukaryotic species, represents a new area for biological regulation in the form of “RNA epigenetics”,12 and has potential functions on the regulation of gene post-transcription and protein translation.13,14 The effects of m6A on RNA depend on the dynamic interaction among its methyltransferase complex (“writers”) of METTL3, METTL14 and WTAP, tow demethylases (“erasers”) FTO and ALKBH5, and binding proteins (“readers”). Several studies have revealed the functions and underlying molecular mechanisms of m6A in various cancer development, progression and treatment. Aberrant m6A modification has been suggested to serve as a biomarker for early-stage cancer diagnosis as well as a new therapeutic target in various cancers.15–18 As most studies focus on tumor-intrinsic oncogenic pathways, potential roles of the mRNA m6A modification in peripheral blood are largely unknown.
This study is to investigate the potential diagnostic and prognostic values of peripheral leukocyte m6A in patients with lung carcinoma, the leading cause of cancer death worldwide that is susceptible to epigenetic regulation.19,20 We also aim to detect m6A modification in leukocyte differentials and characterize their m6A regulatory enzymes.
Materials and Methods
Participants and Blood Sample Collection
This is a randomized case–control, cross-sectional and hospital-based study. A total of 119 newly diagnosed NSCLC patients including 91 lung adenocarcinoma (LUAD) and 28 lung squamous cell carcinoma (LUSC), as well as 74 age- and sex-matched healthy controls (NC) were recruited from the Peking Union Medical College Cancer Hospital, China, between September 2018 and November 2019. Forty-two patients were followed and had the blood samples both at time of first diagnosis and one week after surgery. All the diagnoses were confirmed by histopathology or cytopathology. Clinical information, such as cancer pathological type, stage, differentiation, lymph node metastasis, treatment were reviewed retrospectively. Patients with hypertension, diabetes, myocardial infarction, stroke, renal failure, aneurysm or other serious diseases were excluded. One milliliter of venous blood from each participant was collected in BD Vacutainer Plus K2 EDTA. After removing red blood cells by using red blood cell lysis buffer, leukocytes were harvested and stored at −80°C within two hours to ensure RNA integrity.
This study protocol was in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and approved by the Ethics Committee of Peking Union Medical College Cancer Hospital (grant no: NCC2017G-115). Written informed consent was obtained from all participants of this study.
Biochemical Measurements
Serum tumor biomarkers, including cancer antigen 125 (CA125), carcinoembryonic antigen (CEA), squamous cell carcinoma (SCC) antigen and cytokeratin fragment 211 (Cyfra211) were determined by the direct chemiluminescence method (Beckman).
Total RNA Isolation
Total RNA from blood leukocytes were isolated using TRIzol reagent (Invitrogen) and treated by DNase I (M6101, Promega) to remove genomic DNA. Gel electrophoresis was used for the integrity of RNA measurement, and only intact RNA was used for RNA concentration determination using Nanodrop 2000 (Thermo Fisher). The samples with OD 260/280 nm ratio ≈2 and 260/230 nm values 2.0–2.2 were used for further experiments.
m6A Measurement
m6A modification on RNA was determined in duplicates using the EpiQuick RNA Methylation Quantification Kit (Colorimetric, Epigentek Group, USA). Briefly, 200 ng of total RNA was bound to strip wells using binding solution, incubated at 37 °C for 90 min. After washing three times, capture antibody was added, incubated at room temperature for 60 min, and washed. Detection antibody was then incubated at room temperature for 30 min and washed. The enhancer solution for 30 min and the determined solution for 10 min away from the light were followed. Finally, 100 μL of stop solution was added to quench the enzyme reaction and measurement was performed at wavelength of 450 nm on a microplate reader. The readings were used to calculate the absolute quantification of m6A% by using a formula provided by the manufacturer.
Flow Cytometry Analysis
The m6A modification in leukocyte differentials were quantified by FACSCanto II flow cytometry (BD Biosciences, Shanghai, China) in an observer‐blinded way and using the following antibodies: anti‐CD3 (APC-H7), anti-CD45 (PerCP) and anti-m6A (FITC). Data were analyzed using FlowJo software (Tree Star Inc, Ashland, OR, USA) and BD FACSDiva Software (BD Biosciences).
