Back to Journals » OncoTargets and Therapy » Volume 12
Nucleic Acid Aptamer: A Novel Potential Diagnostic and Therapeutic Tool for Leukemia
Received 20 July 2019
Accepted for publication 14 October 2019
Published 4 December 2019 Volume 2019:12 Pages 10597—10613
DOI https://doi.org/10.2147/OTT.S223946
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Yuan Tan, Yuejin Li, Faqing Tang
Department of Clinical Laboratory, Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha 410013, People’s Republic of China
Correspondence: Faqing Tang
Hunan Cancer Hospital and The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, People’s Republic of China
Tel/Fax +86-731-89762688
Email [email protected]
Abstract: Leukemia immunotherapy has been dominant via using synthetic antibodies to target cluster of differentiation (CD) molecules, nevertheless inevitable cytotoxicity and immunogenicity would limit its development. Recently, increasing reports have focused on nucleic acid aptamers, a class of high-affinity nucleic acid ligands. Aptamers purportedly serve as “chemical antibodies”, have negligible cytotoxicity and low immunogenicity, and would be widely applied for the therapy and diagnosis of various diseases, especially leukemia. In the preclinical applications, nucleic acid aptamers have displayed the augmented specificity and selectivity via recognizing targets on leukemia cells based on unique three-dimensional conformations. As small molecules with nucleic acid characteristics, aptamers need to be chemically modified to resist nuclease degradation, renal clearance and improve binding affinities. Moreover, aptamers can be linked with neoteric detection techniques to enhance sensitivity and selectivity of diagnosis and therapy. In this review, we summarized aptamers’ preparation, chemical modification and conjugation, and discussed the application of aptamers in diagnosis and treatment of leukemia through highly specifically recognizing target molecules. Significantly, the application prospect of aptamers in fusion genes would be introduced.
Keywords: nucleic acid aptamer, diagnosis, therapy, leukemia
Introduction
Leukemia is a hematological malignancy that arises from bone marrow (BM), characterized by the abnormal proliferation of BM precursor cells, contributing to a series of symptoms including anaemia, bleeding, fever and life-threatening infections. Leukemia is composed of four types, acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphoblastic leukemia (CLL), and chronic myeloid leukemia (CML). Routine methods to diagnose leukemia contain morphological examination, cytochemical immunophenotyping, cytogenetics and molecular analysis of BM samples, according to the 2016 revision to WHO classification of myeloid neoplasms and acute leukemia.1 However, the drawbacks of traditional methods like invasion, procedural complications and impractical testing equipment limit the further development of clinical medicine,2 demonstrating the importance of the development of new amplification strategies or diagnostic technologies. The amplification strategies simultaneously detect DNA mutations and copy number variation via amplifying response signal, such as multiplex ligation-dependent probe amplification (MLPA) for monitoring gene aberrations.3,4 As previous researches reveal, conventional therapies for leukemia are composed of immunotherapy, stem cell therapy, chemotherapy, traditional Chinese medicine treatment, targeted therapy and BM transplantation, which markedly improve anti-leukemic efficiency, but still have a poor prognosis and a high fatality rate.5 Among these, the conjugation of chemotherapeutics with antibodies (mostly monoclonal antibodies, mAbs) formulates antibody–drug conjugates to directly deliver targeted drugs to cluster of differentiation (CD) antigens and other external targets, which is a promising strategy for clinical application.6–8 Nevertheless, there is still an inevitable drawback that targets possibly escape from the attack of monoclonal antibodies in the unstable environment of progressive leukemia,9 suggesting that a therapeutic platform with improved therapeutic efficacy and reduced non-specific toxicity is urgently in demand. Fortunately, with the establishment of new therapies, like aptamers-mediated methods, the curative effect of leukemia has been greatly improved.10
Recently, multifunctional nucleic acid aptamers have shown the superiority over monoclonal antibodies and might be excellent alternatives or supplements to monoclonal antibodies in theranostics (therapy and diagnosis) of leukemia.10 Remarkably, the application of aptamers in other hematologic malignancies also shows dramatic prospect. For example, TD05 aptamers were developed against Ramos cells to detect Burkitt’s lymphoma, which were also engineered as drug carriers to therapy diseases.11 Aptamers purportedly serve as “unique antibodies”, consisting of short single strands of DNA or RNA, which are selected by “systemic evolution of ligands by exponential enrichment (SELEX)” including cell-SELEX and protein-SELEX,12 then selectively bind to a wide range of targets including small organic molecules, peptides, proteins, viruses, bacteria, whole cells and even living animals. The interaction of aptamers with target molecules has low immunogenicity, superior stability, high affinity and specificity.12 Therefore, aptamers have been widely applied for the detection and treatment of various diseases, including inflammatory, infections,13 cardiovascular,14 neurodegenerative,15 autoimmune diseases,16 and cancer.17,18 Especially, aptamers-based methods for leukemia therapy and diagnosis have shown the preferable potential when conjugated with drugs, imaging technologies and other detection platforms.19,20 Unfortunately, aptamers need to be chemically modified to significantly overcome nuclease degradation, rapid renal excretion and deficient binding affinity due to the nucleic acid characteristics.21
Herein, we summarized aptamers’ preparation, chemical modification and conjugation, and discussed the application of aptamers in diagnosis and treatment of leukemia through highly specifically recognizing target molecules. Significantly, the application prospect of aptamers in fusion genes would be introduced.
