Back to Journals » OncoTargets and Therapy » Volume 13
Overexpression of LRRC59 Is Associated with Poor Prognosis and Promotes Cell Proliferation and Invasion in Lung Adenocarcinoma
Authors Li D , Xing Y , Tian T, Guo Y, Qian J
Received 9 January 2020
Accepted for publication 7 June 2020
Published 3 July 2020 Volume 2020:13 Pages 6453—6463
DOI https://doi.org/10.2147/OTT.S245336
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Dong Li,1,* Ying Xing,2,* Tiannv Tian,3 Yanan Guo,3 Jing Qian3,4
1Department of Thoracic Surgery, Huzhou Central Hospital, Huzhou, Zhejiang, People’s Republic of China; 2Department of Gastroenterology, The 72nd Army Hospital of the People’s Liberation Army of China, Huzhou, Zhejiang, People’s Republic of China; 3Huzhou University Schools of Nursing and Medicine, Huzhou University, Huzhou, Zhejiang, People’s Republic of China; 4Key Laboratory of Vector Biology and Pathogen Control of Zhejiang Province, Huzhou, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jing Qian
Huzhou University Schools of Nursing and Medicine, Huzhou University, Huzhou, Zhejiang Province 313000, People’s Republic of China
Tel +8613905723707
Email [email protected]
Aim: LRRC59 (leucine-rich repeat-containing protein 59) is a ribosome-binding protein that also interacts with fibroblast growth factors. Limited investigations revealed a possible role of LRRC59 in the aggressive phenotype of breast cancer. However, whether LRRC59 contributes to the progression of lung cancer remains unclear.
Materials and Methods: In this study, an online TCGA-based survival analysis software (GEPIA2) was used to estimate the prognostic value of LRRC59 mRNA expression level for lung cancer. Cell Counting Kit-8 assay, colony-forming assay, cell cycle analysis, and transwell assay were used to assess the biological functions of LRRC59 in lung cancer cells. Then, 94 lung adenocarcinoma (LUAD) patient tissues were collected to examine the expression level of LRRC59 by the tissue microarray (TMA)-based immunohistochemistry staining (IHC). Univariate Kaplan–Meier and multivariate Cox regression analyses were performed to evaluate the prognostic value of LRRC59 protein expression in LUAD.
Results: Higher mRNA level of LRRC59 was significantly associated with worse survival for lung adenocarcinoma, but not for lung squamous cell carcinoma. Knockdown of LRRC59 by shRNA apparently inhibited cell proliferation and colony formation in both H1299 and A549 cells. The G1/S phase arrest induced by LRRC59 depletion was observed in A549 and H1299 cells. Besides, the silencing of LRRC59 decreased cell migrative and invasive abilities. Moreover, TMA-based IHC showed that LRRC59 was highly expressed in LUAD tissues and closely associated with lymph node metastasis (P< 0.001), TNM stage (P< 0.001), and histological differentiation (P=0.007). Further multivariate analysis suggested that LRRC59 overexpression was an independent prognostic factor in LUAD.
Conclusion: LRRC59 may serve as a novel biomarkers and therapeutic target for LUAD clinical practice.
Keywords: LRRC59, lung adenocarcinoma, proliferation, metastasis, prognosis
Introduction
Lung cancer ranks among the most frequent cause of cancer-related deaths with an approximate 5-year overall survival rate of less than 18%. The key reason for poor lung cancer treatment is the inadequate potential biomarkers for predicting and diagnosing non-small cell lung cancer (NSCLC) for molecular targeted or personalized therapies. Lung adenocarcinoma (LUAD) is currently the most common histologic type of lung cancer.1 Therefore, exploring the underlying mechanisms and related molecules during lung adenocarcinoma development and progression seems of great importance.