Bioinformatic Analysis
The whole blood gene expression profiling from 164 patients with malignant lung nodules (MN) and 151 patients with benign lung nodules (BN) were obtained from the Gene Expression Omnibus (GEO) database (GSE108375). Limma packages were used to identify the differentially expressed m6A regulators (P<0.05 and |log2FC|>0.5) and ggplot packages to visualize the results in the form of a volcano plot. The RNA-seq data of m6A regulators in different blood cell types was downloaded from the Human Protein Atlas (HPA) database. Color-coding is based on blood cell type lineages.
Statistical Analysis
All analyses were performed using SPSS 16.0 (SPSS Inc, Chicago, IL, USA). Data for clinical and biological characteristics of patients are expressed as number (%), mean ±SD or median (P25, P75). Comparison of patients and controls involved independent-sample Student's t-test (unpaired), one-way ANOVA and Mann–Whitney U-test for continuous variables. Pearson’s correlation was applied to indicate the relationship between m6A level and other variables. Receiver-operating characteristic (ROC) curves were used to evaluate the sensitivity and specificity. Two-sided P<0.05 indicated statistical significance.
Results
The Level of Leukocyte m6A Methylation is Elevated in Non-small-cell Lung Carcinoma
Baseline demographics of lung cancer patients and controls are listed in Table 1. Overall, the value of leukocyte m6A in NSCLC patients was significantly higher than that of healthy controls (0.085±0.033% vs 0.056%±0.031%, P<0.001) (Table 1, Figure 1A). There was a positive tendency but not statistically significant correlation between the level of m6A and age (Figure 1B). So far, we did not find significant difference in m6A between males and females (Figure 1C). These observations suggest that NSCLC patients displayed abnormal leukocyte m6A level.
 |
Table 1 Baseline Demographics of the Cancer Patients and Controls |
Leukocyte m6A Has a Potential Diagnostic Value in Non-small-cell Lung Carcinoma
We further evaluated potential diagnostic value of leukocyte m6A in NSCLC using ROC analysis, in comparison with other common serum tumor biomarkers (Figure 2, Table 2). In general, leukocyte m6A could significantly discriminate patients with NSCLC from healthy individuals (AUC=0.738, P<0.001) (Figure 2A). Specifically, leukocyte m6A showed a superior diagnostic accuracy in LUSC (AUC=0.963, P<0.001) than SCC and Cyfra211 (Table 2; Figure 2B), with the sensitivity of 100% and the specificity of 85.7% at the optimal cutoff value of 0.043%. In LUAD, m6A level presented better diagnostic accuracy (AUC=0.736, P<0.001) than CEA and CA125, with the sensitivity of 60.6%, and the specificity of 78.0% at the optimal cutoff value of 0.08% (Table 2; Figure 2C). After combining m6A with serum tumor biomarkers, the accuracy rate increased in both subtypes (AUC=0.988 and 0.763, P<0.001). The sensitivities for LUSC and LUAD were 91.3% and 74.6%, and the specificities were 100% and 68% respectively (Table 2; Figure 2B and C).
 |
Table 2 Diagnostic Performance of Leukocyte m6A and Serum Biomarkers in NSCLC by ROC Analysis |
Leukocyte m6A Associates with Clinical Features in Non-small-cell Lung Carcinoma
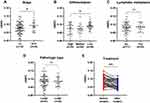
To determine the association of leukocyte m6A with prognostic-related clinical features, information regarding cancer stage, differentiation, pathological type, and lymph node metastasis was reviewed. We also followed 42 patients and examined their leukocyte m6A at first diagnosis and after surgery. The results showed that patients with advanced stage displayed a higher m6A level than those with early stage (P=0.030) (Figure 3A). Furthermore, poorly differentiated NSCLC presented higher m6A level than moderately and well-differentiated NSCLC (P=0.022) (Figure 3B). No statistically significant differences of leukocyte m6A in terms of lymph node metastasis and pathologic type were found (Figure 3C and D). Among the patients who are followed, 27 out of 42 patients showed a remarkable decreased m6A level after surgery (P<0.01), while 12 remained steady and three showed a slight increase (Figure 3E).
