Back to Journals » International Journal of Nanomedicine » Volume 18
Osteoblasts-Derived Exosomal lncRNA-MALAT1 Promotes Osteoclastogenesis by Targeting the miR-124/NFATc1 Signaling Axis in Bone Marrow-Derived Macrophages
Authors Zhang C , Pan L, Zhang H, Ke T, Yang Y, Zhang L, Chen L, Tan J
Received 8 November 2022
Accepted for publication 2 February 2023
Published 15 February 2023 Volume 2023:18 Pages 781—795
DOI https://doi.org/10.2147/IJN.S395607
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Chenyi Zhang,1,* Lai Pan,1,* Haizheng Zhang,1 Ting Ke,1 Yuxuan Yang,1 Lan Zhang,2 Lili Chen,1 Jingyi Tan1
1Department of Periodontology, The Second Affiliated Hospital of Zhejiang University, School of Medicine, Hangzhou, People’s Republic of China; 2Stomatology Department, Zhejiang Hospital, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lili Chen; Jingyi Tan, Email [email protected]; [email protected]
Objective: Emerging studies have explained the crucial role of non-coding RNA (lncRNA) in various pathological progressions. The study was designed to examine the role of lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and miRNA-124 in the differentiation of osteoclasts, to provide new clues or evidences for the pathogenesis of periodontitis.
Methods: We constructed an osteoblast-osteoclast Transwell co-culture system and osteoblast-derived exosomes (OB-exo) intervention model. We assessed the osteoclastogenesis as well as the level of lncRNA-MALAT1 and miRNA-124. The mechanism for lncRNA MALAT1 targeting miR-124 modulating the differentiation of osteoclasts was investigated by cell transfection, quantitative real-time reverse transcription PCR (RT-qPCR), Western blot, and Dual-Luciferase reporter assays.
Results: Osteoblast-derived exosomes were isolated and identified. Co-culture and OB-exo intervention can promote osteoclastogenesis, also significantly up-regulate the expression of MALAT1, while the level of miR-124 is the opposite. Transfection of cells with small interfering RNA (si-MALAT1) and miR-124 mimic decreased the formation of TRAP+ osteoclasts and inhibited the expression of NFATc1. However, the effect was reversed when transfected with miR-124 inhibitor and si-MALAT1. The Dual-Luciferase reporter assay confirmed the binding sites between MALAT1 and miR-124, and miR-124 and NFATc1.
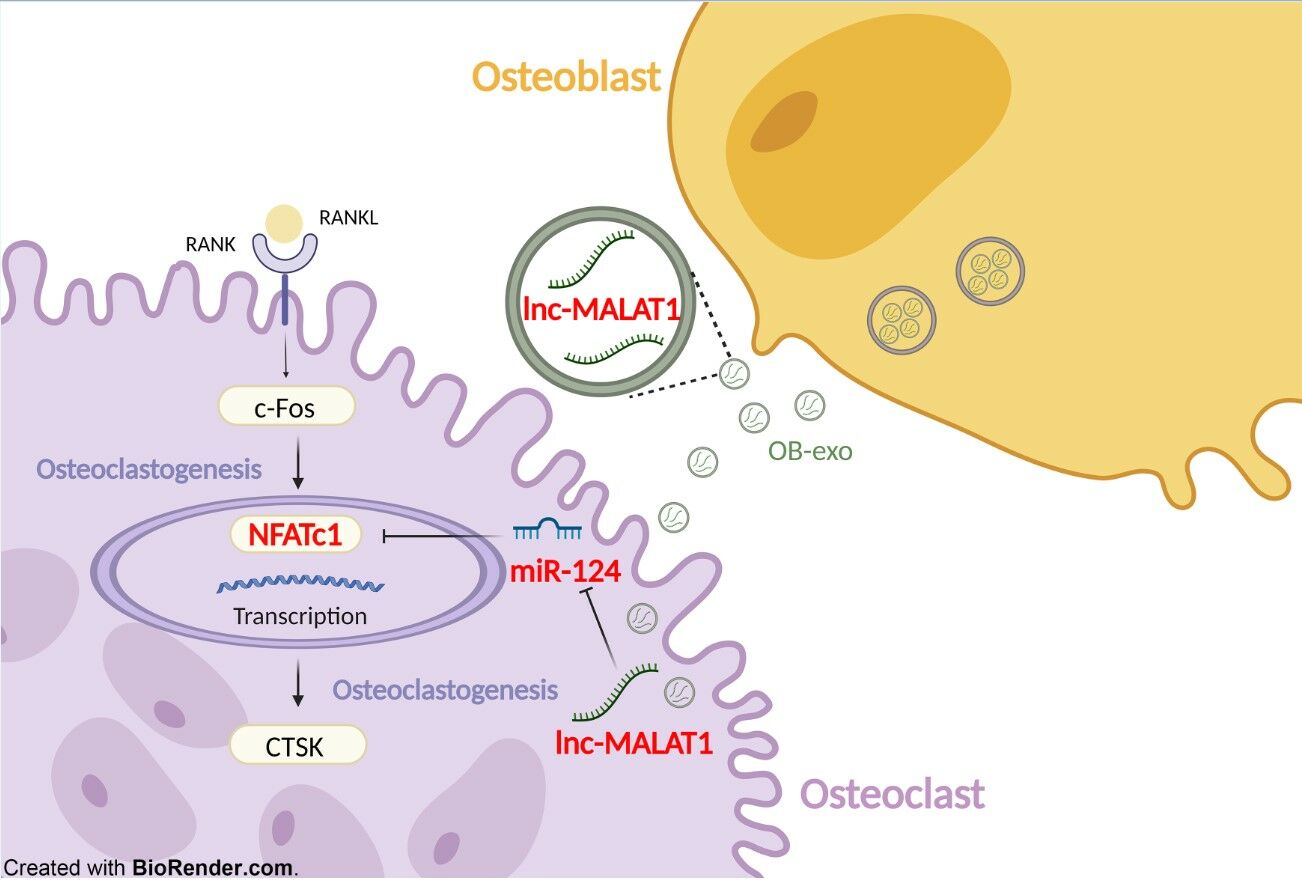
Conclusion: LncRNA MALAT1 functioned as an endogenous sponge by competing for miR-124 binding to regulate NFATc1 expression, accelerating the progression of osteoclastogenesis.
