Back to Journals » Cancer Management and Research » Volume 11
Overexpression of Notch3 is associated with metastasis and poor prognosis in osteosarcoma patients
Authors Tang XF, Cao Y, Peng DB, Zhao GS, Zeng Y, Gao ZR, Lv YF, Guo QN
Received 27 August 2018
Accepted for publication 19 November 2018
Published 8 January 2019 Volume 2019:11 Pages 547—559
DOI https://doi.org/10.2147/CMAR.S185495
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Xue-Feng Tang,1 Ya Cao,1 Dong-Bin Peng,1 Guo-Sheng Zhao,2 Ying Zeng,1,3 Zi-Ran Gao,1 Yang-Fan Lv,1 Qiao-Nan Guo1
1Department of Pathology, Xinqiao Hospital, Army Medical University, Chongqing 400037, China; 2Department of Orthopedic Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, China; 3Department of Pathology, Daping Hospital, Army Medical University, Chongqing 400037, China
Background: Notch signaling abnormalities are associated with the development of various tumors, including hematopoietic and epithelium-derived tumors. However, the role of Notch signaling in tumors originating from mesenchymal cells is unclear. The effect of Notch3 expression on the prognosis of osteosarcoma and its role and mechanism in osteosarcoma cells have never been reported.
Materials and methods: In this study, we performed a clinicopathological analysis of 70 cases of osteosarcoma, with primary focus on survival. Osteosarcoma cell lines MTH and U2OS were used. After knockdown of Notch3 by lentiviral transfection and siRNA, the cell cycle, cell viability, and wound healing capacity were assessed. Subsequently, the Transwell assay was performed, and the expression levels of hairy and enhancer of split-1 (Hes1) and matrix metalloproteinase 7 (MMP7) were detected by RT-PCR and Western blot assay. The expression of MMP7 was also detected after knockdown of Hes1. Animal experiments were performed by injecting the cell lines MTH of Notch3 knockdown into mice tail veins and comparing the development of lung metastasis with the control group.
Results: Comparison of survival curves showed that Notch3 expression significantly impacts patient survival. Additionally, multivariate analysis revealed that Notch3 is an independent prognostic factor for osteosarcoma. In in vivo experiments, osteosarcoma-associated pulmonary metastasis in nude mice was reduced after Notch3 silencing. The expression of downstream effector molecule, Hes1, and that of the invasion and metastasis-associated proteolytic enzyme, MMP7, were reduced, and MMP7 was further decreased by Hes1 knockdown in in vitro experiments.
Conclusion: Notch3 is a prognostic factor for osteosarcoma and might regulate its invasion and metastasis through the downstream target gene Hes1 and effector MMP7.
Keywords: osteosarcoma, prognosis, metastasis, Notch3, Hes1, MMP7
Introduction
Osteosarcoma is the most common primary malignant bone tumor in adolescents,1 with a worldwide incidence of 3.4/1,000,000 per year2 and an estimated frequency of about 2,900 new cases every year in the USA.3 This disease is a heavy burden for children and their parents and brings irreparable psychological trauma and economic losses to patients’ families and society. Approximately 15%–20% of patients present with pulmonary metastases at the time of diagnosis. The prognosis of patients with metastasis is very poor, with an approximate long-term survival rate of 20%–30%, whereas the survival rate of nonmetastatic patients is 65%–70%.4–6 No breakthrough has been made in improving the survival of osteosarcoma patients in the last three decades. The identification of new early diagnostic markers and novel therapeutic targets is urgently needed.7 The effective treatment of osteosarcoma depends on understanding the mechanisms of osteosarcoma invasion and metastasis. Therefore, the study of the underlying mechanism of tumor invasion and metastasis is now a hot topic in osteosarcoma research.