LRRC59 was first described as a ribosome receptor on the ER that also interacts with fibroblast growth factors (FGF1),2–4 it is a tail-anchored ER membrane protein containing a small C-terminal domain facing the ER lumen, a leucine-rich repeat (LRR) domain, and a putative coiled-coil domain facing the cytosol. Zhen et al showed that LRRC59 was explicitly required for the nuclear import of FGF1.5 Also, Pallai et al found that LRRC59 was a novel binding partner of the oncoprotein cancerous Inhibitor of PP2A (CIP2A) and was required for CIP2A nuclear import.6 Recently, Yu et al showed that a fusion transcript of LRRC59-FLJ60017 to be present in prostate carcinomas, ovarian cancer, esophageal adenocarcinoma, non-small cell lung cancer and glioblastoma multiforme.7 Meanwhile, they founded that detection of the fusion transcripts along with high Gleason scores was predictive of prostate carcinoma recurrence. Furthermore, Terp et al demonstrated that the altered expression of LRRC59 was correlated with the metastatic capability of breast cancer cell lines.8 Although the above information suggests that LRRC59 might be related to cancer, the biological functions of LRRC59 are yet to be identified directly in lung cancer.
In the present study, we aimed to investigate the expression pattern and clinical significance of LRRC59 in lung cancer and its critical role in lung cancer cell proliferation and metastasis by silencing in vitro. Our results suggested that LRRC59 is an oncogenic factor that contributes to cancer proliferation and metastasis. LRRC59 may be a novel biomarkers and therapeutic target in lung cancer clinical practice.
Materials and Methods
Online Survival Analysis
Differential expression of LRRC59 in lung cancer and normal tissues was analyzed using the online Gene Expression Profiling and Interactive Analysis (GEPIA2; http://gepia2.cancer-pku.cn/#index) database. GEPIA2 is a newly developed interactive web server for the analysis of RNA sequencing data derived from 9736 tumors and 8,587 healthy samples from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression datasets.9 GEPIA provides customizable functions including tumor/normal differential expression analysis, profile plotting, correlation analysis, patient survival analysis, detection of similar genes, and dimensionality reduction analysis. The |Log2FC| cutoff of the expression of proposed biomarker was 1. The p-value cutoff of the expression of proposed biomarker was 0.01. All boxplot analysis used log2(TPM + 1) for log-scale.
The GEPIA2 survival analysis tool was used to assess the relationship between LRRC59 mRNA expression and lung cancer prognosis based on gene expression levels and uses a Log rank test for hypothesis evaluation. We selected a hazards ratio (HR) based on the Cox PH model and also the 95% confidence interval information to show the 95% confidence interval (CI) as the dotted line. The LRRC59 expression threshold of 50% (median value) was determined to split the LRRC59 high-expression and low-expression cohorts. Therefore, samples with LRRC59 expression levels higher and lower than 50% were applied as the high-expression cohort (cuto-high) and the low-expression cohort (cuto-low), respectively.
Tissue Samples and Cell Culture
A total of 94 newly diagnosed primary LUAD patients (Aged 62±10) were recruited in Huzhou Centre Hospital between July 2004 to April 2009, and their clinicopathological characteristics were retrieved (Table 1). Paired tissue specimens were collected during surgery, including fresh lung tumor and matched adjacent tissues. Two pathologists independently reviewed all pathologic specimens. This study was approved by the Ethics Committee of Huzhou Central Hospital in accordance with the Declaration of Helsinki (approval No.20181102–01), and all the participants provided informed consent. The A549 and H1299 cell lines were obtained from American Type Culture Collection though Shanghai Outdo Biotech Company in china. A549 cells was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Shanghai, China), while H1299 cells was cultured in RPMI-1640 medium (Gibco), containing 10% fetal bovine serum and 1% penicillin/streptomycin, and incubated in a humidified 5% CO2 atmosphere at 37 °C.
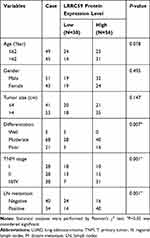 |
Table 1 Association Between LRRC59 Expression and Clinicopathologic of LUAD Patients |
Immunohistochemical Analysis (IHC)
The tissue microarray (TMA) was generated and consisted of 94 LUAD tissues. All stainings were scored based on the staining intensity and extensity of positive cells, the intensity (0=genitive, 1=weak, 2=moderate, and 3=strong) and extensity (0=9% or less of cells stained positive; 1=10%-25%; 2=26% to 50%; 3=51% to 80%; and 4=81% or more) of tumor staining were evaluated. The positive cell density of each core was counted by two independent investigators blind to clinical outcome and knowledge of the clinicopathological data. The final score was calculated by adding the strongest intensity score and the total extensity score (maximum value of 7). The cut-off value of LRRC59 expression was determined by median IHC score. We defined a sample as high LRRC59 expression when the sample had a combined score greater than 4, and a sample was low expression LRRC59 if the combined score ≤4.
