Back to Journals » Research Reports in Clinical Cardiology » Volume 11
Optogenetic Pacing: Current Insights and Future Potential
Received 13 June 2020
Accepted for publication 9 October 2020
Published 3 November 2020 Volume 2020:11 Pages 49—55
DOI https://doi.org/10.2147/RRCC.S242650
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Kones
Airong Li, Rudolph E Tanzi
Genetics and Aging Research Unit, Department of Neurology, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, USA
Correspondence: Airong Li; Rudolph E Tanzi
Genetics and Aging Research Unit, Department of Neurology, Massachusetts General Hospital and Harvard Medical School, 114, 16th Street, Charlestown, MA 02129, USA
Tel +617 724 9397
; +617 726 6845
Fax +617 724 1823
Email [email protected]; [email protected]
Abstract: Optogenetics combines the biological techniques of optics and genetics and uses light to control the activities of living tissues such as neurons and heart. Optogenetic actuators like channelrhodopsin (ChR), halorhodopsin (NpHR), and archaerhodopsin (bacterio-opsin) provide specificity for neuronal or cardiac controls, and the field has made much progress in heart research since its introduction almost a decade ago. This review will provide information about the history, research highlights and clinical applications of optical coherence tomography (OCT) technology. The clinical translation of cardiac optogenetics will be towards human and larger mammalian animal model applications and ultimately optogenetics may have the power to restore normal heart rhythm and greatly improve quality of life.
Keywords: optical coherence tomography, channelrhodopsin, halorhodopsin, archaerhodopsin, Drosophila, heart
Introduction of Optogenetics
Optogenetics is a combined biological technique from both the optics and genetic fields that uses light to control the activities of living tissues such as neurons and heart.
Optical coherence tomography (OCT) provides novel three-dimensional (3D) imaging.1–3 Combined with OCT, optical coherence microscopy (OCM) provides high-resolution imaging.4–10 The image resolution of OCT (~5–10 µm in tissue) and OCM (~1–3 µm) is 50x–100x greater than conventional ultrasound, MRI, or CT. OCT and OCM have been used for medical imaging. Commercially, OCT systems provide a series of applications, such as in interventional cardiology for diagnosis,11 in ophthalmology and optometry for the retina12 and in dermatology to improve diagnosis.13
Specific neuronal or cardiac control are provided by optogenetic actuators such as channelrhodopsin (ChR),14 halorhodopsin (NpHR),15 and archaerhodopsin (bacterio-opsin). ChRs are retinylidene proteins (rhodopsins) that are sensory photoreceptors responded to light.14 NpHR is a chloride ion-specific light-gated ion that responds to green/yellow light,15–18 that senses light in vertebrate retina rhodopsins.18 Bacterio-opsins are a family of receptor proteins in archaea that have light inhibiting action potential.19–22
The History of Optogenetics
In 2002 the American scientist Boris Zemelman and the British scientist Gero Miesenböck used fly rhodopsin photoreceptors to control neural activity23 and in 2005 Peter Hegemann expressed ChR2 in mammalian cells and oocytes, Zhuo-Hua Pan transfected neurons in a manner that allowed them to be electrically active responsive to light24 and Kramer and Isacoff developed organic photoswitches interacting with ion channels.25,26
Also in 2005, Lima and Miesenböck were the first to demonstrate the use of optogenetics control the animal behavior,27 Karl Deisseroth and Feng Zhang were the first to use channelrhodopsin28 and Georg Nagel first reported a single-component light-activated cation channel,28 while Lynn Landmesser and Stefan Herlitze controlled neuronal activity using ChR2 and were also the first to use vertebrate rhodopsin in neurons.29
Alexander Gottschalk and Georg Nagel first used Channelrhodopsin-2 (CHR2) for controlling neuronal activity in 200530 and in the same year they were also the first to make a ChR2 mutant (H134R) for modifying neuronal activity.31
In 2006 Atsushi Miyawaki et al and Roger Tsien et al developed optogenetic recordings called calcium indicators (GECIs)32,33 in flies and zebrafish,34,35 while Nakai et al developed the first GECI to be used in mammals,36 and in 2007 Feng Zhang and Karl Deisseroth with Georg Nagel, Alexander Gottschalkn, Peter Hegemann and Ernst Bamberg were the first to publish optogenetic inhibition research in mammals.37
Awards in OCT field include the inaugural HFSP Nakasone Award to Karl Deisseroth in 2010, the InBev-Baillet Latour International Health Prize to Gero Miesenböck in 2012, the Brain Prize to Ernst Bamberg et al in 2013, and the Else Kröner Fresenius Research Prize to Karl Deisseroth for in 2017.