The Notch signaling pathway is a widespread and highly conserved signal transduction pathway. Its related signaling molecules are involved in the regulation of cell development, differentiation, proliferation, and apoptosis. In mammals, the Notch pathway consists of five extracellular ligands (DLL1, 3, 4, JAG1, and 2), four membrane receptors (Notch 1–4), and DNA-binding proteins.8 Notch signaling plays an important role in various cancer stem cells and is considered to be a contributing factor in the development of osteosarcoma.9
In recent years, multiple studies have shown that Notch3, an important member of the Notch family, is deeply involved in the proliferation, apoptosis, invasion, metastasis, and chemotherapy resistance of tumor cells. Zhang et al found that Notch3 was highly expressed in urothelial carcinoma tissue compared with normal bladder tissue, and the elevated Notch3 expression could promote the growth and resistance to chemotherapy of urothelial carcinoma.10 In pancreatic cancer, reduced Notch3 expression was associated with enhanced tumor cell sensitivity to gemcitabine due to the consequent suppression of phosphatidy linositol 3- kinase/activation of membrane kinases activation.11 Overexpression of Notch3 and pS6 was associated with progression and poor prognosis in epithelial ovarian cancer.12 Zhou et al showed that Notch3 silencing could reduce the invasiveness of hepatocellular carcinoma by regulating matrix metalloproteinase 2 (MMP2) and MMP9 via the extracellular regulated protein kinases 1/2 pathway.13 However, the effect of Notch3 expression on the prognosis, proliferation, migration, and metastasis of osteosarcoma has never been reported.
In this study, we found that Notch3 expression significantly impacted patient survival and was an independent prognostic factor for osteosarcoma. In addition, Notch3 knockdown reduced the proliferation, migration, invasion, and metastasis of osteosarcoma cells in mice. Notch3 may affect the invasiveness and metastasis of osteosarcoma cells by regulating the downstream target gene Hes1 and its effector protein, MMP7.
Materials and methods
Tissue samples and cell lines
For this study, 70 patients with osteosarcoma diagnosed at he Department of Pathology, Xinqiao Hospital, Army Medical University, and Department of Pathology, Daping Hospital, Army Medical University, from January 2008 to December 2015 were selected. All patients had complete pathological data, and their clinical data are shown in Table 1. There were 42 men (60%) and 28 women (40%), with a ratio of 3:2. Patients’ age ranged from 7–70 years (mean =25.1 years), and the peak age of onset was 11–30 years. The clinical and imaging data were collected by reviewing medical records and patient inquiries, the clinical staging was carried out according to the Enneking staging system, and follow-up was also performed. The patients were followed up every 3 months, 6 months, 1 year within 2 years, 2–5 years, and >5 years after discharge, respectively. The morphological diagnosis was performed by two senior pathologists according to the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone.
  | Table 1 Clinical data of 70 patients with osteosarcoma Note: The course of disease is from the onset of symptoms to the time of visiting. Abbreviation: AKP, alkaline phosphatase. |
The human osteosarcoma cell line U2OS and human osteoblast line hFOB1.19 were purchased from the American Type Culture Collection. Osteosarcoma cell line MTH is a malignant transformation of hFOB1.19 osteoblasts, which was obtained and passaged in preliminary experiments by our research group. MTH and U2OS were cultured in DMEM/H complete medium containing 10% FBS and penicillin/streptomycin (100×). Cells were cultured in an incubator set at 37°C, 5% CO2, and 95% humidity.
Immunohistochemistry
Samples from 70 patients were stained via the EnVision two-step immunohistochemical technique, using a Leica Benchmark-ULTRA Autostainer, for four antigens, namely, Notch3, Jagged1, Jagged2, and DLL4. Notch3 was positive in the cytoplasm and cell membrane; DLL4 was positive in the cell membrane; and Jagged1–2 were positive in the cytoplasm in a spotty manner. Positive standard: (1) strong staining; (2) percentage of positive tumor cells >75%. Sections of human breast cancer submitted to the same immunohistochemical protocol were used as a positive control.
Lentivirus production and cell transduction
Exponentially growing osteosarcoma cells were seeded in 6-well plates at a density of 2×105/well. Cells were incubated overnight to reach 30%–50% confluency at the time of transfection. The original culture medium was discarded and replaced with 1 mL serum-free and penicillin–streptomycin-free medium. The lentiviral vectors were taken out from the –80°C refrigerator, thawed in an ice bath or at 4°C, and evenly mixed by gentle pipetting. Subsequently, the optimum multiplicity of infection virus storage solution was added and thoroughly mixed. Polybrene (at the final concentration of 5 µg/mL) was then added to increase the transfection efficiency. Cells were subsequently kept in a 5% CO2 incubator at 37°C. After 24 hours, the medium was replaced with complete culture medium containing 10% FBS. After 2–3 days, fluorescence was observed, and the transfection efficiency was evaluated. Cells were screened with puromycin and passaged for interference efficiency evaluation, cryopreservation, and subsequent experiments.