Keywords: exosome, lncRNA, miRNA-124, osteoclast differentiation
Graphical Abstract:
Introduction
Chronic periodontitis initiated by plaque microorganisms is characterized by progressive destruction of periodontal supporting tissue. The structural stability of alveolar bone depends on the dynamic balance between osteoclasts and osteoblasts under physiological conditions. During inflammation, the hyperactivity of mature osteoclasts leads to resorption of alveolar bone. However, the communication molecules mediating the imbalance of bone remodeling homeostasis and their specific regulatory mechanisms have not been clarified yet.
The all-round osteoblast-osteoclast crosstalk plays an important role in triggering periodontal bone remodeling. In recent years, the importance of exosomes in cell–cell interaction has received increasing attention. Exosomes are widely distributed microvesicles with a diameter ranging from 30 to 150nm containing proteins, microRNAs, long non-coding RNAs, etc. Exosomes can be recognized and internalized by recipient cells, releasing and transferring contained active components to regulate a variety of signaling pathways, thus affecting physiological and pathological behaviors. Li et al1 showed that osteoclast-derived exosomes could be transferred into osteoblasts to inhibit activity of osteoblasts in vitro and new bone formation in vivo, therefore having the potential to be a therapeutic target for diseases related to the reduction of bone formation. It is suggested that osteoblast-osteoclast crosstalk may mediate remodeling of bone microenvironment via exosomes rich in bioactive components.
lncRNAs are a series of long non-coding RNAs enriched in exosomes containing more than 200 nucleotides, acting as messenger to regulate biological activities such as cell proliferation, migration and differentiation through corresponding signaling pathways. At present, lncRNA-OIP5-AS1, Linc01133 derived from exosomes have been reported involving in the osteogenic differentiation of human periodontal ligament cells and osteoblasts.2 As a structurally conserved lncRNA extensively involved in immune response, MALAT1 has been found to be active in periodontal bone microenvironment: MALAT1 expression was abnormally up-regulated in gingival fibroblasts from patients with chronic periodontitis,3 as well as in gingival cells treated with LPS, furtherly activated the production and secretion of pro-inflammatory, participating in the occurrence and development of periodontitis. This led to our way of thinking: MALAT1 may have a certain correlation with periodontitis and its specific regulatory mechanism. In addition, lncRNAs are particularly characterized by spongy function, which can compete with mRNA to bind microRNAs (miRNAs), participating in biological activities such as cell cycle and differentiation. Studies by Yang et al4 have shown that MALAT1 has a binding site to miR-34c, acting as a sponge for it to up-regulate SATB2 expression in osteoblasts and the activity of alkaline phosphatase (ALP), ultimately promoting osteogenic activity and mineralization. Yet, the role of MALAT1 in osteoclasts has been poorly reported, which is precisely the focus of our study.
MicroRNAs are a series of micro non-coding RNAs with a length of 22–28 nucleotides, which could typically bind to the 3’UTR region of target gene, thereby regulating its expression level. MicroRNAs function widely in cell proliferation, differentiation and apoptosis. miR-144,5 miR-316 and miR-1407 have been confirmed in emerging studies to be highly involved in the differentiation of osteoblasts and osteoclasts, suggesting that miRNAs in bone microenvironment were important modulators in mediating bone remodeling and regeneration. Currently, it has been reported that miR-124 is a potent functional factor in the inflammatory bone microenvironment, which was down-regulated in metastatic bone tissue and closely correlated with bone resorption.8 In the in vitro experiment, the ability of miR-124 to inhibit the survival and differentiation of osteoclast precursor cells was confirmed by miR-124 mimic and inhibitor. Therefore, we consider that miR-124 is a kernel regulatory in the osteoblast-osteoclast intercellular crosstalk based on its importance in mediating the homeostatic dysregulation of inflammatory alveolar bone remodeling.