Optogenetics was chosen as the Method of the Year 2010 and the Breakthroughs of the Decade by the prominent research journals Nature Methods and Science, respectively (https://www.medinc.co.uk/optogenetics-breakthrough-of-the-decade-by-dr-zulfiquar/). The application of the first Drosophila heart study using OCT in 2015 was featured on the Discovery channel and in the Boston Globe (Light - powered hearts? https://www.bostonglobe.com/lifestyle/2015/10/25/light-powered-hearts/ETWV7DZU6pwMNm1P59TLGL/story.html).
Drosophila Heart Has Alterations in Development
The Drosophila heart has marked morphological and functional changes during development as seen through a longitudinal study of various development stages. The heart beat is reduced dramatically when the fly is in the pupal stage and stops beating during pupae 2 (Figure 1). These data show that a circadian clock gene dCry affected heart38 in heart development and functioning.38
 |
Figure 1 (A) Heart remodeling during Drosophila lifecycle were by OCM sections. (B) Heart variations at different developmental stages were by M-mode images. Reproduced from Alex A, Li AR, Zeng XX, et al. A circadian clock gene, cry, affects heart morphogenesis and function in drosophila as revealed by optical coherence microscopy. PLoS One. 2015;10(9):e013723. Creative Commons license and disclaimer available from: http://creativecommons.org/licenses/by/4.0/legalcode.38 Scale bars represent 500 µm. |
Blue Light Optogenetic Pacing in Drosophila Melanogaster
An integrated ultrahigh resolution OCM imaging and optogenetic pacing system used to non-invasively monitor Drosophila heart in response to optical stimulations have clearly shown mCherry fluorescence signal in the heart of a ChR2-mCherry transgenic fly compared to a wild-type control fly (Figure 2) 39 and using these transgenic Drosophila models, the fly heart showed successfully pacing.39
 |
Figure 2 Optogenetic pacing in Drosophila. (A) mCherry fluorescence signal was clearly observed in the heart of an adult 24B-GAL4; UAS-H134R-ChR2 fly specimen. No fluorescence signal was observed from the wild-type control fly (24B-GAL4/+). (B) M-mode OCM image and measurements of heart chamber size showing successful pacing of a pupa heart using blue light pulses. Alex A, Li A, Tanzi RE, Zhou C. Optogenetic pacing in Drosophila melanogaster. Sci Advan. 2015;1:e1500639. Reprinted with permission from Alex et al. Optogenetic pacing in Drosophila melanogaster.Sci. Adv. 2015;1:e1500639. Distributed under CC BY-NC.39 |
Red Light Optogenetic Pacing in Drosophila Melanogaster
Optogenetic fly models are sensitive to the fact that red light is absorbed less strongly than blue light to increase the excitability of the heart tissue and flies expressing ReaChR were able to be tachypaced under red light stimulation (Figure 3).40,41
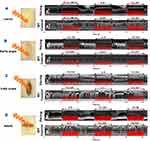 |
Figure 3 Flies are optogenetically cardiac paced by the ReaChR-expressing Drosophila using red light. ReaChR and WT flies of M-mode images were acquired during pacing at the larval (A), early pupal (B), late pupal (C), and adult (D) stages. Reproduced from Men J, Li A, Jerwick J, Li Z, Tanzi RE, Zhou C. Non-invasive red-light optogenetic control of Drosophila cardiac function. Comm Biol. 2020;3:336. Creative Commons license and disclaimer available from: http://creativecommons.org/licenses/by/4.0/legalcode. 40 Methods was used from reference41 to create this image. |
Optogenetic Control of Cardiac Arrhythmia
Optogenetics has the capability to treat the cardiac conduction system, restore pacemaking ability and terminate cardiac arrhythmias.42 Cardiac tissue exposed to optogenetic tools can provide mechanistic insights into arrhythmia.43 Shift light tuned behavior through photosensitive ion channels and pumps (opsins) by optogenetic methods and pacing of cardiac preparations have now been successful in several experimental models.44 The opsins induce reliable, precise stimulation or silencing of electrophysiological activity in the cardiac cells.45 ChR2 expressed in cardiomyocytes can sensitively activate Ca2+ signaling properties45 and ChR2 expression in transgenic mice controlled heart muscles in vivo.42 Stimulation of Gs-signaling in cardiomyocytes and the whole heart by optogenetics was documented in the light-sensitive Gs-protein coupled receptor in mice cardiac tissue.46 Self-sustained spiral waves in heart can be manipulated precisely to influence cardiac function and overall dynamics in cardiac excitable media.47 Studies have demonstrated that near-infrared (NIR) light has the ability for tissue-penetration and NIR had the potential to manipulate cardiovascular diseases non-invasively.48 The red-shifted opsins achieved greater tissue depths than conventional blue-sensitive channel-rhodopsins.45 These studies have increased our understanding of cardiac physiology.