One control and three shNotch3 interference lentiviral vectors were designed and constructed by Cyagen Biosciences (Guangzhou) Co., Ltd, Guangzhou, China. In this experiment, screened shNotch3-1 and shNotch3-2 were used as interference target series for osteosarcoma cell transfection to silence Notch3 gene expression.
The following sequences were used for control and shNotch3 cells:
Scrambled shRNA, 5′-CCTAAGGTTAAGTCGCCCTCG-3′;
Notch3-1 shRNA1, 5′-GGTGATCGGCTCGGTAGTAAT-3′;
Notch3-2 shRNA2, 5′-TTTGTAACGTGGAGATCAATG-3′;
Notch3-3 shRNA3, 5′-TCTGCAAGGACCGAGTCAATG-3′.
Hes1 knockdown by siRNA in osteosarcoma cells
Exponentially growing MTH and U2OS cells in good growing condition were digested with 0.25% trypsin to prepare the single cell suspension, which was then inoculated into 6-well plates at the density of 3×105 cells/well. The density was elevated by 30%–50% at the time of transfection. Five microliters siRNA, 5 µL Lipofectamine 2000 transfection reagent, and 490 µL Opti-MEM were mixed and incubated at room temperature for 10 minutes. The original medium was discarded, and the above mixture was supplemented with 0.5 mL serum-free and penicillin–streptomycin-free medium, mixed thoroughly, and added to each well. The six-well plates were placed in a 5% CO2 incubator, set at 37°C and 95% humidity, for 6 hours. Subsequently, the culture medium was replaced with DMEM complete medium containing 10% FBS. Cells were collected after 48 hours of culture.
The siRNA-Hes1 target series was designed and constructed by Guangzhou RiboBio Co., Ltd, Guangzhou, China. In this experiment, screened siHes1-2 and siHes1-3 were used as interference target series for osteosarcoma cell transfection to silence Hes1 gene expression.
The following sequences were used:
Hes1-1 siRNA, 5′-CAACACGACACCGGATAAA-3′;
Hes1-2 siRNA, 5′-CACGACACCGGATAAACCA-3′;
Hes1-3 siRNA, 5′-CGACACCGGATAAACCAAA-3′.
Cell cycle detection by flow cytometry
Cells were prepared and divided into tubes, each containing 1–5×105 cells. Cells were fixed with ice-cold 70% alcohol, stored overnight at –20°C, washed, and the supernatant was discarded. Then, cells were resuspended in 1 mL PBS, well mixed with 1 mL DNA extraction buffer, and incubated at room temperature for 5 minutes. After centrifugation, the supernatant was discarded, and the cells were resuspended with 1 mL DNA dye, incubated in the dark, at room temperature, for 30 minutes and shaken well. The cell cycle was detected by flow cytometry. The number of prepared cells in each sample was 1×105, and the cell number for the final analysis was 104 cells.
Cell viability assessment by the cell counting kit 8 (CCK8) method
Exponentially growing shNotch3 and control cells were digested to prepare single cell suspensions. Cells were then counted and incubated in 96-well plates, at the density of 2,000 cells/well. Each group included four replicates, and plates were kept in an incubator overnight. Prior to detection, the original culture medium in each well was replaced by medium containing 10% CCK8 reagent (100 µL/well). A blank control was also established. Cells were cultured at 37°C for 1 hour, and the optical density at 450 nm was continuously measured for 4 days using a microplate reader.
Wound healing assay
Osteosarcoma cells in good growing condition were selected, and three scratches were evenly made in each well using a 200 µL pipette tip moving perpendicularly to the bottom of the plate. After washing three times with PBS buffer, serum-free DMEM was added to the cells. The scratch width at 0 hour was recorded under a microscope, and the 6-well plates were stored in an incubator at 37°C for 24 hours. Then, the “wound gap” was measured, and the healing distance in each group was calculated.