Quantitative Real-Time Polymerase Chain Reaction
mRNA transcripts were quantified by quantitative real-time polymerase chain reaction (RT-qPCR). Briefly, RNAs from lung cancer cell lines were isolated with RNAiso Trizol (Invitrogen, Shanghai, China). cDNA was reversed transcribed using the PrimeScriptTM RT reagent kit (Takara, Shiga, Japan). Then, cDNA templates were amplified by real-time RT-PCR using the SYBR Green PCR Kit (Takara) by the ABI7500 System (Applied Biosystems, USA). β-actin was used as an internal control. The relative expression levels of mRNA were calculated by the 2−ΔΔCt method. The primer (10μM for each gene) sequences were as follows: LRRC59, 5′-TGACTACTCTACCGTCGGATTT-3′(for), 5′-TTCAGGTCCAACCACTTCAGG-3′(rev); β-actin, 5′-ACGCCAACACAGTGCTGTCTG-3′(for), 5′-GGCCGGACTCGTCATACTCC-3′(rev).
Short Hairpin RNA Lentivirus Transduction
The short hairpin RNA (shRNA) lentivirus transduction approach was used to knockdown LRRC59 gene expression. Lentivirus vector and two short hairpin RNA (shRNA) targeting LRRC59 were purchased from Genepharma Shanghai. Targeting LRRC59 sequences were listed as followed: shRNA#1:5ʹ-CCTGGATCTGTCTTGTAATAA-3ʹ; shRNA#2:5ʹ-GCAGTGTAAAGCAGTGTGCAAA-3ʹ. shCon:5ʹ-AATACGGCGATGTGTCAGG-3ʹ, The shRNAs were packaged into lentiviruses by cotransfecting with packaging plasmids pMD2G and pSPAX2 into 293T cells. Forty-eight hours after transfection, the supernatant was collected for infection of A549/H1299 cells. Infected cells were selected with media containing puromycin (2 μg/mL) for 3 days.
Immunoblot Analysis
Lung cancer cell was lysed for 30 min on ice in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% Na deoxycholate, 0.1% SDS, 1 mM Na3VO4, 1 mM PMSF, 10 mg/mL leupeptin, 10 mg/mL aprotinin) that contained 1mM PMSF. Whole-cell lysates containing 50 µg of proteins, were separated by 10% SDS-PAGE gel electrophoresis gels, the proteins were transferred to a 0.45-μm hydrophobic PVDF transfer membrane (Millipore) using a semidry transfer apparatus (Bio-Rad). The transfer ran at 17V for 30min, and blocked for 1h at room temperature in a 5% BSA solution in 1×TBST. Afterwards, the membrane was rinsed and immunoblotted with specific primary antibodies. Antibodies for β-actin from Cell Signaling Technology (USA), the antibody for LRRC59 was purchased from Abcam (ab127912). And incubations of the appropriate secondary antibody (Cell signaling Technology) for 1h at room temperature. Antibodies were used at 1:1000 dilutions according to the manufacturer’s instructions. Immunoreactive proteins were detected using an enhanced chemiluminescent (ECL) reagent (Thermo Scientific, Rockford, USA) and images were captured using Azure Biosystem C600 (Azure Biosystems, CA, USA).
CCK-8 Assay
Cell viability was assessed by Cell Counting Kit 8(CCK8, Dojindo, Japan). Briefly, cells were seeded in 96-well plate at a density of 1,000 cells/well and incubated at 37 °C for 24 to 96 h;10 μL CCK-8 solution was added to each well, followed by incubation for 3h. The absorbance value of each well was measured at 450 nm. Each sample was assayed in four duplicates.
Colony-Formation Assay
LRRC59 knockdown and control cells were digested by trypsin and seeded in a 6-well plate at a density of 300 cells/well. All the cells were cultured for ten days. Then the clones were washed by PBS, fixed in 4% paraformaldehyde, stained with 0.1% crystal violet and counted by Image J. Data represent the average of three independent experiments.