Rapid antitachycardia pacing produced by an electric shock can resynchronize the heart and terminate arrhythmias such as atrial fibrillation (AF).49 Electric shock functions in electrical defibrillation in mice cardiomyocytes50–52 and atrial neonatal rat cardiomyocytes.53 Aging may significantly reduce heart rate via electrical pacing in Drosophila, similar to that seen in elderly humans. Age was also associated with an increase in rhythm disturbances.54 In Drosophila, NpHR stops the heart rate from beating in relation to light intensity.55
During Drosophila metamorphosis glutamatergic neurons provide extensive innervation to the adult heart. Muscles of the first abdominal cardiac chamber showed pacemaker action potentials.56
Optogenetic pacing of adult hearts may characterize the effects in flies.57 KCNQ1 in humans is related to myocardial repolarization and KCNQ1 mutant Drosophila showed abnormal contractions and fibrillations.57
Of the genes identified in Drosophila genetic screens, mutants in a fly orthologue of epidermal growth factor (EGF) rhomboid 3 enlarged cardiac chambers.58 Proper EGFR signaling maintains adult cardiac function.58 A mutation in the Notch ortholog weary (wry) results in dilated cardiomyopathy.59 Insulin-IGF receptor signaling regulates the age-dependent changes in cardiac function60.
Multimodal and multisite pacing studies showed chronic stability and excellent biocompatibility in small animals.61
Clinical Applications of OCT in Heart
Coronary vasculature and coronary graft assessment are primary applications of OCT in cardiovascular medicine. Clinically, several systems have become commercially available.
OCT can assist patients who have stable coronary artery disease for a more detailed lumen segmentation. On the other hand, in patients with acute coronary syndrome, intraluminal thrombus can be detected at 100% with OCT in comparison to coronary angioscopy62–65 which detected plaques in 79% and stenosis in 24% of patients.66
The Future of Optogenetics
Optogenetics can control and monitor the biological function of cells, tissues or organs. The field has made significant progress in heart research from its inception almost a decade ago. This review has provided information on the introduction, history, research highlights and clinical applications of OCT technology.
The direction of clinical translation of cardiac optogenetics in human application appears to be towards larger mammalian animal models and tools such as safe and stable opsin expression in heart.43 Because optogenetics may restore normal heart rhythm to increase the overall quality of life67–69 and action potential duration of ChR2- or NpHR can be modulated in opsin-expressing rat cardiomyocytes,70 optogenetics may potentially play an important therapeutic role in treating heart diseases.
OCT will likely be of great assistance in Drosophila genetic screens that can be designed to identify additional cardiovascular-related genes and may also be valuable in assessing pre-clinical drug development cardiotoxicity, which account for approximately 20% of withdrawal of drug development.71,72 Electrophysiology measures used to detect cardiotoxicity are often low throughput67,73 and efficient high throughput screening tools that significantly reduce cost are needed.71,72 Overall, it is clear that optogenetics has the potential for use in evaluating cardiotoxicity through high throughput and automation.
Acknowledgments
This work was supported by the NIH (R15EB019704 to A.L., R03AR063271 to A.L., and R01AG014713 and R01MH060009 to R.E.T., the NSF1455613 to A.L.), the Cure Alzheimer’s Fund (to R.E.T.).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography. Science. 1991;254(5035):1178–1181. doi:10.1126/science.1957169
2. Fujimoto JG, Pitris C, Boppart SA, Brezinski ME. Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia. 2000;2(1–2):9–25. doi:10.1038/sj.neo.7900071
3. Fujimoto JG. Optical coherence tomography for ultrahigh resolution in vivo imaging. Nat Biotechnol. 2003;21(11):1361–1367. doi:10.1038/nbt892
4. Izatt JA, Hee MR, Owen GM, Swanson EA, Fujimoto JG. Optical coherence microscopy in scattering media. Opt Lett. 1994;19(8):590–592. doi:10.1364/OL.19.000590
5. Kempe M, Thon A, Rudolph W. Resolution limits of microscopy through scattering layers. Opt Commun. 1994;110(5–6):492–496. doi:10.1016/0030-4018(94)90237-2
6. Izatt JA, Kulkarni MD, Wang H-W, Kobayashi K, Sivak MV
7. Kempe M, Rudolph W. Analysis of heterodyne and confocal microscopy for illumination with broad-bandwidth light. J Mod Opt. 1996;43(10):2189–2204.