Transwell assay
Appropriate amounts of Matrigel were moved into EP tubes on ice and mixed with double volume of DMEM. Subsequently, 20 µL Matrigel working solution was added in 8 µm pore-sized Transwell chambers (Millipore, Billerica, MA, USA). shNotch3 and CON cells were transacted for 48 hours, then digested, and centrifuged. Cells were resuspended into single cells with serum-free DMEM and counted using a counting plate. Subsequently, 100 µL diluted cell suspension was incubated in the upper chamber at the density of 5×104 cells/well and 600 µL DMEM containing 15% FBS was added in the lower chamber. The upper chambers were taken out at 8 and 24 hours of incubation, respectively, after which the cells were washed with PBS buffer and fixed with 4% paraformaldehyde for 10 minutes. After washing with PBS buffer, the lower bottom was stained with crystal violet. Cells were observed under an inverted microscope, and eight visual fields were randomly selected, recorded, and counted.
Reverse transcription PCR
The PCR primer sequences of Notch3, hairy and enhancer of split-1 (Hes1), and MMP7 were determined according to the literature. The stably expressed human housekeeping gene, GAPDH, was selected as internal reference and synthesized by Shanghai Invitrogen, Shanghai, China (Table 2). RNA extraction, concentration detection, and mRNA reverse transcription were sequentially performed. Real-time quantitative PCR solutions were prepared, and the detection was performed according to the standard protocol for real-time quantitative PCR. PCR results were analyzed by the built-in software of the Bio-Rad Thermal Cycler.
  | Table 2 Primer sequences |
Western blot assay
Proteins were extracted with protein lysate from Pierce, and protein concentration was determined by the bicinchoninic acid method. Electrophoresis, transfer, blocking, and antibody incubation were then sequentially performed. Antibodies used for Western blot assay included Notch3, Hes1, and MMP7. Bands were detected with the Pierce photographic developer in a Bio-Rad gel imager, and GAPDH served as an internal reference. Related protein expression was analyzed with the Image Lab analyzing software.
Animal experiments
For this experiment, 32 4-week-old male nude mice were selected. The mouse body weight was 13–15 g, and the animals were aged 5 weeks when the experiments were performed. The nude mice were randomly divided into four groups, and the osteosarcoma cells (MTH shNotch3, MTH CON, U2OS shNotch3, and U2OS CON cell lines) were injected into the tail veins. After 1 month, the mice were sacrificed, and their lungs were sectioned for HE staining. Using an optical microscope, the number of pulmonary metastatic lesions was observed and recorded. Half of the tumor tissues were preserved in liquid nitrogen for subsequent real-time quantitative PCR experiments.
Statistical analysis
Statistical analysis was performed using the SPSS19.0 and GraphPad Prism5.0 software. Differences between two groups or among multiple groups were assessed by the RxC table chi-squared test (number of cases >40) or Fisher’s exact test (<40 cases). Survival analysis was performed for 54 patients who had complete follow-up data. Survival curves were plotted using the Kaplan–Meier method. Univariate prognostic analysis was performed using the log-rank test, and multivariate prognostic analysis was performed using the Cox proportional hazards regression model. Experimental data were expressed as mean ± SD (M ± SD). The significance test level was α=0.05. When P<0.05, the difference was considered statistically significant.
Results
Notch3 is a prognostic factor for osteosarcoma
The expression rate of Notch3 was 68.57% (48/70). The expression rates of the three ligands, DLL4, Jagged1, and Jagged21, were 8.57% (6/70), 8.57% (6/70), and 0 (0/70), respectively (Supplementary materials Table S1, Figure S1–3).