Aptamers’ Generation and Optimization
Aptamers’ Isolation and Constitution
The nucleic acid aptamers are isolated by “SELEX” including protein-SELEX and cell-SELEX. In protein-SELEX, the purified or recombinant protein functions as a target for SELEX, and the procedure is simpler compared to cell-SELEX, but some selected aptamers binding to purified membrane proteins fail to recognize targets in whole cells,22 and the instability of protein structure also affects its recognition function, which limit aptamers’ application in medical fields.23 In cell-based SELEX, the selected targets are embedded on viable cells, therefore the used cells must be available, cultivable and stable. Moreover, the cell-based SELEX does not need complicated purification steps.24 The whole cell-SELEX process usually contains the following steps: Firstly, designing an initial single-stranded DNA oligonucleotide pool composed of 1014–1015 random sequences. Next, random sequences fold into different secondary and tertiary structures and are incubated with immobilized targets to form aptamers-target complexes, which are used for positive selection. Then, the unbound sequences are washed off during the partitioning step. Next, the target-bound sequences are amplified by PCR in DNA aptamers or RT-PCR in RNA aptamers to create an enriched library for the next round of selection. Lastly, the enriched aptamers' sequences are frequently identified by high-throughput sequencing methods. This selection process is repeated 10–15 cycles, until the specific sequences of the target are enriched in the pool.25 Remarkably, the more advanced cell-ExSELEX (genetic alphabet Expansion for SELEX) technique has been developed to generate unnatural base (UB)-containing DNA aptamers with higher affinity and specificity, which is based on hydrophobic UB pairs (UBPs) between 7-(2-thienyl)imidazo[4,5-b]pyridine (Ds) and 2-nitro-4-propynylpyrrole (Px).26
Aptamers’ Chemical Modification
The nucleic acid aptamers consisting of ssDNA or RNA need to be chemically modified to prevent degradation and enhance aptamers’ biological and physicochemical stability. In current, there are the following modifications: 1) 2′-substitutions on the sugar ring: 2ʹ-OH moieties of RNA aptamers are heavily modified with fluorine (2ʹ-F), Methoxy (2ʹ-OMe), amine (2ʹ-NH2) and locked nucleic acids (LNAs).27–29 Unlocked nucleic acid (UNA), 2ʹ-deoxy-2ʹ-fluoro-D-arabinonucleic acid (2ʹ-F ANA) and 2ʹ-O-methyl RNA analogues are used to modify 2ʹ-position of the DNA aptamers’ sugar ring.30–32 2) Terminal 3′–3′ and 5′–5′ internucleotide linkage: Aptamers are modified by 3′-inverted dT and then extend chain in the standard 3′→5′ pattern, which needs controlled pore glass (CPG) modified with 5′-hydroxyl of the first nucleoside.33–35 3) 3′-biotin conjugates: 3′-biotin is similar to 3′-inverted dT modification.35 4) Modifications on phosphodiester linkage: Replacing DNA phosphodiester linkage with triazole linkages,36 methylphosphonate or phosphorothioate dramatically enhances resistance to nuclease hydrolysis.37–39 5) Mirror Image L-DNA: Based on natural D-form DNA aptamers, L-enantiomeric oligonucleotide aptamers (Spiegelmers) are chemically synthesized. L-DNA aptamers, like NOX-A12 and NOX-H94, have been proven to be more stable.40,41