In this study, we aim to investigate whether osteoblasts could interfere the differentiation of osteoclast precursor cells by secreting exosomes carrying lncRNA-MALAT1, and the role of osteoblast-derived exosomes (OB-exo) in the progression of periodontitis. It was found that MALAT1 was highly expressed in OB-exo, and delivered into osteoclast precursor cells as components of OB-exo, inhibiting the expression of miR-124, thereby up-regulating the level of osteoclastic differentiation marker NFATc1 and promoting osteoclasts precursor cell differentiation. Our study provides evidence for the communication and crosstalk between osteoblasts and osteoclasts and active bone resorption in periodontitis microenvironment.
Materials and Methods
Cell Culture
Zhejiang University’s Second Affiliated Hospital obtained approval for the study from its experimental animal ethics committee (2018–223) and the animal experiments were performed following Laboratory animal-Guideline for ethical review of animal welfare (GBT 35892–2018).9 Bone marrow cells were flushed from femurs and tibias of C57BL/6 male mice aged 6–8 weeks with α-MEM (Gibco, USA) containing 10% fetal bovine serum (FBS; Bovogen, Australia) and 1% penicillin–streptomycin (TBD, China). The cells were cultured with 50 ng/mL macrophage colony-stimulating factor (M-CSF; Novoprotein, China) for 3 days. The last remained adherent cells were bone marrow-derived macrophages (BMMs) defined as osteoclast precursors cells and used for the following in vitro studies. The induction medium for osteoclasts composed of α-MEM, 50ng/mL M-CSF and 50ng/mL RANKL (Novoprotein). Every alternate day, the medium was changed until multinucleated cells appeared under microscope.
The mouse macrophage cell line RAW264.7 was purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China), then cultured with α-MEM (Gibco) containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (TBD). RAW264.7 cells were treated with 50 ng/mL RANKL to induce osteoclastogenesis.
Calvarial osteoblasts were harvested from calvaria of 1-day-old C56BL/6 mice as described previously.10 Briefly, we cut calvarial bones into small fragments and cultured them in dishes with control medium consisted of 90% H-DMEM (Sigma, USA) and 10% fetal bovine serum (Gibco). Depending on the density of cells, the medium was renewed 2–3 times per week, while further study was conducted on the third to sixth passages. The osteogenic induction medium (Ind) consisted of control medium, 50 μg/mL
All cells were cultured at 37℃ in a humidified 5% CO2 atmosphere.
Co-Culture
After pre-induction, bone marrow-derived macrophages were incubated at 3× 105 cells per well in the lower chamber of a six-well transwell plate, while calvarial osteoblasts (2× 105 cells per well) planted in the upper chamber. The induction medium was composed of 90% α-MEM, 10% FBS, 50ng/mL RANKL, 50ng/mL M-CSF, 50 μg/mL
Exosome Isolation and Identification
Calvarial osteoblasts supernatants were collected and centrifuged at 300g for 10min, 2000g for 20min, 10,000g for 40min, 3000g for 20min at 4°C, excluding cell debris and collecting the supernatants each time. Next, the supernatants were centrifuged at 100,000g for 70min to remove the containing proteins. The pallet was collected and resuspended in phosphate-buffered saline (PBS; TBD), then the centrifugation process of 100,000g repeated for 70min once. The calvarial osteoblasts exosomes (OB-exo) were stored at −80°C.
The identification of exosomes was achieved using transmission electron microscopy (TEM) as well as nanoparticle tracking analysis (NTA). RNA was extracted from exosomes using the Trizol reagent (Invitrogen, USA), while Lysis Buffer and 1% Protease Inhibitor (Sigma) were used to extract proteins for further analysis. The concentration was finally detected.
To investigate the effect of OB-exo on osteoclastogenesis, the BMM cells were treated with 0, 5, 20 µg/mL OB-exo added to the induction medium.
Staining and Cell Identification
Alkaline phosphatase (ALP) staining as well as Alizarin red S (ARS) staining were performed to identify osteogenic differentiation. After osteogenic induction for 7 days, the medium was removed and the cells were softly washed twice with PBS. Then, paraformaldehyde (4%) was put into wells to fix the cells for 20min at room temperature. ALP staining was performed using a BCIP/NBT staining kit (Beyotime, China) following the instructions. The ARS staining was performed with 0.1% alizarin red S (Sigma) after 14 days’ incubation. In order to quantify the calcium nodules, the alizarin red was dissolved with 100 mM cetylpyridinium chloride solution, then the optical density value at 562 nm was measured using spectrophotometry (Bio-Rad, USA).
To identify osteoclastogenesis, tartrate-resistant acid phosphatase (TRAP) staining was performed with Leukocyte Acid Phosphatase Kit (Sigma-Aldrich, USA). Cells stained purple and had more than three nuclei were considered as TRAP+ osteoclasts. Images were acquired by Leica light microscope (Solms, Germany), then the maximum diameter and area of TRAP+ osteoclasts were recorded.