The survival time of 54 patients ranged between 1 and 78 months, with a median survival time of 18 months and mean survival time of 41.68 months. The 1-year, 2-year, and 5-year survival rates were 87%, 66%, and 30%, respectively. Survival curves were generated with the parameters of gender (male/female), age (≤18 years and >18 years), tumor size (maximum tumor size <5 cm/≥5 cm), Enneking staging (stage I–IIA/IIB–III), amputation (yes/no), chemotherapy (yes/no), preoperative chemotherapy (yes/no), and Notch3 expression (positive/negative). Comparison results showed significant differences in the survival curves of Enneking staging, preoperative chemotherapy, and Notch3 expression. The mean survival times of Notch3+ (n=30) and Notch3– group (n=24) were 32.77 and 48.47 months, respectively. The 1-year, 2-year, and 5-year survival rates for Notch3+/– were 65.9%/87.5%, 42.0%/83.3%, and 18.4%/41.4%, respectively. The difference in survival time of Notch3+/– group was statistically significant (P=0.017).
Results of multivariable prognostic analysis (gender, age, tumor size, Enneking staging, amputation, chemotherapy, preoperative chemotherapy, and Notch3 expression) showed that Enneking staging, preoperative chemotherapy, and Notch3 expression were independent prognostic factors for osteosarcoma (Figure 1, Table 3).
  | Table 3 Cox regression analysis Note: *P<0.05; **P<0.01. |
The Notch3+ group had higher metastatic rates and metastatic patients had lower survival rates
Gender, age, staging, tumor size, serum alkaline phosphatase, recurrence, and metastasis were compared between the Noch3+ (n=33) and Notch3– (n=37) groups. Results showed that serum alkaline phosphatase (P=0.013), recurrence (P=0.060), and metastasis rates (P=0.005) differed significantly between the two groups.
Pulmonary metastases occurred in 17 patients, of which 12 cases (70.59%) expressed Notch3 in the tumor cells. Of the 17 patients, 16 were followed-up for 2–57 months. Of these, 13 patients died due to tumor progression. The survival time of the 16 patients ranged from 2–27 months, with a median survival time of 12.5 months and a mean survival time of 17.13 months. The 1-year, 2-year, and 5-year survival rates were 54.2%, 21.7%, and 0%, respectively.
Expression and identification of Notch3 knockdown
Lentiviral vectors were used to silence Notch3 expression in MTH and U2OS osteosarcoma cell lines. Results of real-time PCR revealed that Notch3 mRNA levels were significantly inhibited in the shNotch3-1 and shNotch3-2 groups compared with the control group (Figure 2A). Western blotting results also showed that the protein levels significantly declined compared with the control group (Figure 2B). Therefore, the shNotch3-1 and shNotch3-2 interference series were selected for the subsequent experiments.
  | Figure 2 Changes in the Notch3 mRNA (A) and protein (B) expressions after lentiviral transfection. Note: *P<0.05; **P<0.01. |
Notch3 knockdown decreased the proliferation of osteosarcoma cells
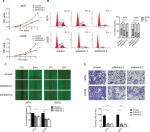
The CCK8 assay results showed that the proliferation rate of shNotch3 cells decreased significantly compared with the control group from the second day of detection (Figure 3A). Results of cell cycle detection demonstrated that osteosarcoma cells showed G1-phase cell cycle arrest after the knockdown of Notch3 expression (Figure 3B).
Notch3 knockdown inhibited osteosarcoma cell migration
The “wound healing” test was used to investigate the effect of shNotch3 on the migration ability of osteosarcoma cells. The wound healing distances of shNotch3-1 and shNotch3-2 MTH cells were 44.3% ± 8.6% and 47.3% ± 3.2% of the control group at 24 hours, while those of shNotch3-1 and shNotch3-2 U2OS cells were 46% ± 3.3% and 37.8% ± 8.0% of the control group, respectively, suggesting that Notch3 knockdown attenuated the migration of osteosarcoma cells (Figure 3C).
Notch3 knockdown inhibited osteosarcoma cell invasion
Transwell invasion assay was used to investigate the effect of shNotch3 on the invasiveness of osteosarcoma cells. At 24 hours, the number of invading cells in the MTH control, shNotch3-1, and shNotch3-2 MTH groups was 45.4±4.98, 10.4±2.3, and 8.0±3.6, respectively, while the number of invading cells in the U2OS control, U2OS shNotch3-1, and shNotch3-2 groups was 39.2±4.8, 6.6±4.0, and 6.6±2.4, respectively, suggesting that Notch3 knockdown attenuated the invasiveness of osteosarcoma cells (Figure 3D).