Cell Cycle Analysis
The analysis of cell cycle was used by Cell Cycle Analysis kit (BD Biosciences, Shanghai, China) and measured according to their protocols. In brief, cells were harvested at 96 h after infection and resuspended in PBS containing 1% FBS. Cells (1 × 106 to 3 × 106) were fixed with 70% ethanol and washed twice before incubating for 15 min at 37°C with propidium iodide (PI)/RNase A solution (5 μg/mL PI/250 μg/mL RNase A in PBS/1% FBS). The incorporation of PI was quantified by FACScan (BD FACS Canto II). The percentage of cells in each phage of the cell cycle was assessed using Modfit LT software.
Apoptosis Analysis
To detect apoptosis, the expression of Annexin V/propidium Iodide (PI) was evaluated using the Annexin V-FITC Apoptosis Detection kit (BD pharmingen, USA). Briefly, cells were gently detached, then harvested and washed with cold PBS. Cells (7 × 104) were then suspended in 200 µL binding buffer containing Annexin V, and incubated for 10 min at room temperature. After centrifugation (300 g, 3 min), cells pellet was resuspended in 200 µL binding buffer containing 5 µg/mL propidium iodide, and stained with Annexin V-FITC and PI. Then cells were analyzed using flow cytometry in three different experiments (BD FACS Canto II).
Transwell Assay
Cell migration ability of A549/H1299 cells was estimated by transwell assay using Transwell chamber with pore size of 8.0 μm (Millipore) according to the manufacturer’s instructions. Cell invasion ability was performed by using Transwell plates coated with Matrigel (BD Biosciences, USA). 1× 105 constructed cell clones were suspended in serum-free medium and plated on transwell chambers. The medium containing 10% FBS was added to the lower chamber as chemoattractant. After 24 h, the chambers were stained with 1% crystal violet solution for 15 min and immersed in PBS for 10 min. Then, the cells in the lower chamber were observed and counted under an inverted microscope. The values are expressed as the mean cell numbers under five random fields of view (200×). Three independent experiments were conducted for the same conditions.
Statistic Analysis
All data were analyzed using SPSS pack 26.0 statical software and Graphpad Prism 5.0 software. Survival analyses Bivariate comparisons of clinicopathlogical features between patients with high or low LRRC59 scores were performed using χ2-test. The association of multiple prognostic factors with cancer-specific survival was accessed using univariate and multivariate Cox proportional hazards regression model analysis. Survival analyses were performed using Kaplan–Meier curves and Log rank test. Data are presented as mean±standard deviation of at least three independent experiments. Between-groups comparisons were performed using Student’s t-tests, P<0.05 was considered to indicate a statistically significant difference.
Results
High LRRC59 mRNA Level Was Associated with Worse Prognosis in LUAD
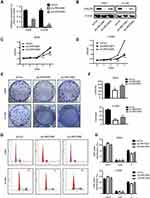
To evaluate the relationship between LRRC59 and lung cancer, analysis using the GEPIA2 online tool indicated an upregulated trend of LRRC59 mRNA expression in lung adenocarcinoma and lung squamous cell carcinomas compared with that in normal samples (Figure 1A). Moreover, high mRNA expression of LRRC59 was significantly associated with worse survival in lung cancer patients (Figure 1B). Interestingly, the higher mRNA expression level of LRRC59 was associated only with poor OS for LUAD, but not for lung squamous cell carcinoma (Figure 1C and D).