OB-exo was suspended with PBS and 1:50 diluted PKH-67 (Sigma, in Diluent C) to label it. BMMs were cultured with medium added with PKH-67-labeled OB-exo. To observe the cytoskeleton and nuclei, cells were fixed and stained for Phalloidin and DAPI staining. The observation of uptake of exosomes by osteoclasts was under oil or light microscope.
Cell Transfection
A chemically synthesized miRNA mimic, inhibitor and corresponding negative control (TSINGKE, China) were used to overexpress or inhibit miR-124. LncRNA-MALAT1 expression was suppressed using small interfering (si) RNA, targeted to the lncRNA-MALAT1 sequence (GenePharma, China). The efficiency of transfection was detected by real-time RT-qPCR. BMMs were transfected with miR-124 mimic (10nM), inhibitor, miR-NC, si-MALAT1 (20nM) and si-NC with Lipofectamine 2000 (Invitrogen, CA). One day later, the cells were harvested to detect miR-124 and lncRNA-MALAT1 level. The sequences are listed in Table 1.
 |
Table 1 Sequence for Cell Transfection |
Dual-Luciferase Reporter Assay
Luciferase reporters were performed by cloning wild-type-lncRNA-MALAT1, mutant-lncRNA-MALAT1, wild-type-NFATc1-3′UTR, or mutant- NFATc1-3′UTR into psicheck2 vectors. Lipofectamine 3000 (Invitrogen) was used to transfect luciferase reporter plasmids into RAW264.7 cells together with miR-124-3p mimic, inhibitor or miR-NC. After transfection for 48h, activities of firefly luciferase and renilla luciferase were measured separately following the protocols. Firefly luciferase activity was normalized using Renilla luciferase for each sample. Each assay was repeated in three independent experiments.
Cell Viability Assay
Cells were seeded and treated with OB-exo at different concentration in 96-well plates. We measured the cell viability by Cell Counting Kit 8 (CCK-8, Yeasen, China) according to manufacturer instructions. Briefly, 10μL CCK-8 solution was added to wells containing 100μL culture medium at appointed time and incubated at 37 °C with 5% CO2 atmosphere for 2h in dark. Spectrophotometer (Bio-Rad) was used to measure the optical density (OD) values at 450 nm.
Quantitative Real-Time Reverse Transcription PCR (RT-qPCR)
Extract total RNA using Trizol reagent (Invitrogen) on ice 4 days after adding exosomes or after transfection. miR-124-3p of BMMs was extracted by miRNeasy Serum/Plasma kit (QIAGEN, USA). Reverse transcription was performed with 1μg of total RNA in a final volume of 20μL using the Hifair II Strand cDNA Synthesis Kit (Yeasen, China). Each mRNA or miRNA was normalized to the GAPDH or U6 levels, respectively. The expression level of target genes was quantified using a SYBR Green Master Mix kit (Yeasen) on ABI 7500 Fast real-time PCR system (Applied Biosystems, Thermo Fisher Scientific). The primers sequences are listed in Table 2.
 |
Table 2 Primer Sequence for RT-qPCR |
Western Blot Analysis
Cells were washed twice with cold PBS, followed by lysis buffer (Thermo Fisher Scientific, USA) and 1% Protease Inhibitor (Sigma) to extracted proteins. BCA kit (Thermo Fisher Scientific, USA) was used to quantify the concentration of proteins. Proteins were separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Bio-Rad, USA). The membranes were blocked using 5% milk for 1h at room temperature and then incubated overnight with primary antibodies against NFATc1 (DF6446, Affinity, China), c-Fos (26192-1-AP, Proteintech, USA), GAPDH (60004-1-Ig, Proteintech), MMP-9 (ab228402, Abcam, UK), Alix (ET1705-74, HUABIO, China), CD63 (ab217345, Abcam), CD9 (ET1601-9, HUABIO) at 4°C. After three times of washing in TBST (Tris-buffered saline and Tween 20), membranes were incubated with alpaca anti-rabbit antibody (HUABIO, China) and peroxidase conjugated goat anti-mouse antibody (Fudebio, China) at room temperature for 1 h. An ECL kit (Fudebio, China) was used to visualize protein bands after washing. ImageJ software (National Institutes of Health, MD, USA) was used for further analysis.
Statistical Analysis
Each experiment was repeatedly performed three times. Values were presented as the mean ± standard deviation (SD). One-way analysis of variance and Student’s t-test were used to perform comparisons between groups with GraphPad Prism. P value <0.05 was considered statistically significant.
Results
Identification of Osteoblasts and Osteoclasts
BMMs were isolated and stimulated with 50ng/mL RANKL as well as 50ng/mL M-CSF for 4 days continuously, then TRAP staining was performed. The formation of purple stained multinucleated osteoclasts was observed under a light microscope (Figure 1A). Number of TRAP+ cells increased sharply compared with the control group. DAPI staining showed that the mature osteoclasts were multinucleated. The relative expression of osteoclastic markers such as NFATc1, c-Fos, CTSK was up-regulated compared with the NC group detected by real-time PCR and Western blot (Figure 1B). Osteoblasts were identified by ALP and ARS staining (Figure 1C).