Identify m6A Modification in Leukocyte Differentials
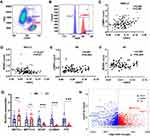
To explore the reason of elevated leukocyte m6A in lung cancer, we conducted a flow cytometry analysis with m6A antibody in leukocyte differentials in both NSCLC and NC blood. To our surprise, m6A modification was only detected in lymphocytes and monocytes, both of which show a high fluorescence intensity value, while this modification seemed deficient in neutrophils (Figure 4A and B). We further explored the Human Protein Atlas (HPA) database and found the m6A writers (METTL3, METTL14, WTAP) and erasers (FTO, ALKBH5) were highly expressed in lymphocytes, especially in T cells, medially in monocyte and dendric cells and low in granulocytes, which was consistent with our observations (supplementary Figure 1). We then performed a correlation analysis of the counts of lymphocytes and monocytes with m6A both in NSCLC and NC. It turned out that m6A level was positively correlated with the number of lymphocytes (r=0.299, P<0.001), while negatively with monocytes in NSCLC (r=−0.227, P=0.01) (Figure 4C and D). However, we did not find similar correlations in the NC group (Figure 4E and F).
After detecting the expression of METTL3, METTL14, WTAP, FTO and ALKBH5 in leukocyte in both NSCLC and NC groups, we found that the core methyltransferase, METTL3, was upregulated in NSCLC (P<0.01), while the two m6A demethylase, FTO and ALKBH5, were downregulated in NSCLC (P<0.001) (Figure 4G). We further analyzed the expression of 22 reported m6A regulators in the blood of patients with benign or malignant lung nodules (GSE108375). It was demonstrated that the expression of m6A regulators were largely changed in two groups. METTL14 and RBM15, two m6A writers, were significantly upregulated in the blood of LC patients. FTO, ALKBH5, the m6A erasers, together with several m6A readers, were significantly downregulated in LC blood. Therefore, elevated leukocyte m6A in NSCLC might result from high level of m6A methyltransferase complex and low level of FTO and ALKBH5 in cancer status.
Discussion
“Epigenetic cancer epidemiology” studies aim to identify biomarkers of cancer risk or prevention, especially when abnormal epigenetic patterns can be detected in easily accessible tissues such as peripheral blood.21 Previously, we and others have demonstrated that epigenetic variability, especially DNA methylation, may contribute to the risk of cancer development.2,22,23 Currently, there were many commercially available biomarkers for various tumor screening, diagnosis, monitoring therapeutic effectiveness or tumor prognosis. Few showed satisfactory utility for NSCLC. Here, we investigated the dynamic changes of m6A level in peripheral blood from individuals with NSCLC, which add a new epigenetic biomarker that may contribute to cancer screening and diagnosis.
N6-methyladenosine (m6A), as the most abundant internal modification of RNA in eukaryotic cells, affects multiple aspects of RNA metabolism, ranging from pre-RNA processing, mRNA stability and translation.16 Emerging evidence suggests that m6A-modulating proteins represent a double-edged sword for cancer by targeting genes for oncogenic protein expression, cancer cell proliferation, survival, tumor initiation, and progression.17,24–26 Clarifying the molecular mechanisms that mediate these m6A modifications in RNA and identifying the aberrant expression of m6A regulatory factors in clinical biopsy specimens could contribute to the early diagnosis of cancer, prediction of cancer prognosis, and development of novel therapeutic approaches.24,27
This current investigation is the first study, to the best of our knowledge, to indicate the dynamic changes in leukocyte m6A and its association with NSCLC development. m6A showed a better performance in ROC curve analysis as compared with common serum tumor biomarkers such as CEA, CA125, SCC and Cyfra211. In particular, m6A showed a superior diagnostic accuracy in squamous cell lung carcinoma than biomarkers, SCC and Cyfra211 with the sensitivity of 100% and the specificity of 85.7%, which highlights the potential utility of m6A as a screening, monitoring, or diagnostic biomarker for NSCLC. Notably, elevated leukocyte m6A in NSCLC was associated with tumor stage and differentiation and was reduced to the control level after surgery. The phenomenon is consistent with the results of several studies based on the dysregulation of tumor-tissue–resident m6A.28–30
Accumulating evidence has supported the view that cancer is a systemic disease. Like DNA methylation, RNA methylation changes in peripheral blood might reflect the abnormal methylation status of the whole body. This random and global process might not be limited to a certain area or cell type, not just in foci where tumors are localized.31,32 In fact, it has been known that cancer can induce distant changes on myeloid cells function, mobilization, and differentiation, well before clinically evident metastasis develops.33 In this study, we identified elevated leukocyte m6A were mainly from lymphocytes and monocytes, and they had a strong correlation in the NSCLC group, but not in the NC group. Furthermore, the HPA database showed that CD4+ and CD8+ T cells, the two effective T cells in a cancer response, displayed a high level of ALKBH5 and FTO. Effective CD4+ and CD8+ T cells were even higher in the expression of the demethylases than their naïve counterparts. In contrast, regulatory T (T-reg) cells, which play a major role in immunosuppressive tumor microenvironment, displayed a high level of m6A methyltransferase. These results imply that elevated leukocyte m6A modification in cancer might be related to the disfunction of immune-related cells.