 |
Figure 1 The prognostic value of the mRNA expression of LRRC59 by online tool GPEIA2 (http://gepia2.cancer-pku.cn). (A) LRRC59 mRNA expression levels in lung adenocarcinoma and lung squamous cell carcinomas are higher than normal tissues. The signature score is calculated by mean value of log2(TPM + 1) of LRRC59 in lung cancer and normal tissues. The |Log2FC| cutoff of the expression of proposed biomarker was 1. The p-value cutoff of the expression of proposed biomarker was 0.01. The red box indicates the tumor samples while the gray one represents the normal tissues. (B) Prognostic HRs of LRRC59 in all lung cancer. Survival curves of LRRC59 in lung adenocarcinoma (C) and lung squamous cell carcinoma (D). Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TPM, transcripts per million; HR, hazard ratio. |
Knockdown of LRRC59 Inhibited Lung Cancer Cell Proliferation and Metastasis
To explore the function of LRRC59 in lung cancer, we generated LRRC59 knockdown cells by transducing LRRC59 shRNAs into H1299 and A549 cells. As shown in Figure 2A and B, shRNA#1 led to a substantial decrease of LRRC59 expression in H1299 and A549 cells at the RNA and protein levels, while shRNA#2 inhibited LRRC59 expression in A549 cells not well as shRNA#1. Cell proliferation was tested by CCK-8 assay. The results showed that the proliferation of H1299 and A549 cells in the shLRRC59 group was significantly impaired compared with that in the control group (p<0.001, Figure 2C and D). Colony‐formation assays revealed that silencing LRRC59 (shLRRC59) dramatically inhibited the size and number of colonies in both cell lines, compared with the cells in the shCon group (Figure 2E and F).
As proliferation-suppression phenotypes were observed after depletion of LRRC59 by RNA interference, cell cycle distribution was further detected by flow cytometry. Consistently, A significant increase in the G1 phase was observed in A549 and H1299 cells transduced with shLRRC59s compared with control cells (Figure 2G and H).
Furthermore, the effects of LRRC59 on the migrated and invasive ability of lung cancer cells were assessed by Transwell assay. As shown in Figure 3, the silencing of LRRC59 in A549 and H1299 cells caused a significant reduction in cell migration (Figure 3A and B) and cell invasion (Figure 3C and D). These data indicated that LRRC59 plays a vital role in cancer cell migration, invasion, and tumor progression.
LRRC59 Depletion Induced the Apoptosis of A549 Cells, but Not P53-Deficient H1299 Cells
To estimate the effect of LRRC59 knockdown on apoptosis, Annexin V‐FITC/PI double staining and flow cytometry was performed in A549 and H1299 cells (Figure 4). Our result showed that the apoptosis rate of LRRC59 knockdown in A549 cells was more significantly higher than that in control cells, while silencing LRRC59 expression has a slight effect on apoptosis in H1299 cells.
LRRC59 Serves as an Independent Factor for the Prediction of Worse Survival in LUAD
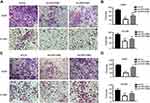
Our results revealed that LRRC59 is an oncogenic factor that contributes to cancer proliferation and metastasis. The online survival tool analysis (GEPIA2) also indicated that LUAD patients with high mRNA levels of LRRC59 had poorer prognosis. To further confirm that LRRC59 could be an independent risk factor of worse outcomes in LUAD patients, the expression and localization of LRRC59 were analyzed by TMA-based IHC. The expression of LRRC59 was mainly localized in the cancer cell cytoplasm (Figure 5A–C); Whereas, LRRC59 expression in the nuclei was sparse. Additionally, we compared 94 lung adenocarcinoma specimens with paired adjacent normal tissue in a large microarray, the average staining score of tumor tissue (avg=5.47) was significantly higher than adjacent tissue (avg=1.48, P< 0.001, Figure 5D).
Then, the association between LRRC59 expression and the clinicopathological variables of 94 patients was analyzed. The level of LRRC59 expression was divided into low group (n=38) and high group (n=56) according to the cutoff value stated in methods mentioned before. The data are summarized in Table 1. High LRRC59 expression was closely associated with lymph node metastasis (P<0.001), TNM stage (P<0.001), histological differentiation (P=0.007), while there was no correlation with age (P=0.078), gender (P=0.495), tumor size (P=0.147).
Furthermore, we employed Kaplan–Meier analysis to evaluate the relationship between LRRC59 expression and OS in LUAD patients. The results showed that patients with high LRRC59 expression displayed a shorter overall survival than those of low LRRC59 expression (P=0.02) (Figure 5E), which was similar to that of the GEPIA2 online prediction. Importantly, multivariate Cox regression analysis indicated that high expression of LRRC59 protein was an independent prognostic factor of LUAD (Table 2).
 |
Table 2 Univariate and Multivariate Cox Proportional Hazards Analysis of LRRC59 Expression and OS for LUAD Patients |
Discussion
LUAD is one of the most aggressive and rapidly mortal tumor types with overall survival less than five years. The poor prognosis of LUAD is mainly due to late diagnosis and indistinct mechanism of cancer development. The identification of novel therapeutic targets in LUAD is urgently needed.
