Back to Journals » OncoTargets and Therapy » Volume 12
Osteopontin Mediates Cetuximab Resistance via the MAPK Pathway in NSCLC Cells
Authors Cui J, Wang J, Lin C, Liu J, Zuo W
Received 24 August 2019
Accepted for publication 5 November 2019
Published 26 November 2019 Volume 2019:12 Pages 10177—10185
DOI https://doi.org/10.2147/OTT.S228437
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Jian Cui,1,* Jun Wang,2,* Chao Lin,3 Jixiang Liu,1 Wei Zuo1
1Department of Respiratory and Critical Medicine, The First Affiliated Hospital of Nanchang University, Nanchang 330006, People’s Republic of China; 2Department of Rehabilitation Medicine, The First Affiliated Hospital of Nanchang University, Nanchang 330006, People’s Republic of China; 3Department of General Practice, The First Affiliated Hospital of Nanchang University, Nanchang 330006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Zuo
Department of Respiratory and Critical Medicine, The First Affiliated Hospital of Nanchang University, 17 Yongwai Zheng Street, Nanchang, Jiangxi Province 330006, People’s Republic of China
Email [email protected]
Background: Non-small cell lung cancer (NSCLC) is the most common type of lung cancer. The high expression of osteopontin (OPN) is an important factor that aggravates drug resistance and causes a poor prognosis in this disease. Therefore, understanding the molecular mechanism of OPN is critical for the treatment and prognosis of NSCLC.
Methods: We used bioinformatics analysis to verify the expression of OPN in normal lung tissues and lung cancer tissues. Then we overexpressed and knocked down OPN in cell lines to detect cell proliferation, migration, invasion, and effects on signaling pathways. Finally, malignant progression and drug resistance induced by OPN were investigated by the wound healing assay, transwell assay, clone formation assay, and Western blot analysis.
Results: We verified that OPN was upregulated in NSCLC tissues, and its overexpression induced NSCLC cell proliferation, migration, and invasion via the mitogen-activated protein kinase (MAPK) pathway. Furthermore, overexpression of OPN reduced the sensitivity of NSCLC cells to cetuximab by upregulating MAPK pathway-related proteins. These results suggested that OPN promoted malignant progression and mediated drug resistance via the MAPK signaling pathway in NSCLC cells.
Conclusion: This study reveals the important role of OPN in NSCLC cells, making it a potential target for improving chemotherapy efficiency in patients with NSCLC.
Keywords: NSCLC, OPN, MAPK, cetuximab resistance
Introduction
Non-small cell lung cancer (NSCLC), the most common type of lung cancer, is a heterogeneous disease with a complex molecular and genomic profile.1,2 Although most patients are treated with surgery and chemotherapy, their 5-year survival remains poor,3 mostly due to metastasis and chemotherapy resistance.4 Therefore, improving the effective therapeutic modalities for NSCLC is an area of active investigation.
Recently, monoclonal antibodies have become new therapeutic agents for the treatment of malignant disease. Cetuximab (CTX), a chimeric mouse human antibody, recognizes the extracellular domain of epidermal growth factor receptor (EGFR) and inhibits the binding of activating ligands to the receptor, representing an alternative approach to EGFR targeting.5–7 Clinical trials have been conducted to elucidate the benefits of adding CTX to chemotherapy for NSCLC, and cumulative data have been mixed.8 Therefore, identifying novel biomarkers that predict metastasis and drug resistance is important for the treatment of patients with NSCLC.
Osteopontin (OPN), also known as secreted phosphoprotein 1 (SPP1), is a phosphorylated glycoprotein that regulates cell adhesion, migration, and invasion via binding to cell surface receptors including cluster of differentiation 44 and αvβ3 integrin.9,10 OPN is overexpressed and positively correlated with drug resistance in numerous types of cancer including breast, prostate, and ovarian cancers.11–13 Studies have shown that high OPN expression is related to tumor staging, lymph node invasion, and tumor growth, promotes cisplatin/radiation resistance and is associated with a poor survival rate in lung cancer patients.14–17 Despite numerous studies indicating that OPN may be involved in multiple pathological processes in NSCLC, the correlation between OPN expression levels and CTX resistance in NSCLC remains unknown.