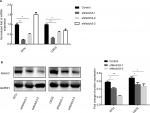
Notch3 knockdown inhibited the expression of Hes1 and MMP7
Results of qRT-PCR and Western blotting showed that both the mRNA and protein expression levels of Hes1 and MMP7 were significantly reduced by Notch3 knockdown (Figure 4).
  | Figure 4 Notch3 knockdown inhibited Hes1 and MMP7 mRNA (A) and protein (B) expressions. Note: *P<0.05; **P<0.01. Abbreviation: MMP7, matrix metalloproteinase 7. |
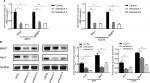
Expression and identification of Hes1 knockdown
siRNA was used to silence the expression of Hes1 in MTH and U2OS osteosarcoma cell lines. Results of real-time PCR revealed that Hes1 mRNA levels were significantly inhibited in the siHes1-2 and siHes1-3 groups compared with the control group (Figure 5A). Results of Western blot assay showed that the protein levels in the two groups also significantly declined compared with the control group (Figure 5B). Therefore, the siHes1-2 and siHes1-3 interference series were selected for the subsequent experiments.
Hes1 knockdown inhibited MMP7 expression
Results of qRT-PCR and Western blotting showed that both the mRNA and protein expression levels of MMP7 were significantly reduced by Hes1 knockdown (Figure 5C, D).
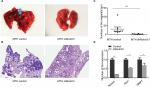
Notch3 knockdown reduced pulmonary metastasis in vivo
There were no pulmonary metastases in the U2OS control and U2OS shNotch3 groups. The pulmonary metastasis rate was 100% (8/8) in the MTH control nude mice and 50% (4/8) in the shNotch3 group (Table 4). The number of lung metastatic lesions in the nude mice of the MTH shNotch3 group was significantly decreased (P<0.0001). Compared with the control group, Notch3, Hes1, and MMP7 mRNA expressions in Notch3 knockdown group were significantly decreased (Figure 6).
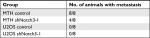  | Table 4 Pulmonary metastasis rates in nude mice transplanted with MTT and U2OS cells |
Discussion
To date, there are very few available studies on the role of Notch3 in osteosarcoma, and the regulation of invasion and metastasis by Hes1 and MMP7 has never been reported. Notch signaling is a key pathway involved in the occurrence and development of osteosarcoma.14–16 However, most available studies have focused on the functions of Notch1 and Notch2. There are few reports on the effect and mechanism of Notch3 in osteosarcoma. Although it is well established that the MMP family is involved in cancer invasion and metastasis, most available studies have focused on MMP2 and MMP9. In the development of osteosarcoma, tumor cells could secrete high levels of MMP7 to promote metastasis by degrading type IV collagen, reducing intercellular adhesion, and degrading the extracellular matrix. It has been reported that the downregulation of HOXB7 could significantly inhibit the expression of MMP2 and MMP7 in osteosarcoma cell line MG63.17 Noncoding RNA, MALAT1, inhibited the invasiveness of U2OS cells by reducing the expression of β-catenin, MMP7, and c-Myc.18 Our previous studies also revealed that TSSC3 downregulation triggered a series of signal transductions and upregulated MMP7 to induce osteosarcoma cell migration and invasion by upregulating mesenchymal markers and suppressing epithelial markers.19 Meanwhile, MMP7 was also positively associated with vascular endothelial growth factor (VEGF), promoting tumor angiogenesis.20,21 However, the role of Notch3 in the invasion and metastasis of osteosarcoma and its association with MMP7 are yet to be reported.
This study demonstrated that Notch3 was a prognostic factor for osteosarcoma and regulated tumor cell migration and metastasis. Results of the retrospective clinicopathological study and survival analysis showed that Notch3 expression exerted a significant impact on the survival curves, and Notch3 was an independent prognostic factor for osteosarcoma in multivariate prognostic analysis. In vivo experiments revealed that Notch3 downregulation reduced the pulmonary metastasis rate of osteosarcoma in nude mice.