Recently, Yu et al showed that a fusion transcript of LRRC59-FLJ60017 among seven other fusion transcripts is strongly associated with prostate cancer prognosis A combination of these fusion transcripts in prostate cancer with Gleason’s grading was predictive of prostate carcinoma recurrence. In this study, we found that LRRC59 is widely upregulated in LUAD specimens and correlates with TNM stage, lymph node metastasis, histological differentiation, and independently predicts shorter overall survival in LUAD patients. Combined with the predictive results of biological information, we speculate that LRRC59 might play a tumor-promoting role in the LUAD process.
Although the altered expression of LRRC59 proteins has been reported in a variety of human cancers,8,9 the primary function of LRRC59 is largely unknown at present. LRRC59 was initially identified as an intracellular binding partner of Fibroblast Growth Factor 1 (FGF1) and required for the nuclear import of FGF1.5 It is well known that FGF1 is wildly involved in cell proliferation, survival, migration and invasion,10 which is easily hijacked and rewired by cancer cells to foster tumor growth, survival, and metastasis. Our data showed that knockdown of LRRC59 significantly inhibited the proliferation and induced G1/S phase arrest of LUAD cells. Moreover, we found that silencing LRRC59 induced the apoptosis of A549 cells, but not p53-deficient H1299 cells. Consistently, previous reports have shown that intracellular FGF1 inhibits both p53-dependent apoptosis and cell growth arrest via an intracrine pathway.10–13 However, we did not detailed the FGF1 pathways in our study, whether LRRC59 modulates LUAD progression via FGF1 pathway remains further exploration.
Previous studies have shown that the upregulation of LRRC59 was correlated with the metastatic capability of breast cancer cell lines,7,8,14 the silencing of LRRC59 caused an evident transition from a mesenchymal phenotype toward an epithelial one, and strongly impaired wound closure. Consistently, we found that LRRC59 is widely upregulated in LUAD specimens and positively correlates with lymph node metastasis. Moreover, a significant decrease in migration and invasion were observed in both A549 and H1299 cells with LRRC59 knockdown. Thus, we preliminarily determined that LRRC59 is a poor prognostic factor for LUAD, and might serve a key role in LUAD metastasis.
Recently, LRRC59 is attracting more attention as a potential oncogene involved in nuclear import of FGF1 and the cancerous inhibitor of PP2A (CIP2A)6. FGF1 and CIP2A is over-expressed in a variety of cancers,15–18 including lung cancer. In addition, LRRC59 gene amplification or protein overexpression is observed in several kinds of cancers.7–9,14 In this study, we showed that LRRC59 is widely upregulated in LUAD specimens and associated with poor prognosis. This is the first research on LRRC59 effects on prognosis of LUAD. We hope this study can trigger more interest in LRRC59 function and targeted therapy in LUAD. Unfortunately, this research focused on clinical study of the correlation between LRRC59 and LUAD prognosis, and did not determine whether LRRC59 modulates LUAD progression via FGF1 or CIP2A pathway. More experiments are needed to elucidate downstream signaling pathways initiated by LRRC59 as well as the entire signaling network of LRRC59. In addition, the animal model is an essential tool for studying the role of biomarkers in lung cancer progression. We hope that our results in vitro can trigger further investigation of the role of LRRC59 in LUAD in vivo.
Conclusion
In conclusion, we initially reported that LRRC59 was overexpressed in LUAD tissues compared with adjacent nontumorous lung tissues, and it was significantly associated with TNM stage, lymph node metastasis, histological differentiation and unfavorable prognosis of LUAD. In addition, knockdown of LRRC59 could substantially inhibit lung cancer cell proliferation, colony formation, migration, and invasion by in vitro study. Together, our findings indicate that LRRC59 may represent a potential biomarker and a promising therapeutic target for LUAD.