In fact, several studies have explored the issue recently. Li et al have reported m6A RNA modification is required not only for T helper cell differentiation and proliferation, but also for T cells to properly exit the naïve state.34 Especially, m6A mRNA methylation sustains T-regs suppressive role, and depletion of METTL3 in T-regs results in its disfunctions and instability.35 Coinciding with our analysis based on online data, these studies imply m6A could target a group of genes encoding components of principle signaling pathways in distinct T cell subtypes, thereby regulating the differentiation of naïve T cells and sustaining the suppressive functions of T-regs. This may explain in our study why the leucocytes m6A is associated with clinicopathological parameters that reflect the malignancy of a tumor, and why m6A level tends to return to normal levels after receiving treatment. Role of the m6A regulatory enzymes on blood immune cells and antitumor immune responses, especially on distinct subtype of T cells, will be the focus of our future work.
In conclusion, for the first time we determined the elevated level of leukocyte m6A methylation in NSCLC and explored its diagnostic value as a surrogate biomarker. We cannot establish the temporal causality between RNA methylation and risk of cancer because this was a retrospective, cross-sectional case–control study. Nevertheless, our study provides evidence linking the risk role of m6A level in blood with malignancies and identify its localization on immune cells. Specifically, we have identified leukocyte m6A as a potential biomarker for NSCLC screening, diagnosis, development, and treatment.
Abbreviations
NSCLC, non-small-cell lung carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; ROC, receiver-operating characteristic; m6A, N6-methyladenosine; CEA, carcinoembryonic antigen; SCC, squamous cell carcinoma; CA 125, cancer antigen 125; Cyfra211, cytokeratin fragment 211.
Data Sharing Statement
All datasets generated for this study are included in the manuscript.
Ethics Statement
This study protocol was in accordance with the ethical guidelines of the 1975 Declaration of Helsinki and approved by the Ethics Committee of Peking Union Medical College Cancer Hospital (grant no: NCC2017G-115).
Acknowledgment
We thank Laura Smales (BioMed Editing, Toronto, Canada) for critical reading and editing of the manuscript.
Author Contributions
Conception and design, PY, ZD and CW; investigation, PY, LX, LK and GY; formal analysis, PY, YZ and XX; validation, PY and XD; writing—original draft preparation, PY and ZD; writing—review and editing, LX, LK, GY, XX, GY, XD, YZ, XD, ZD and CW; Supervision, CW. All the authors have made a substantial contribution to the work reported and have reviewed or revised the manuscript. The authors have agreed on all versions of the article before submission, during revision, the final version accepted for publication, and any significant changes introduced at the proofing stage. The authors agree on the journal to submit and agree to be accountable for all aspects of the work.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences (No. 2019-I2M-2-002); and by National Key R&D Program of China (No. 2018YFC1315000/2018YFC1315002).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi:10.1016/j.cell.2012.06.013
2. Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics. 2019;11(1):62. doi:10.1186/s13148-019-0656-7
3. Bhat SA, Majid S, Wani HA, Rashid S. Diagnostic utility of epigenetics in breast cancer - A review. Cancer Treat Res Commun. 2019;19:100125. doi:10.1016/j.ctarc.2019.100125
4. Baglietto L, Ponzi E, Haycock P, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer. 2017;140(1):50–61. doi:10.1002/ijc.30431
5. FitzGerald LM, Naeem H, Makalic E, et al. Genome-wide measures of peripheral blood dna methylation and prostate cancer risk in a prospective nested case-control study. Prostate. 2017;77(5):471–478. doi:10.1002/pros.23289
6. Li L, Zheng H, Huang Y, et al. DNA methylation signatures and coagulation factors in the peripheral blood leucocytes of epithelial ovarian cancer. Carcinogenesis. 2017;38(8):797–805. doi:10.1093/carcin/bgx057
7. Flotho C, Sommer S, Lubbert M. DNA-hypomethylating agents as epigenetic therapy before and after allogeneic hematopoietic stem cell transplantation in myelodysplastic syndromes and juvenile myelomonocytic leukemia. Semin Cancer Biol. 2018;51:68–79.