Results of in vitro experiments showed that Notch3 knockdown decreased the expressions of downstream effector molecule Hes1 and invasion and metastasis-associated proteolytic enzyme MMP7. Hes1 downregulation further inhibited MMP7 expression. These results suggested that Notch3 might regulate the invasion and metastasis of osteosarcoma by targeting Hes1 and MMP7. Ligand binding results in proteolytic cleavage of Notch receptors to release the signal-transducing Notch intracellular domain (NICD). NICD migrates into the nucleus and associates with the nuclear proteins of the RBP-Jκ family. RBP-Jκ, when complexed with NICD, acts as a transcriptional activator, and the RBP-Jκ–NICD complex activates expression of primary target genes of Notch signaling such as the Hes1. Hes1 is a basic helix-loop-helix type of transcriptional repressor and suppresses expression of downstream target genes.22
The first report of the role of Notch signaling in tumors came from T-lymphocyte leukemia.23 Subsequent studies have shown that Notch has dual roles of promoting or inhibiting cancer24 and was closely related to the vascular microenvironment and tumor angiogenesis.25 The importance of Notch signaling abnormalities in hematopoietic and epithelium-derived tumors has been well documented, but their role in tumors originating from mesenchymal cells is still unclear. Recent studies have shown that Notch signaling abnormalities were associated with osteosarcoma tumor growth. The inhibition of Notch signaling by γ-secretase could affect the growth of osteosarcoma cells through cell cycle arrest.26 Curcumin might inhibit the growth of osteosarcoma cells under hypoxic conditions by inhibiting Notch1 signaling.27 Downregulation of Notch-1 by curcumin could be a promising treatment option for osteosarcoma.28 The Notch signaling pathway has also been involved in antioxidative stress, migration, invasion, and increased VEGF expression of osteosarcoma, in in vitro experiments.9 In addition, the Notch pathway is known to interact with other important signaling pathways, such as Wnt and TFGβ.29 These studies collectively indicate that the Notch pathway could be a possible therapeutic target for osteosarcoma.
Conclusion
Notch3 is a prognostic factor for osteosarcoma and regulates the invasion and metastasis of osteosarcoma by targeting the downstream target gene Hes1 and effector MMP7.
Ethics approval and consent to participate
All human tissues experiments were approved by the Institutional Ethics Committee of Third Military Medical University (Army Medical University. Chongqing, China). Written informed consent for the experimental studies was obtained from the patients or their guardians. All the animal care and experimental procedures were conformed to the Institutional Animal Care and Use Committee of Xinqiao Hospital, Third Military Medical University, according to the Guide for the Care Use of Laboratory Animals.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 81672653) and the Chongqing Research Program of Basic Research and Frontier Technology (Grant No. cstc2015jcyjBX0067).
Disclosure
The authors report no conflicts of interest in this work.
References
Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. | ||
Misaghi A, Goldin A, Awad M, et al. Osteosarcoma: a comprehensive review. SICOT J. 2018;4:12. | ||
Hameed M, Dorfman H. Primary malignant bone tumors--recent developments. Semin Diagn Pathol. 2011;28(1):86–101. | ||
Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16(5):543–556. | ||
Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325. | ||
Ozger H, Eralp L, Atalar AC, et al. [Survival analysis and the effects of prognostic factors in patients treated for osteosarcoma]. Acta Orthop Traumatol Turc. 2007;41(3):211–219. Turkish. | ||
Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16(18):2727–2736. | ||
Bagheri L, Pellati A, Rizzo P, et al. Notch pathway is active during osteogenic differentiation of human bone marrow mesenchymal stem cells induced by pulsed electromagnetic fields. J Tissue Eng Regen Med. 2018;12(2):304–315. | ||