Some studies suggest that 1–1.5% of all genes are hypoxia-regulated, and OPN is one of these genes.18 The downstream effects of tumor hypoxia on hypoxia-inducible factor 1-alpha activation and angiogenesis may contribute to resistance to radiation and CTX.19,20 Therefore, the aim of the present study was to assess the effects of OPN expression levels on NSCLC cell proliferation, invasion, and CTX resistance and to investigate the potential mechanism by in vitro biological experiments. Our findings indicate that OPN facilitates the oncogenesis and CTX resistance of NSCLC by modulating the mitogen-activated protein kinase (MAPK) signaling pathway.
Materials and Methods
Cell Lines and Cell Culture
The human NSCLC cell line, A549, was purchased from Guangzhou Cellcook Biotech Co., Ltd. (Guangzhou, China). Cells were maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Gaithersburg, MD, USA), supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin at 37°C with 5% CO2 in a humidified incubator.
Lentivirus Transduction
GFP-expressing LV-OPN and sh-OPN lentivirus were constructed by Shanghai GeneChem Co., Ltd. (Shanghai, China). Empty GFP-expression lentivirus was used as the negative control (NC). A549 cells were pretreated with LV-OPN or sh-OPN at a multiplicity of infection of 10. The supernatant was removed after 16 h and replaced with complete culture medium. Quantitative PCR (qPCR) and Western blotting were conducted to verify the transduction efficiency.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Dojindo, Japan) was used to determine cell viability. Cells at a density of 5×103 cells/well in 96-well plates were incubated for 24 h. Then medium containing CCK-8 solution (10 mL CCK-8 in 100 mL medium) was added to each well at the same time daily for 3 days. The optical density values were detected at an absorbance of 450 nm. All experiments were performed in triplicate.
Wound Healing Assay
Cells at a density of 5×106 cells/well were seeded in 6-well plates and cultured overnight to allow the cells to reach 100% confluence. A sterilized pipette tip was used to scratch a fine line with the same width on the cell layer. Cells were washed, and 2 mL serum-free medium was added to each well. Photos were taken at 0 and 18 h after scratching.
Cell Migration and Invasion Assays
Cells at a density of 5×104 cells/well were placed in the upper chamber with or without pre-coated Matrigel (8 μm; BD Biosciences, Franklin Lakes, NJ, USA) of 24-well plates in serum-free media, whereas medium containing 10% FBS was added to the lower chamber. After 24 h incubation, cells that remained in the upper chamber were removed with cotton wool and those that invaded into the lower surface were fixed in 4% paraformaldehyde and stained with Giemsa. The number of cells was counted under a microscope. Each experiment was repeated three times.
Clone Formation Assay
Cells at a density of 5×103 cells/well were seeded in 10 cm diameter tissue culture dishes. After colonies formed, the cells were stained with 0.1% Crystal Violet. The colonies with ≥50 cells were counted using a microscope.
Quantitative PCR
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions and then reverse transcribed into cDNA with the Superscript III First-Strand Synthesis System (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed using the SYBR Green Real-Time PCR Master Mix and StepOnePlus Real-Time PCR system (Thermo Fisher Scientific). The relative expression of OPN was determined by the 2−ΔΔCt method using GAPDH as an internal control for normalization. The following primers were used: GAPDH, 5′-GGAGCGAGATCCCTCCAAAAT-3ʹ (forward) and 5′-GGCTGTTGTCATACTTCTCATGG-3ʹ (reverse). OPN, 5ʹ-GATATAGATACTGATAGATCTAGATATG-3ʹ (forward) and 5ʹ-CTCTCTTGTCTAGTCTGTATGTTC-3ʹ (reverse).
Western Blotting
A total of 30 μg protein was separated via a 10% SDS-PAGE gel and electrotransferred onto polyvinylidene difluoride membrane membranes. Then the membranes were blocked in 5% BSA for 1 h at room temperature and subsequently incubated overnight at 4°C with the following primary antibodies: OPN (1:1000; Abcam, Cambridge, MA, USA), mitogen-activated protein kinase kinase (MEK1/2, 1:1000; Cell Signaling Technology [CST], Danvers, MA, USA), phosphorylated MEK1/2 (p-MEK1/2, 1:1000; CST), extracellular signal-regulated kinase 1/2 (ERK1/2, 1:1000; CST), phosphorylated ERK1/2 (p-ERK1/2, 1:1000; CST), and GAPDH (1:1000; Proteintech, Rosemont, IL, USA). After three washes with Tris-Buffered Saline with Tween 20, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. Protein bands were detected on a Bio-Rad Image Lab System.