8. Li B, Gan A, Chen X, et al. Diagnostic performance of DNA hypermethylation markers in peripheral blood for the detection of colorectal cancer: a meta-analysis and systematic review. PLoS One. 2016;11(5):e0155095. doi:10.1371/journal.pone.0155095
9. Zhang FF, Cardarelli R, Carroll J, et al. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6(5):623–629. doi:10.4161/epi.6.5.15335
10. Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41(1):126–139. doi:10.1093/ije/dyq154
11. Zhang L, Liang Y, Li S, et al. The interplay of circulating tumor DNA and chromatin modification, therapeutic resistance, and metastasis. Mol Cancer. 2019;18(1):36. doi:10.1186/s12943-019-0989-z
12. He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6(12):863–865. doi:10.1038/nchembio.482
13. Maatz H, van Heesch S, Kreuchwig F, et al. Epigenetics and control of RNAs. Methods Mol Biol. 2017;1488:217–237.
14. Zhu LY, Zhu YR, Dai DJ, Wang X, Jin HC. Epigenetic regulation of alternative splicing. Am J Cancer Res. 2018;8(12):2346–2358.
15. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79(7):1285–1292. doi:10.1158/0008-5472.CAN-18-2965
16. Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into m(6)A RNA enzymes. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):222–229. doi:10.1016/j.bbagrm.2018.10.014
17. Wang S, Chai P, Jia R, Jia R. Novel insights on m(6)A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17(1):101. doi:10.1186/s12943-018-0847-4
18. Wu X, Sang L, Gong Y. N6-methyladenine RNA modification and cancers. Am J Cancer Res. 2018;8(10):1957–1966.
19. Q-V Á, M-P S. Epigenetics of lung cancer: a translational perspective. Cellular Oncol. 2019;42(6):739–756.
20. Duruisseaux M, Esteller M. Lung cancer epigenetics: from knowledge to applications. Semin Cancer Biol. 2018;51:116–128. doi:10.1016/j.semcancer.2017.09.005
21. Verma M, Rogers S, Divi RL, et al. Epigenetic research in cancer epidemiology: trends, opportunities, and challenges. Cancer Epidemiol Biomarkers Prev. 2014;23(2):223–233. doi:10.1158/1055-9965.EPI-13-0573
22. Zhang S, Zhao B, Zhou A, et al. mA demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606.e596. doi:10.1016/j.ccell.2017.02.013
23. Zhang D, Ai D, Tanaka H, Hammock BD, Zhu Y. DNA methylation of the promoter of soluble epoxide hydrolase silences its expression by an SP-1-dependent mechanism. Biochim Biophys Acta. 2010;1799(9):659–667. doi:10.1016/j.bbagrm.2010.09.006
24. Luo J, Liu H, Luan S, He C, Li Z. Aberrant regulation of mRNA m(6)A modification in cancer development. Int J Mol Sci. 2018;19:9. doi:10.3390/ijms19092515
25. Chi HC, Tsai CY, Tsai MM, Lin KH. Impact of DNA and RNA methylation on radiobiology and cancer progression. Int J Mol Sci. 2018;19:2. doi:10.3390/ijms19020555
26. Ianniello Z, Fatica A. N6-methyladenosine role in acute myeloid leukaemia. Int J Mol Sci. 2018;19:8. doi:10.3390/ijms19082345
27. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613.
28. Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. doi:10.1016/j.celrep.2017.02.059
29. Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–1083. doi:10.1038/s41556-018-0174-4
30. Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19(1):326. doi:10.1186/s12885-019-5538-z
31. Al-Moghrabi N, Nofel A, Al-Yousef N, et al. The molecular significance of methylated BRCA1 promoter in white blood cells of cancer-free females. BMC Cancer. 2014;14:830. doi:10.1186/1471-2407-14-830
32. Barciszewska AM. Global DNA demethylation as an epigenetic marker of human brain metastases. Biosci Rep. 2018;38:5. doi:10.1042/BSR20180731
33. Wang L, Simons DL, Lu X, et al. Breast cancer induces systemic immune changes on cytokine signaling in peripheral blood monocytes and lymphocytes. E Bio Medicine. 2020;52:102631. doi:10.1016/j.ebiom.2020.102631
34. Li HB, Tong J, Zhu S, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548(7667):338–342. doi:10.1038/nature23450
35. Tong J, Cao G, Zhang T, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28(2):253–256. doi:10.1038/cr.2018.7
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.