Mu X, Isaac C, Greco N, et al. Notch signaling is associated with ALDH activity and an aggressive metastatic phenotype in murine osteosarcoma cells. Front Oncol. 2013;3:143. | ||
Zhang H, Liu L, Liu C, et al. Notch3 overexpression enhances progression and chemoresistance of urothelial carcinoma. Oncotarget. 2017;8(21):34362–34373. | ||
Yao J, Qian C. Inhibition of Notch3 enhances sensitivity to gemcitabine in pancreatic cancer through an inactivation of PI3K/Akt-dependent pathway. Med Oncol. 2010;27(3):1017–1022. | ||
Liu Z, Yun R, Yu X, et al. Overexpression of Notch3 and pS6 is associated with poor prognosis in human ovarian epithelial cancer. Mediators Inflamm. 2016;2016:5953498. | ||
Zhou L, Zhang N, Song W, et al. The significance of Notch1 compared with Notch3 in high metastasis and poor overall survival in hepatocellular carcinoma. PLoS One. 2013;8(2):e57382. | ||
Tao J, Jiang MM, Jiang L, et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell. 2014;26(3):390–401. | ||
Ma Y, Ren Y, Han EQ, et al. Inhibition of the Wnt-β-catenin and Notch signaling pathways sensitizes osteosarcoma cells to chemotherapy. Biochem Biophys Res Commun. 2013;431(2):274–279. | ||
Chen S, Lee BH, Bae Y. Notch signaling in skeletal stem cells. Calcif Tissue Int. 2014;94(1):68–77. | ||
Yang Y, Chen J, Chen Q. Upregulation of HOXB7 promotes proliferation and metastasis of osteosarcoma cells. Mol Med Rep. 2017;16(3):2773–2778. | ||
Zhang JH, Kang XH, Lu P, et al. [Expression of long non-coding RNA MALAT1 in osteosarcoma and its effect on invasiveness and metastatic potential of osteosarcoma cells]. Zhonghua Bing Li Xue Za Zhi. 2016;45(8):561–565. Chinese. | ||
Lv YF, Dai H, Yan GN, et al. Downregulation of tumor suppressing STF cDNA 3 promotes epithelial-mesenchymal transition and tumor metastasis of osteosarcoma by the Wnt/GSK-3β/β-catenin/Snail signaling pathway. Cancer Lett. 2016;373(2):164–173. | ||
Ito TK, Ishii G, Saito S, et al. Degradation of soluble VEGF receptor-1 by MMP-7 allows VEGF access to endothelial cells. Blood. 2009;113(10):2363–2369. | ||
Kisand K, Kerna I, Kumm J, et al. Impact of cryopreservation on serum concentration of matrix metalloproteinases (MMP)-7, TIMP-1, vascular growth factors (VEGF) and VEGF-R2 in Biobank samples. Clin Chem Lab Med. 2011;49(2):229–235. | ||
Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. | ||
Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. | ||
Previs RA, Coleman RL, Harris AL, et al. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res. 2015;21(5):955–961. | ||
Pallares J, Rojo F, Iriarte J, et al. Study of microvessel density and the expression of the angiogenic factors VEGF, bFGF and the receptors Flt-1 and FLK-1 in benign, premalignant and malignant prostate tissues. Histol Histopathol. 2006;21(8):857–865. | ||
Tanaka M, Setoguchi T, Hirotsu M, et al. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100(12):1957–1965. | ||
Wang Z, Zhang K, Zhu Y, et al. Curcumin inhibits hypoxia-induced proliferation and invasion of MG-63 osteosarcoma cells via downregulating Notch1. Mol Med Rep. 2017;15(4):1747–1752. | ||
Li Y, Zhang J, Ma D, et al. Curcumin inhibits proliferation and invasion of osteosarcoma cells through inactivation of Notch-1 signaling. FEBS J. 2012;279(12):2247–2259. | ||
Shimizu T, Nakagawa K. [Novel signal transduction pathways: the molecular basis for targeted cancer therapies in Hedgehog/Notch/Wnt pathway]. Nihon Rinsho. 2015;73(8):1342–1348. Japanese. |
Supplementary materials
We supplemented the expression of the Jagged1, Jagged2, and DLL4 in osteosarcoma.
The expression rate of Notch3 was 68.57% (48/70). The expression rates of the three ligands, DLL4, Jagged1, and Jagged21, were 8.57% (6/70), 8.57% (6/70), and 0 (0/70), respectively.
  | Table S1 Expression of 4 antigens in 70 osteosarcoma cases |
  | Figure S1 Jadded1+ tumor cells in case 48. |
  | Figure S2 Jadded2– tumor cells in case 8. |
  | Figure S3 DLL4+ tumor cells in case 36. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.