Co-Culture Promotes Osteoclastogenesis
To investigate the effect of calvarial osteoblasts on osteoclastogenesis, we plated BMMs in the lower chamber of a six-well Transwell plate, while osteoblasts in the upper chamber (Figure 2A). Co-culture group could find more osteoclasts formation compared with the control group and the group added with exosome release inhibitor GW4869 after stimulation. Compared to the control group and GW4869 group, the maximum diameter, quantity and covering area of OCs in the co-culture group were significantly increased (Figure 2B). To further investigate the effect on osteoclastogenesis, real-time PCR and Western blot were used. The results showed that gene and protein level of NFATC1 and CTSK, which were essential for osteoclastogenesis were consistently up-regulated in the co-culture group (Figure 2C and D). In consideration of OB-exo’s transmission between chambers, it is suggested OB-exo may be the key molecule of communication between OBs and OCs promoting osteoclastogenesis.
Effect of OB-Exo on Osteoclastogenesis
Representative exosomes were observed using transmission electron microscopy (Figure 3A), and the results of NTA indicated that the diameter of particles was mostly concentrated at 110 nm which was considered as OB-exo (Figure 3C). Cytomembrane characteristic proteins such as Alix, CD63 and CD9 were confirmed at high level in osteoblasts-derived exosomes (Figure 3B). In addition, exosomes labeled with PKH-67 could be absorbed by BMMs demonstrated by immunofluorescence assay. Fluorescence images showed that OB-exo was accumulated around nuclei (Figure 3D).
In the co-culture system, exosomes act as an important biological messenger involving in transmitting between chambers to promote communication between osteoblasts and osteoclasts. In order to verify that exosomes indeed have a crucial role in the differentiation of BMMs, OB-exo was isolated by ultracentrifugation and added to the induction medium at the concentration of 0, 5, 20µg/mL to act upon the BMMs. BMMs were additionally incubated for 4 days and stained for TRAP. The number, maximum diameter, covering area of TRAP+ OCs was significantly higher than those of the RANKL group (Figure 3E). Measured cell viability using CCK-8 (Figure 3F). Results of real-time PCR and Western blot indicated that compared with RANKL group, all concentrations of exosomes groups could obviously up-regulate the expression of NFATc1 and CTSK (Figure 3G and H). The up-regulation gradually became stronger as OB-exo concentration increased. These results indicated that OB-exo could stimulate osteoclast differentiation. In addition, we found that the gene expression of c-Fos did not change with OB-exo stimulation. Therefore, we hypothesized that OB-exo promotes differentiation through NFATC1 and CTSK pathway.
Knockdown of lncRNA-MALAT1 Suppressed Osteoclastogenesis in vitro
Total RNA was harvested from exosomes of the OB-NC and OB-Ind groups to measure the level of lncRNA-MALAT1. As expected, the level of MALAT1 sharply increased in the OB-Ind group compared with the OB-NC group (Figure 4A). As a long noncoding RNA, LncRNA-MALAT1 is widely expressed in exosomes of cells such as endothelial progenitor cells, neuroblastoma cells and so on, strongly related to cell damage and inflammation. Our early studies respectively measured the expression of MALAT1 in co-cultured and exosome-stimulated BMM cells. The results showed that the formation of osteoclasts was positively correlated with high MALAT1 expression as well as low miR-124 level (Figure 4B).
As mentioned above, we hypothesized that MALAT1 might regulate osteoclast differentiation and formation. Knockdown of MALAT1 was preformed using siRNA, and the efficiency was verified by real-time PCR (Figure 4C). As expected, TRAP staining indicated that mature multinucleated osteoclasts formation was reduced by inhibition of MALAT1 expression (Figure 4D).
LncRNA-MALAT1 Functions as a Sponge by Binding to miR-124
LncRNA can bind to miRNA through complementary sequences to downregulate its expression. To illustrate the underlying mechanism of lncRNA-MALAT1 promoting osteoclastogenesis, we performed bioinformatics analysis using StarBase and the results showed LncRNA-MALAT1 may contain possible binding sites for miR-124. To investigate the binding relation between MALAT1 and miR-124, the potential binding sequences on MALAT1 were mutated from GUGCCUU to CACGGAA, and followed Dual-Luciferase reporter assays was carried out in RAW264.7 cells (Figure 5A and B). miR-124 mimic reduced the luciferase activity compared with the NC group (P<0.05) and MALAT1 mutated group (P<0.01). What’s more, we measured the expression of miR-124 after transfection of si-MALAT1. Not unfortunately, the expression of miR-124 was effectively up-regulated by knockdown of MALAT1 (P<0.05, Figure 5C). Taken together, the findings investigated that lncRNA-MALAT1 may act as a sponge for miR-124 by binding with it.