In Vitro Drug Treatments
For gene expression studies involving drug treatment, A549 cells were pretreated with LV-OPN lentivirus for 16 h and then treated with 10 µM U0126 ((MEK inhibitor) and 10 µM SCH772984 (ERK1/2 inhibitor) for 12 h. In clone formation experiments, CTX (Erbitux, Merck, Darmstadt, Germany) were administered at different concentrations (0, 1, 5, 10, 20 μg/mL), every 2 days. In CCK-8 experiments, CTX was administered at a concentration of 10 μg/mL for 24 h.
Analysis of Clinical Data
We analyzed the mRNA level of OPN in The Cancer Genome Atlas (TCGA) Lung Adenocarcinoma cohort, which was downloaded from TCGA. A heatmap was used to illustrate gene expression in clinical samples using the Oncomine online database (http://www.oncomine.com).
Statistical Analysis
Statistical analyses were performed using SPSS version 23.0 (SPSS Inc., Chicago, IL, USA) and derived from at least three independent replicates. Data were calculated by one-way analysis of variance and presented as the mean ± standard error of the mean. P values less than 0.05 were considered statistically significant.
Results
Elevated Expression of OPN in Human NSCLC
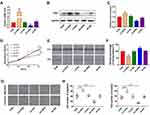
To explore the expression level of OPN in human NSCLC, we compared the mRNA expression of OPN between paired normal and tumor tissues in TCGA database. As bioinformatics analysis revealed, OPN was significantly upregulated in the tumor tissues (Figure 1A). Furthermore, we verified this result through the Oncomine database.21–25 As expected, the elevated expression of OPN was observed in NSCLC relative to normal lung tissues (Figure 1B).
 |
Figure 1 Elevated expression of the OPN gene in human NSCLC tissues. (A) Relative expression of OPN mRNA in 125 human NSCLC tissues and 37 normal tissues based on TCGA data. (B) Heatmap of OPN (also known as SPP1) gene expression in clinical NSCLC samples and normal tissues based on Oncomine data. ****P<0.0001. (1. Lung Adenocarcinoma vs Normal Bhattacharjee Lung, Proc Natl Acad Sci USA, 2001;21 2. Lung Adenocarcinoma vs Normal Hou Lung, PLoS One, 2010;22 3. Lung Adenocarcinoma vs Normal Landi Lung, PLoS One, 2008;23 4. Lung Adenocarcinoma vs Normal Selamat Lung, Genome Res, 2012;24 5. Lung Adenocarcinoma vs Normal Su Lung, BMC Genomics, 2007.25) |
Overexpression of OPN Induces Cell Proliferation, Migration, and Invasion in NSCLC in Vitro
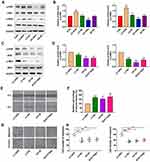
To evaluate the effects of OPN levels on malignant biological properties in NSCLC cells, the lentiviral vector was used to overexpress or silence the OPN gene in A549 cells. Following transfection, qPCR and Western blotting were performed to examine OPN expression. The results showed that OPN was upregulated in overexpressed cells and downregulated in silenced cells (Figure 2A–C). Then CCK-8, wound healing, and transwell assays were performed in the stably transfected A549 cell lines to detect cell proliferation, migration, and invasion, respectively. CCK-8 assays indicated that the overexpression of OPN significantly promoted the proliferation of A549 cells (Figure 2D). Wound healing assays showed that migration ability of the LV-OPN group was significantly higher than that of other groups (Figure 2E and F). Transwell assays revealed that migration and invasion were markedly enhanced in cell lines transfected with LV-OPN compared with LV-NC, whereas the opposite results were found in the silenced group (Figure 2G and H). These results indicated that OPN had positive effects on the malignant biological properties of NSCLC cells.
OPN Promotes a Malignant Phenotype via the MAPK Pathway
It is well known that the MAPK pathway participates in regulating the invasion and metastasis of NSCLC;26 thus, we explored the potential effects of OPN on the MAPK pathway. Western blotting showed that OPN overexpression increased p-MEK and p-ERK in A549 cells, and silencing of OPN had the opposite results (Figure 3A and B). Upon treatment with U0126 (MEK1/2 inhibitor) and SCH772984 (ERK1/2 inhibitor), p-MEK and p-ERK levels were decreased, respectively, compared to the LV-OPN group (Figure 3C and D), and wound healing distances were shorter (Figure 3E and F). The migration and invasion of cells overexpressing OPN were inhibited by U0126 and SCH772984 (Figure 3G and H). These results suggested that malignant behaviors might be induced by OPN in NSCLC cells by activating the MAPK/ERK pathway.