Acknowledgments
This work was supported by grants from the Technology Project of Medical & Health Science in Zhejiang province (2019RC286).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Siegel R, Xu J, et al. Cancer statistics. CA Cancer J Clin. 2010;60(5):277–300. doi:10.3322/caac.20073
2. Skjerpen CS, Wesche J, Olsnes S. Identification of ribosome-binding protein p34 as an intracellular protein that binds acidic fibroblast growth factor. J Biol Chem. 2002;277(26):23864–23871. doi:10.1074/jbc.M112193200
3. Ichimura T, Ohsumi T, Shindo Y, et al. Isolation and some properties of a 34‐kDa‐membrane protein that may be responsible for ribosome binding in rat liver rough microsomes. FEBS Lett. 1992;296(1):7–10.
4. Ohsumi T, Ichimura T, Sugano H, et al. Ribosome-binding protein p34 is a member of the leucine-rich-repeat-protein superfamily. Biochem J. 1993;294(2):465.
5. Zhen Y, Sorensen V, Skjerpen CS, et al. Nuclear import of exogenous FGF1 requires the ER-protein LRRC59 and the importins Kpnalpha1 and Kpnbeta1. Traffic. 2012;13(5):650–664. doi:10.1111/j.1600-0854.2012.01341.x
6. Pallai R, Bhaskar A, Barnett-Bernodat N, et al. Leucine-rich repeat-containing protein 59 mediates nuclear import of cancerous inhibitor of PP2A in prostate cancer cells. Tumour Biol. 2015;36(8):6383–6390. doi:10.1007/s13277-015-3326-1
7. Yu YP, Liu P, Nelson J, et al. Identification of recurrent fusion genes across multiple cancer types. Sci Rep. 2019;9:9.
8. Terp MG, Lund RR, Jensen ON, et al. Identification of markers associated with highly aggressive metastatic phenotypes using quantitative comparative proteomics. Cancer Genomics Proteomics. 2012;9(5):265–273.
9. Zefang T, Boxi K, Chenwei L, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):556–560. doi:10.1093/nar/gkz430
10. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–129. doi:10.1038/nrc2780
11. Yan PY, Ding Y, Chen Z. Novel fusion transcripts associate with progressive prostate cancer. Am J Pathol. 2014;184(10):2840–2849. doi:10.1016/j.ajpath.2014.06.025
12. Bouleau S, Grimal H, Rincheval V, et al. FGF1 inhibits p53-dependent apoptosis and cell cycle arrest via an intracrine pathway. Oncogene. 2005;24(53):7839–7849. doi:10.1038/sj.onc.1208932
13. Pirou C, Montazer-Torbati F, Jah N, et al. FGF1 protects neuroblastoma SH-SY5Y cells from p53-dependent apoptosis through an intracrine pathway regulated by FGF1 phosphorylation. Cell Death Dis. 2017;8(8):e3023. doi:10.1038/cddis.2017.404
14. Rodriguez-Enfedaque A, Bouleau S, Laurent M, et al. FGF1 nuclear translocation is required for both its neurotrophic activity and its p53-dependent apoptosis protection. Biochim Biophys Acta Bioenerg. 2009;1793(11):1719–1727. doi:10.1016/j.bbamcr.2009.09.010
15. Li J, Wei Z, Li H, et al. Clinicopathological significance of fibroblast growth factor 1 in non–small cell lung cancer. Hum Pathol. 2015;46(12):1821–1828. doi:10.1016/j.humpath.2015.07.022
16. Vaarala MH, Väisänen M-R, Ristimäki A. CIP2A expression is increased in prostate cancer. J Exp Clin Cancer Res. 2010;29(1):136. doi:10.1186/1756-9966-29-136
17. Böckelman C, Lassus H, Hemmes A, et al. Prognostic role of CIP2A expression in serous ovarian cancer. Br J Cancer. 2011;105(7):989–995. doi:10.1038/bjc.2011.346
18. Teng HW, Yang SH, Lin JK, et al. CIP2A is a predictor of poor prognosis in colon cancer. J Gastrointest Surg. 2012;16(5):1037–1047. doi:10.1007/s11605-012-1828-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.