NFATc1 Was a Target Gene of miR-124
StarBase was utilized to predict the potential target genes of miR-124, and we found osteoclastic marker NFATc1 was a candidate. To investigate whether miR-124 directly binding to the mRNA 3′-UTR of NFATc1, the potential binding sequences on NFATc1 mRNA were mutated and then luciferase activity was evaluated. As expected, the luciferase activity of the NFATc1 WT group was reduced by miR-124 mimic compared with the NC group (P<0.001, Figure 6A and B), while there was no obvious change after cotransfected with miR-124 mimic and the mutant NFATc1 mRNA.
What’s more, we calculated the expression of miR-124 and lnc-MALAT1 after transfecting miR-124 mimic, inhibitor and miR-NC. The expression of MALAT1 was impaired by overexpressing miR-124 companied with reduced osteoclast differentiation, while it was increased after transfection of miR-124 inhibitor (Figure 6C). In view of the binding site between miR-124 and NFATc1, real-time PCR (Figure 6D) and Western blot (Figure 6E) were performed in order to confirm that miR-124 affected osteoclastogenesis via NFATc1. The results also illustrated the negative regulation of miR-124 on NFATc1 and NFATc1-dependent CTSK. TRAP staining suggested the negative effect of miR-124 on osteoclastogenesis in terms of maximum diameter, covering area and number of TRAP+ OCs (Figure 6F).
Lnc-MALAT1 Up-Regulated NFATc1 Expression and Osteoclastogenesis Through miR-124
Compared with the si-MALAT1 transfection group, TRAP staining showed cells cotransfected with si-MALAT1 and miR-124 inhibitor formed more osteoclasts (Figure 7A), suggesting miR-124 may be a downstream of lncRNA-MALAT1 which could reverse its effect. To test whether MALAT1 could regulated NFATc1 expression via targeting miR-124, si-MALAT1 and miR-124 inhibitor were cotransfected into cells. Compared with the si-NC group, both mRNA (Figure 7B) and protein level (Figure 7C) decreased in the si-MALAT1 group. Nevertheless, miR-124 inhibitor significantly reversed the inhibitory effect of si-MALAT1 on NFATc1 (P<0.05) and CTSK (P<0.001) expression, as well as osteoclastogenesis, suggesting that miR-124 was identified as downstream of lncRNA-MALAT1. Altogether, lnc-MALAT1 promotes osteoclast differentiation via miR-124/NFATc1 axis.
Discussion
Periodontal diseases are basically a homeostasis imbalance of bone-related cells in inflammatory bone microenvironment, affecting periodontal supporting tissues. The process of bone reconstruction and resorption is dependent on various factors including abundant communication between corresponding cells. Although antioxidants for scavenging reactive oxygen species (ROS) could partly reduce bone resorption, some of their clinical application was limited due to low solubility and bioavailability.11,12 Unlike antioxidants, it has been demonstrated that exosomes secreted by various bone-derived cells are carriers of active components regulating bone remodeling,1,4,5 thus revealing new pathways of osteoblast-osteoclast crosstalk. At present, more and more exosomes have been isolated and found to play a role in the communication between cells physiologically and pathologically, which may have the potential to be promising biomarker, novel therapeutic agent and delivery carrier for further use.13,14
Co-culture system are widely used to study cell–cell interactions and could be divided into two types: direct communication and indirect co-culture. Indirect co-culture is constructed by Transwell system excluding the effect of direct contact and communication between cells, and focus on the role of paracrine pathway between cells. We realized osteoblast-osteoclast co-culture through Transwell system and found the trends of osteoclasts formation was enhanced, which could be suppressed by the exosomes formation inhibitor GW4869. Based on this, we speculate the importance of osteoblast-derived exosomes in the osteoblast-osteoclast crosstalk.
Exosomes in the paracrine pathway, as widespread microvesicles, have been shown to be actively involved in the etiology of bone metabolism-related diseases.15 Due to its special structure and way of secretion, exosomes could transmit the contained components to recipient cells through membrane fusion, affecting cells’ function and transducing signals. The expression of osteoclastic-related differentiation markers was up-regulated and the tendency of osteoclast formation was significantly enhanced after OB-exo treatment, from which we inferred that OB-exo was enriched with some substances accelerating the differentiation of osteoclast precursor cells. Further isolated RNA from OB-exo and detected its components, we found that it was enriched in evolutionarily conserved lncRNA-MALAT1. Meanwhile, the expression of MALAT1 was monitored to be up-regulated in the osteoblast-osteoclast co-culture system and the osteoclasts treated with OB-exo, accompanied by increasing formation of mature TRAP+ multinucleated osteoclasts. However, after silencing MALAT1 expression with siRNA, the formation of mature osteoclasts decreased, suggesting that MALAT1 enriched in OB-exo has the effect of promoting osteoclastic differentiation. Li et al3 showed that MALAT1 was overexpressed in gingival fibroblasts from patients with chronic periodontitis. This finding led us to think that lncRNA-MALAT1 may be highly involved in the regulation of periodontitis-related physiological activities and the homeostasis in bone remodeling. Emerging studies showed that lncRNA is an essential active partner of osteogenic activity,16 while the effect of lncRNA- OIP5-AS1,17 Linc0113318 and lncRNA-MALAT14 in osteogenic differentiation of osteoblasts and periodontal ligament stem cells have been reported. However, the role of lncRNA in osteoclastic differentiation has rarely been reported, while our results complemented the importance of lncRNA shuttled by osteoblasts exosome in bone remodeling, providing new possibilities for pathogenesis of periodontitis.