8. Kempe M, Rudolph W, Welsch E. Comparative study of confocal and heterodyne microscopy for imaging through scattering media. J Optical Soc Am Optics Image Sci Vision. 1996;13(1):46–52. doi:10.1364/JOSAA.13.000046
9. Clark AL, Gillenwater A, Alizadeh-Naderi R, El-Naggar AK, Richards-Kortum R. Detection and diagnosis of oral neoplasia with an optical coherence microscope. J Biomed Opt. 2004;9(6):1271–1280. doi:10.1117/1.1805558
10. Aguirre AD, Hsiung P, Ko TH, Hartl I, Fujimoto JG. High-resolution optical coherence microscopy for high-speed, in vivo cellular imaging. Opt Lett. 2003;28:21. doi:10.1364/OL.28.002064
11. Yonetsu T, Kakuta T, Lee T, et al. Assessment of acute injuries and chronic intimal thickening of the radial artery after transradial coronary intervention by optical coherence tomography. Eur Heart J. 2010;31(13):1608–1615. doi:10.1093/eurheartj/ehq102
12. Povazay B, Hermann B, Hofer B, et al. Wide-field optical coherence tomography of the choroid in vivo. Invest Ophthalmol Vis Sci. 2009;50(4):1856–1863. doi:10.1167/iovs.08-2869
13. Ulrich M, Themstrup L, de Carvalho N, et al. Dynamic optical coherence tomography in dermatology. Dermatology. 2016;232(3):298–311. doi:10.1159/000444706
14. Nagel G, Ollig D, Fuhrmann M, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296(5577):2395–2398. doi:10.1126/science.1072068
15. Zhang F, Wang LP, Brauner M, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–639. doi:10.1038/nature05744
16. Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One. 2007;2(3):e299. doi:10.1371/journal.pone.0000299
17. Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36(1–4):129–139. doi:10.1007/s11068-008-9027-6
18. Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of Parkinsonian neural circuitry. Science. 2009;324(5925):354–359. doi:10.1126/science.1167093
19. Inoue K, Ito S, Kato Y, et al. A natural light-driven inward proton pump. Nat Commun. 2016;7:13415. doi:10.1038/ncomms13415
20. Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2011;9(1):90–95. doi:10.1038/nmeth.1782
21. El-Gaby M, Zhang Y, Wolf K, Schwiening CJ, Paulsen O, Shipton OA. Archaerhodopsin selectively and reversibly silences synaptic transmission through altered pH. Cell Rep. 2016;16(8):2259–2268. doi:10.1016/j.celrep.2016.07.057
22. Enami N, Yoshimura K, Murakami M, Okumura H, Ihara K, Kouyama T. Crystal structures of archaerhodopsin-1 and −2: common structural motif in archaeal light-driven proton pumps. J Mol Biol. 2006;358(3):675–685. doi:10.1016/j.jmb.2006.02.032
23. Zemelman BV, Nesnas N, Lee GA, Miesenbock G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci U S A. 2003;100(3):1352–1357. doi:10.1073/pnas.242738899
24. Bi A, Cui J, Ma YP, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50(1):23–33. doi:10.1016/j.neuron.2006.02.026
25. Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7(12):1381–1386. doi:10.1038/nn1356
26. Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2(1):47–52. doi:10.1038/nchembio756
27. Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121(1):141–152. doi:10.1016/j.cell.2005.02.004
28. Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–1268. doi:10.1038/nn1525
29. Abdel Ghany WA, Nada M, Mahran MA, et al. Combined anterior and posterior lumbar rhizotomy for treatment of mixed dystonia and spasticity in children with cerebral palsy. Neurosurgery. 2016;79(3):336–344. doi:10.1227/NEU.0000000000001271
30. Li X, Gutierrez DV, Hanson MG, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102(49):17816–17821. doi:10.1073/pnas.0509030102
31. Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15(24):2279–2284. doi:10.1016/j.cub.2005.11.032
32. Miyawaki A, Llopis J, Heim R, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388(6645):882–887. doi:10.1038/42264
33. Kerr R, Lev-Ram V, Baird G, Vincent P, Tsien RY, Schafer WR. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26(3):583–594. doi:10.1016/S0896-6273(00)81196-4
34. Fiala A, Spall T, Diegelmann S, et al. Genetically expressed cameleon in Drosophila melanogaster is used to visualize olfactory information in projection neurons. Curr Biol. 2002;12(21):1877–1884. doi:10.1016/S0960-9822(02)01239-3
35. Higashijima S, Masino MA, Mandel G, Fetcho JR. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol. 2003;90(6):3986–3997. doi:10.1152/jn.00576.2003
36. Ji G, Feldman ME, Deng KY, et al. Ca2+-sensing transgenic mice: postsynaptic signaling in smooth muscle. J Biol Chem. 2004;279(20):21461–21468. doi:10.1074/jbc.M401084200
37. Wang J, Zhu LS, Xie H, Song Y, Sun RL, Zhang FD. [Degradation dynamics of POPs atrazine in soils under long-term located fertilization conditions]. Huan Jing Ke Xue. 2007;28(12):2821–2826. Chinese.