OPN Confers Resistance to CTX via the MAPK Pathway
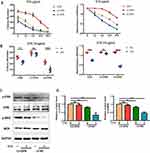
To identify the underlying mechanisms of the effects of OPN on drug resistance, we tested the effects of OPN on the efficacy of CTX in vitro. Treatment of stably transfected cell lines with CTX showed that CTX inhibited cell growth in the control and sh-OPN groups, whereas the LV-OPN group remained refractory to CTX up to 10 μg/mL. As expected, after 24 h of treatment with CTX, the colony number and cell viability did not significantly change in the LV-OPN group (Figure 4A and B). Thus, overexpression of OPN was sufficient to limit the efficacy of cetuximab, as the concentration and time changed. After 24 h of treatment with CTX, Western blotting showed that p-MEK and p-ERK levels were significantly decreased in the LV-NC group. Because stimulation with OPN was shown to activate the MAPK pathway, p-MEK and p-ERK levels did not significantly change in the LV-OPN group despite treatment with CTX (Figure 4C and D). Taken together, these data show that OPN can confer resistance to CTX by activating the MAPK pathway.
Discussion
Lung cancer is the most common malignant tumor worldwide, and NSCLC accounts for more than 80% of pathological types.27,28 Despite advances in surgery, chemotherapy, radiation therapy, and targeted therapy, the overall survival rate of NSCLC has not significantly improved.29 Several clinical trials have added CTX to chemotherapy in patients with NSCLC, but the results have been mixed.30–32 Drug resistance is an important factor leading to chemotherapy failure.33 Therefore, it is important to investigate the cellular mechanisms of drug resistance and to identify potential treatment strategies that significantly improve the efficacy of chemotherapy.
OPN is overexpressed in numerous types of cancer.34 In this study, we found that OPN was also overexpressed in NSCLC by analyzing TCGA database, and further verified this result with the Oncomine database. Several lines of evidence have shown that aberrant OPN expression is associated with the proliferation and metastasis of various tumors.35,36 These studies consistently suggest that OPN overexpression may be a biomarker for poor prognosis in cancer patients. A previous study found that aberrant OPN expression only affected the invasion of A549 cells in vitro but had no effect on cell migration.37 In contrast, our study showed that OPN overexpression induced the proliferation, migration, and invasion of NSCLC cells in vitro. In addition, one study indicated that OPN enhances drug resistance to cisplatin in human SCLC cells.38 However, the involvement of OPN in CTX resistance in NSCLC cells has rarely been reported. In our study, we presented evidence that OPN overexpression was sufficient to limit the efficacy of CTX over time and concentration. We demonstrated that OPN levels were correlated with the malignant biological properties of NSCLC and the efficacy of CTX in NSCLC cells.
The MAPK signaling pathway participates in the regulation of proliferation, migration and apoptosis in many cancer types, including lung cancer.39,40 Some studies have indicated that OPN increases cell migration via the MAPK signaling pathway in human prostate cancer and human chondrosarcoma cells.41,42 In this study, we investigated the role of MAPK signaling pathways in the migration and invasion of NSCLC cells and found that the levels of p-MEK and p-ERK were significantly increased after OPN overexpression, which were dramatically changeover by MAPK pathway inhibitors simultaneously. Similarly, the levels of p-MEK and p-ERK in the control group were significantly reduced after CTX treatment, while no significant changes were observed in the OPN overexpression group. Hence, we deduced that OPN may be crucial in NSCLC development and drug resistance via activating the MAPK signaling pathway. These data are not consistent with a previous study, which demonstrated that the DNA damaging reagents bleomycin and doxorubicin induced OPN expression through the ERK pathway in A549 cells.43 It seems that MAPK signaling pathway exerts different effects on NSCLC with different drugs. This discrepancy needs to be further explored.
This study had a couple of limitations. We did not use a large number of complete clinical samples to verify the prognostic value of OPN. In addition, regarding the mechanism, we only studied the changes of markers in the MAPK pathway, and the direct mechanism underlying OPN regulation of MAPK was not clarified. In vitro experiments are needed to verify our conclusions.
In summary, our study showed that OPN promotes malignant progression and confers drug resistance via the MAPK signaling pathway in NSCLC cells. Thus, OPN may be a promising therapeutic target for the treatment of NSCLC. Specific silencing of OPN may be a future direction for developing a novel therapeutic strategy for NSCLC patients.