At present, the specific mechanism of MALAT1 in promoting osteoclastic differentiation is yet unclear. We detected that MALAT1 has a potential binding site with miR-124 by Starbase bioinformatics prediction, which may be actively involved in osteoclasts formation. Our study showed the up-regulation of MALAT1 in osteoclasts was accompanied by the decreased miR-124 level, whether in co-culture system or OB-exo intervention model. Next, we further verified the significant negative association between miR-124 and MALAT1 by using si-MALAT1 and miR-124 inhibitor. miR-124, which could inhibit the differentiation of bone marrow-derived macrophages into mature osteoclasts,19 is thought to be a molecule strongly associated with bone resorption and participate in the process of tumor-associated bone metastasis.8 Dual-Luciferase reporter assay has demonstrated a practical and effective binding site between miR-124 and MALAT1, suggesting that MALAT1 functions through sponge miR-124, promoting the differentiation of osteoclasts. Several emerging studies showed that lncRNAs can base-pair with miRNAs, thereby acting as competing endogenous to sponge miRNAs.20 The research of Ni et al21 showed that lncSOX2OT was highly enriched in exosomes derived from peripheral blood of patients with small cell lung cancer and lncSOX2OT could shuttled into adjacent cells to activate the downstream RAC1-related pathway by competitively consuming miRNA-194-5p, thus ultimately promoted the occurrence of bone metastasis. The above study is consistent with our experimental results suggesting that lncRNAs enriched in exosomes are key to triggering a chain reaction of bone metabolic homeostasis disorders, while its function depends on the ability of sponging miRNAs to activate the downstream pathway.
In this study, we found that the expression of NFATc1 and its downstream marker CTSK was significantly decreased after overexpressing miR-124 or knocking down MALAT1, and the results of TRAP staining were consistent with it: the formation of mature multinucleated osteoclasts was reduced. The binding site between NFATc1 and miR-124 was validated by Dual-Luciferase reporter assay, suggesting that miR-124 targets the 3′ UTR region of NFATc1 mRNA to inhibit osteoclasts differentiation. Gong et al22 reported results similar to our experiment, indicating that miR-124 may interact with NFATc1 to inhibit osteoclasts activity, which may cause dynamic imbalance in the bone microenvironment. NFATc1, as the most powerful transcription factor gene stimulated by RANKL23 and a downstream target of c-Fos,8 is a specific marker of osteoclasts differentiation, can reverse the symptoms of osteosclerosis in c-Fos-deficient mice,24 thus playing an important role in the differentiation of osteoclast precursor cells. Our study clarifies that osteoblasts-derived exosomes could shuttle into osteoclast precursor cells and promote osteoclasts formation via the lncRNA-MALAT1/miR-124/NFATc1 axis, thereby accelerating this process of osteoclasts differentiation in a c-Fos-independent manner. We expect our finding may provide ideas for the construction of a controllable nanoplatform where exosomes work together with antioxidants to inhibit the hyper-activation of osteoclasts and treat periodontitis. However, our studies did not verify the function of exosomes derived from MALAT1-knockout osteoblasts, which may have an inhibition effect on osteoclast precursor cells. Moreover, the level of lnc-MALAT1 in gingival crevicular fluid and saliva is currently unclear, which have potential to be sensitive early markers of periodontitis and bone resorption and deserved further study.
Conclusion
Taken together, our results indicate that exosomes isolated from the osteoblasts are capable of accelerating alveolar resorption via osteoclastogenesis. The effect is at least partially mediated by lnc-MALAT1 enriched in OB-exo and promote osteoclasts differentiation via suppressing the activation of the miR-124/NFATc1 axis, which provides a basis for the dynamic imbalance in the periodontal bone microenvironment and the occurrence and development of periodontitis. Therefore, our findings suggest that lnc-MALAT1 may be a potential bone anabolic therapeutic target to re-establish bone homeostasis and alleviate periodontitis.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant number 81800972) and Medical Healthy Scientific Technology Project of Zhejiang Province (grant numbers 2022498513 and 2022511252).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
All authors declare that there are no conflicts of interest associated with this study.