38. Alex A, Li AR, Zeng XX, et al. A circadian clock gene, cry, affects heart morphogenesis and function in drosophila as revealed by optical coherence microscopy. PLoS One. 2015;10(9):e0137236. doi:10.1371/journal.pone.0137236
39. Alex A, Li A, Tanzi RE, Zhou C. Optogenetic pacing in Drosophila melanogaster. Sci Advan. 2015;1:e1500639. doi:10.1126/sciadv.1500639
40. Men J, Li A, Jerwick J, Li Z, Tanzi RE, ZhouC. Non-invasive red-light optogenetic control of Drosophila cardiac function. Comm Biol. 2020;3:336.
41. Dong Z, Men J,Yang Z, et al. FlyNet 2.0: drosophila Heart 3D (2D + time) segmentation in optical coherence microscopy images using a convolutional long short-term memory neural network. Biomed Opt Express. 2020;11(3):1568–1579.
42. Jiang C, Li HT, Zhou YM, Wang X, Wang L, Liu ZQ. Cardiac optogenetics: a novel approach to cardiovascular disease therapy. Europace. 2018;20(11):1741–1749.
43. Boyle PM, Karathanos TV, Trayanova NA. Cardiac optogenetics: 2018. JACC Clin Electrophysiol. 2018;4(2):155–167. doi:10.1016/j.jacep.2017.12.006
44. O’Shea C, Holmes AP, Winter J, et al. Cardiac optogenetics and optical mapping – overcoming spectral congestion in all-optical cardiac electrophysiology. Front Physiol. 2019;10:182. doi:10.3389/fphys.2019.00182
45. Ferenczi EA, Tan X, Huang CL. Principles of optogenetic methods and their application to cardiac experimental systems. Front Physiol. 2019;10:1096. doi:10.3389/fphys.2019.01096
46. Makowka P, Bruegmann T, Dusend V, et al. Optogenetic stimulation of Gs-signaling in the heart with high spatio-temporal precision. Nat Commun. 2019;10(1):1281. doi:10.1038/s41467-019-09322-7
47. Steinbeck JA, Choi SJ, Mrejeru A, et al. Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson’s disease model. Nat Biotechnol. 2015;33(2):204–209. doi:10.1038/nbt.3124
48. Rao P, Wang L, Cheng Y, et al. Near-infrared light driven tissue-penetrating cardiac optogenetics via upconversion nanoparticles in vivo. Biomed Opt Express. 2020;11(3):1401–1416.
49. Adgey AA, Spence MS, Walsh SJ. Theory and practice of defibrillation: (2) defibrillation for ventricular fibrillation. Heart. 2005;91(1):118–125. doi:10.1136/hrt.2003.019927
50. Crocini C, Ferrantini C, Coppini R, et al. Optogenetics design of mechanistically-based stimulation patterns for cardiac defibrillation. Sci Rep. 2016;6:35628. doi:10.1038/srep35628
51. Burton RA, Klimas A, Ambrosi CM, et al. Optical control of excitation waves in cardiac tissue. Nat Photonics. 2015;9(12):813–816. doi:10.1038/nphoton.2015.196
52. Majumder R, Feola I, Teplenin AS, de Vries AA, Panfilov AV, Pijnappels DA. Optogenetics enables real-time spatiotemporal control over spiral wave dynamics in an excitable cardiac system. Elife. 2018;7.