Acknowledgments
This study was supported by grants from the Education Department of Jiangxi Province (GJJ180008).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31(8):1039–1049. doi:10.1200/JCO.2012.45.3753
3. Coate LE, Shepherd FA. Maintenance therapy in advanced non-small cell lung cancer: evolution, tolerability and outcomes. Ther Adv Med Oncol. 2011;3(3):139–157. doi:10.1177/1758834011399306
4. Zhan J, Wang P, Li S, et al. HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients. Theranostics. 2019;9(7):2084–2099. doi:10.7150/thno.29463
5. Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354(6):567–578. doi:10.1056/NEJMoa053422
6. Rosenthal DI, Harari PM, Giralt J, et al. Association of human papillomavirus and p16 status with outcomes in the IMCL-9815 phase III registration trial for patients with locoregionally advanced oropharyngeal squamous cell carcinoma of the head and neck treated with radiotherapy with or without cetuximab. J Clin Oncol. 2016;34(12):1300–1308. doi:10.1200/JCO.2015.62.5970
7. Borghaei H, Langer CJ, Millenson M, et al. Phase II study of paclitaxel, carboplatin, and cetuximab as first line treatment, for patients with advanced non-small cell lung cancer (NSCLC): results of OPN-017. J Thorac Oncol. 2008;3(11):1286–1292. doi:10.1097/JTO.0b013e318189f50e
8. Moran T. Is more the better?-Cetuximab in non-small cell lung cancer patients. Transl Lung Cancer Res. 2018;7(Suppl 3):S195–S197. doi:10.21037/tlcr.2018.04.14
9. Weber GF, Lett GS, Haubein NC. Osteopontin is a marker for cancer aggressiveness and patient survival. Br J Cancer. 2010;103(6):861–869. doi:10.1038/sj.bjc.6605834
10. Anborgh PH, Mutrie JC, Tuck AB, Chambers AF. Role of the metastasis-promoting protein osteopontin in the tumour microenvironment. J Cell Mol Med. 2010;14(8):2037–2044. doi:10.1111/jcmm.2010.14.issue-8
11. Bandopadhyay M, Bulbule A, Butti R, et al. Osteopontin as a therapeutic target for cancer. Expert Opin Ther Targets. 2014;18(8):883–895. doi:10.1517/14728222.2014.925447
12. Pang H, Cai L, Yang Y, Chen X, Sui G, Zhao C. Knockdown of osteopontin chemosensitizes MDA-MB-231 cells to cyclophosphamide by enhancing apoptosis through activating p38 MAPK pathway. Cancer Biother Radiopharm. 2011;26(2):165–173. doi:10.1089/cbr.2010.0838
13. Hsieh IS, Huang WH, Liou HC, Chuang WJ, Yang RS, Fu WM. Upregulation of drug transporter expression by osteopontin in prostate cancer cells. Mol Pharmacol. 2013;83(5):968–977. doi:10.1124/mol.112.082339
14. Donati V, Boldrini L, Dell’Omodarme M, et al. Osteopontin expression and prognostic significance in non-small cell lung cancer. Clin Cancer Res. 2005;11(18):6459–6465. doi:10.1158/1078-0432.CCR-05-0541
15. Ouyang X, Huang Y, Jin X, et al. Osteopontin promotes cancer cell drug resistance, invasion, and lactate production and is associated with poor outcome of patients with advanced non-small-cell lung cancer. Onco Targets Ther. 2018;11:5933–5941. doi:10.2147/OTT
16. Hu Z, Lin D, Yuan J, et al. Overexpression of osteopontin is associated with more aggressive phenotypes in human non-small cell lung cancer. Clin Cancer Res. 2005;11(13):4646–4652. doi:10.1158/1078-0432.CCR-04-2013
17. Wang M, Han J, Marcar L, et al. Radiation resistance in KRAS-mutated lung cancer is enabled by stem-like properties mediated by an osteopontin-EGFR pathway. Cancer Res. 2017;77(8):2018–2028. doi:10.1158/0008-5472.CAN-16-0808
18. Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi:10.1038/nrc704
19. Viloria-Petit A, Crombet T, Jothy S, et al. Acquired resistance to the antitumor effect of epidermal growth factor receptor-blocking antibodies in vivo: a role for altered tumor angiogenesis. Cancer Res. 2001;61(13):5090–5101.
20. Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem. 2006;281(36):25903–25914. doi:10.1074/jbc.M603414200
21. Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi:10.1073/pnas.191502998
22. Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. doi:10.1371/journal.pone.0010312
23. Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. doi:10.1371/journal.pone.0001651
24. Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7):1197–1211. doi:10.1101/gr.132662.111
25. Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi:10.1186/1471-2164-8-140
26. Wei CH, Wu G, Cai Q, et al. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol. 2017;10(1):125. doi:10.1186/s13045-017-0493-0
27. Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2012;10(10):1236–1271. doi:10.6004/jnccn.2012.0130
28. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
29. Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725–737. doi:10.1038/nrc.2017.87
30. Herbst RS, Kelly K, Chansky K, et al. Phase II selection design trial of concurrent chemotherapy and cetuximab versus chemotherapy followed by cetuximab in advanced-stage non-small-cell lung cancer: Southwest Oncology Group study S0342. J Clin Oncol. 2010;28(31):4747–4754. doi:10.1200/JCO.2009.27.9356
31. Kim ES, Moon J, Herbst RS, et al. Phase II trial of carboplatin, paclitaxel, cetuximab, and bevacizumab followed by cetuximab and bevacizumab in advanced nonsquamous non-small-cell lung cancer: SWOG S0536. J Thorac Oncol. 2013;8(12):1519–1528. doi:10.1097/JTO.0000000000000009
32. Lynch TJ, Patel T, Dreisbach L, et al. Cetuximab and first-line taxane/carboplatin chemotherapy in advanced non-small-cell lung cancer: results of the randomized multicenter phase III trial BMS099. J Clin Oncol. 2010;28(6):911–917. doi:10.1200/JCO.2009.21.9618
33. Salama JK, Vokes EE. New radiotherapy and chemoradiotherapy approaches for non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1029–1038. doi:10.1200/JCO.2012.44.5064
34. Coppola D, Szabo M, Boulware D, et al. Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res. 2004;10(1 Pt 1):184–190. doi:10.1158/1078-0432.CCR-1405-2
35. Ortiz-Martinez F, Perez-Balaguer A, Ciprian D, et al. Association of increased osteopontin and splice variant-c mRNA expression with HER2 and triple-negative/basal-like breast carcinomas subtypes and recurrence. Hum Pathol. 2014;45(3):504–512. doi:10.1016/j.humpath.2013.10.015
36. Hao C, Wang Z, Gu Y, Jiang WG, Cheng S. Prognostic value of osteopontin splice variant-c expression in breast cancers: a meta-analysis. Biomed Res Int. 2016;2016:7310694. doi:10.1155/2016/7310694
37. Sun BS, You J, Li Y, Zhang ZF, Wang CL. Osteopontin knockdown suppresses non-small cell lung cancer cell invasion and metastasis. Chin Med J (Engl). 2013;126(9):1683–1688.
38. Gu T, Ohashi R, Cui R, et al. Osteopontin is involved in the development of acquired chemo-resistance of cisplatin in small cell lung cancer. Lung Cancer. 2009;66(2):176–183. doi:10.1016/j.lungcan.2009.02.004
39. Herner A, Sauliunaite D, Michalski CW, et al. Glutamate increases pancreatic cancer cell invasion and migration via AMPA receptor activation and Kras-MAPK signaling. Int J Cancer. 2011;129(10):2349–2359. doi:10.1002/ijc.v129.10
40. Zhu J, Wang L, Liao R. Long non-coding RNA SDPR-AS affects the development of non-small cell lung cancer by regulating SDPR through p38 MAPK/ERK signals. Artif Cells Nanomed Biotechnol. 2019;47(1):3172–3179. doi:10.1080/21691401.2019.1642904
41. Robertson BW, Bonsal L, Chellaiah MA. Regulation of Erk1/2 activation by osteopontin in PC3 human prostate cancer cells. Mol Cancer. 2010;9:260. doi:10.1186/1476-4598-9-260
42. Chen YJ, Wei YY, Chen HT, et al. Osteopontin increases migration and MMP-9 up-regulation via alphavbeta3 integrin, FAK, ERK, and NF-kappaB-dependent pathway in human chondrosarcoma cells. J Cell Physiol. 2009;221(1):98–108. doi:10.1002/jcp.21835
43. Kato A, Okura T, Hamada C, et al. Cell stress induces upregulation of osteopontin via the ERK pathway in type II alveolar epithelial cells. PLoS One. 2014;9(6):e100106. doi:10.1371/journal.pone.0100106
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.