References
1. Li D, Liu J, Guo B, et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat Commun. 2016;7(1):10872. doi:10.1038/ncomms10872
2. Li B, Xu H, Han H, et al. Exosome-mediated transfer of lncRUNX2-AS1 from multiple myeloma cells to MSCs contributes to osteogenesis. Oncogene. 2018;37(41):5508–5519. doi:10.1038/s41388-018-0359-0
3. Li J, Wang M, Song L, Wang X, Lai W, Jiang S. LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway. J Periodontal Res. 2020;55(2):182–190. doi:10.1111/jre.12700
4. Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging. 2019;11(20):8777–8791. doi:10.18632/aging.102264
5. Zhang D, Wu Y, Li Z, et al. MiR-144-5p, an exosomal miRNA from bone marrow-derived macrophage in type 2 diabetes, impairs bone fracture healing via targeting Smad1. J Nanobiotechnology. 2021;19(1):226. doi:10.1186/s12951-021-00964-8
6. Ge J, Guo S, Fu Y, et al. Dental follicle cells participate in tooth eruption via the RUNX2-MiR-31-SATB2 loop. J Dent Res. 2015;94(7):936–944. doi:10.1177/0022034515578908
7. Wang N, Liu X, Tang Z, et al. Increased BMSC exosomal miR-140-3p alleviates bone degradation and promotes bone restoration by targeting Plxnb1 in diabetic rats. J Nanobiotechnology. 2022;20(1):97. doi:10.1186/s12951-022-01267-2
8. Cai WL, Huang WD, Li B, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer. 2018;17(1):9. doi:10.1186/s12943-017-0746-0
9. Guidelines for the ethical review of laboratory animal welfare. Standardization administration of China Beijing ICP 09001239. GB/T35892-2018. Available from: http://www.gb688.cn/bzgk/gb/newGbInfo?hcno=9BA619057D5C13103622A10FF4BA5D14.
10. Doolittle ML, Ackert-Bicknell CL, Jonason JH. Isolation and culture of neonatal mouse calvarial osteoblasts. Methods Mol Biol. 2021;2230:425–436.
11. Ahmadian E, Eftekhari A, Kavetskyy T, Khosroushahi AY, Turksoy VA, Khalilov R. Effects of quercetin loaded nanostructured lipid carriers on the paraquat-induced toxicity in human lymphocytes. Pestic Biochem Physiol. 2020;167:104586. doi:10.1016/j.pestbp.2020.104586
12. Chodari L, Dilsiz AM, Vahedi P, et al. Targeting mitochondrial biogenesis with polyphenol compounds. Oxid Med Cell Longev. 2021;2021:4946711. doi:10.1155/2021/4946711
13. Santonocito S, Polizzi A, Palazzo G, Isola G. The emerging role of microRNA in periodontitis: pathophysiology, clinical potential and future molecular perspectives. Int J Mol Sci. 2021;22(11):5456. doi:10.3390/ijms22115456
14. Nik MKN, Awang R, Mohamad S, Shahidan W. Plasma- and saliva exosome profile reveals a distinct MicroRNA signature in chronic periodontitis. Front Physiol. 2020;11:587381. doi:10.3389/fphys.2020.587381
15. Murphy C, Withrow J, Hunter M, et al. Emerging role of extracellular vesicles in musculoskeletal diseases. Mol Aspects Med. 2018;60:123–128. doi:10.1016/j.mam.2017.09.006
16. Li H, Zheng Q, Xie X, et al. Role of exosomal non-coding RNAs in bone-related diseases. Front Cell Dev Biol. 2021;9:811666. doi:10.3389/fcell.2021.811666
17. Wang S, Duan Y. LncRNA OIP5-AS1 inhibits the lipopolysaccharide-induced inflammatory response and promotes osteogenic differentiation of human periodontal ligament cells by sponging miR-92a-3p. Bioengineered. 2022;13(5):12055–12066. doi:10.1080/21655979.2022.2067291
18. Li Q, Zhou H, Wang C, Zhu Z. Long non-coding RNA Linc01133 promotes osteogenic differentiation of human periodontal ligament stem cells via microRNA-30c / bone gamma-carboxyglutamate protein axis. Bioengineered. 2022;13(4):9602–9612. doi:10.1080/21655979.2022.2054912
19. Tang L, Yin Y, Liu J, Li Z, Lu X. MiR-124 attenuates osteoclastogenic differentiation of bone marrow monocytes via targeting Rab27a. Cell Physiol Biochem. 2017;43(4):1663–1672. doi:10.1159/000484027
20. Han JJ, Wang XQ, Zhang XA. Functional interactions between lncRNAs/circRNAs and miRNAs: insights into rheumatoid arthritis. Front Immunol. 2022;13:810317. doi:10.3389/fimmu.2022.810317
21. Ni J, Zhang X, Li J, et al. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12(7):662. doi:10.1038/s41419-021-03928-w
22. Gong M, Liang T, Jin S, et al. Methylation-mediated silencing of miR-124 facilitates chondrogenesis by targeting NFATc1 under hypoxic conditions. Am J Transl Res. 2017;9(9):4111–4124.
23. Takayanagi H. Mechanistic insight into osteoclast differentiation in osteoimmunology. J Mol Med. 2005;83(3):170–179. doi:10.1007/s00109-004-0612-6
24. Matsuo K, Galson DL, Zhao C, et al. Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J Biol Chem. 2004;279(25):26475–26480. doi:10.1074/jbc.M313973200
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