53. Bingen BO, Engels MC, Schalij MJ, et al. Light-induced termination of spiral wave arrhythmias by optogenetic engineering of atrial cardiomyocytes. Cardiovasc Res. 2014;104(1):194–205. doi:10.1093/cvr/cvu179
54. Paternostro G, Vignola C, Bartsch DU, Omens JH, McCulloch AD, Reed JC. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001;88(10):1053–1058. doi:10.1161/hh1001.090857
55. Stanley CE, Mauss AS, Borst A, Cooper RL. The effects of chloride flux on drosophila heart rate. Methods Protoc. 2019;2:3. doi:10.3390/mps2030073
56. Dulcis D, Levine RB. Glutamatergic innervation of the heart initiates retrograde contractions in adult Drosophila melanogaster. J Neurosci. 2005;25(2):271–280. doi:10.1523/JNEUROSCI.2906-04.2005
57. Ocorr K, Reeves NL, Wessells RJ, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007;104(10):3943–3948. doi:10.1073/pnas.0609278104
58. Yu L, Lee T, Lin N, Wolf MJ. Affecting Rhomboid-3 function causes a dilated heart in adult Drosophila. PLoS Genet. 2010;6(5):e1000969. doi:10.1371/journal.pgen.1000969
59. Kim IM, Wolf MJ, Rockman HA. Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ Res. 2010;106(7):1233–1243. doi:10.1161/CIRCRESAHA.109.213785
60. Luong N, Davies CR, Wessells RJ, et al. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4(2):133–142.
61. Gutruf P, Yin RT, Lee KB, et al. Wireless, battery-free, fully implantable multimodal and multisite pacemakers for applications in small animal models. Nat Commun. 2019;10(1):5742. doi:10.1038/s41467-019-13637-w
62. Chamie D, Bezerra HG, Attizzani GF, et al. Incidence, predictors, morphological characteristics, and clinical outcomes of stent edge dissections detected by optical coherence tomography. JACC Cardiovasc Interv. 2013;6(8):800–813. doi:10.1016/j.jcin.2013.03.019
63. Bezerra HG, Attizzani GF, Sirbu V, et al. Optical coherence tomography versus intravascular ultrasound to evaluate coronary artery disease and percutaneous coronary intervention. JACC Cardiovasc Interv. 2013;6(3):228–236. doi:10.1016/j.jcin.2012.09.017
64. Fujino Y, Bezerra HG, Attizzani GF, et al. Frequency-domain optical coherence tomography assessment of unprotected left main coronary artery disease – a comparison with intravascular ultrasound. Catheter Cardiovasc Interv. 2013;82(3):E173–83. doi:10.1002/ccd.24843
65. Terashima M, Kaneda H, Suzuki T. The role of optical coherence tomography in coronary intervention. Korean J Intern Med. 2012;27(1):1–12. doi:10.3904/kjim.2012.27.1.1
66. Saito Y, Kobayashi Y. Update on antithrombotic therapy after percutaneous coronary intervention. Intern Med. 2020;59(3):311–321. doi:10.2169/internalmedicine.3685-19
67. Ambrosi CM, Entcheva E. Optogenetics’ promise: pacing and cardioversion by light? Future Cardiol. 2014;10(1):1–4. doi:10.2217/fca.13.89
68. van Weerd JH, Christoffels VM. The formation and function of the cardiac conduction system. Development. 2016;143(2):197–210. doi:10.1242/dev.124883
69. Joshi J, Rubart M, Zhu W. Optogenetics: background, methodological advances and potential applications for cardiovascular research and medicine. Front Bioeng Biotechnol. 2019;7:466. doi:10.3389/fbioe.2019.00466
70. Park SA, Lee SR, Tung L, Yue DT. Optical mapping of optogenetically shaped cardiac action potentials. Sci Rep. 2014;4:6125. doi:10.1038/srep06125
71. Piccini JP, Whellan DJ, Berridge BR, et al. Current challenges in the evaluation of cardiac safety during drug development: translational medicine meets the Critical Path Initiative. Am Heart J. 2009;158(3):317–326. doi:10.1016/j.ahj.2009.06.007
72. Klimas A, Ambrosi CM, Yu J, Williams JC, Bien H, Entcheva E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat Commun. 2016;7:11542. doi:10.1038/ncomms11542
73. Entcheva E, Bub G. All-optical control of cardiac excitation: combined high-resolution optogenetic actuation and optical mapping. J Physiol. 2016;594(9):2503–2510.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