Aptamers are easy to be excreted rapidly through renal clearance due to the unique characteristics of small nucleic acid molecules. Some methods have been designed to extend half-life: 1) 5′-End with cholesterol: Cholesterol is added to the 5′-end of aptamer to form a cholesterol-oligonucleotide (cholODN) conjugate.42 2) 5′-End with Dialkyl Lipids: Dialkylglycerol (DAG) is linked to the 5′-end of aptamer.43 3) 5′-End PEGylation: Clinical trials have demonstrated that PEGylated aptamers have not only increased blood residence but also enhanced targeted delivery of drugs.44,45 4) Gold nanoparticles: The bioavailable nanomaterial is used to modify aptamers for reducing renal clearance.46
Considerably, there are several kinds of aptamer-modification strategies to augment binding affinity and target selectivity: 1) Modifications on bases slow off-rate modified aptamers (SOMAmers): 5-BzdU [5-(N-benzylcarboxyamide)-2-deoxyuridine]-modified aptamer and SOMAmers containing 5-(N-benzylcarboxamide)-2-deoxyuridine (Bn-dU) or 5-[N-(1-naphthylmethyl) carboxamide]-2-deoxyuridine (Nap-dU) have a strong affinity and stability.47 2) Based on crystal structure modifications: Crystal-modified aptamer prominently inhibits ATX activity that is a recognized therapeutic target for a number of diseases.48 3) Nuclear magnetic resonance (NMR) spectroscopy-guided aptamer optimization: Aptamer complexes formed by NMR-guided optimization could actually improve binding affinity.49,50 4) Phosphorodithioate (PS2) modification: PS2 substitution on RNA aptamers dramatically improves target binding affinity by ∼ 1000-fold by inducing the fit rearrangement of the PS2-containing nucleotide.38,51 5) Deoxyuridines modification: 5′-position modified deoxyuridines also improve aptamer-binding affinity by promoting additional hydrophobic interactions between aptamers and their cognate targets.52 6) The hydrophobic base Ds modification: The data have indicated that the incorporation of unnatural base Ds can yield aptamers with greatly augmented affinities.53
Aptamers’ Conjugations
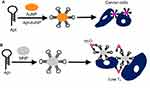
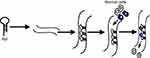
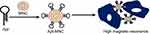
The sensitivity and selectivity of aptamers for the diagnosis and treatment of leukemia have been significantly improved after being combined with novel detection techniques. 1) Aptamer-nanoparticle: Aptamer–gold nanoparticle (Apt-AuNP) complexes selectively bind to target cells, generating discernible color changes from red to violet (Figure 1A). Moreover, aptamer-nanoparticle bioconjugates could enhance the intracellular delivery of drugs to target cells.54 By contrast, aptamer-magnetic nanoparticle (Apt-MNP) has a peculiar character, which leads to a decrease in adjacent water T2 by binding to target cells (Figure 1B), then detected by magnetic resonance imaging (MRI) or relaxometry.19 2) Microfluidic devices: Aptamers are immobilized on the surface of a microfluidic channel. The specific immobilized aptamers effectively capture target cells (Figure 2).55 When combined with nanochannel-ion channel hybrid, the captured cells dramatically block the ionic flow, resulting in the obvious variation of I-V property detected by electrochemical linear sweep voltammetry technique with greatly enhanced sensitivity.56 3) Quantum dots (QDs) probes: QDs-aptamer complexes could not only emit a visible fluorescence spectrum in the presence of target cells but also differentiate different types of leukemic cells, which is suitable for efficient cancer detection and targeted imaging (Figure 3).57 4) Molecular Beacon (MB): MB is a dual-labeled single-stranded DNA with a fluorophore at 5ʹ end and a quencher at the 3ʹ end (Figure 4). The fluorescent imaging of MB probes can be extended to the rapid detection of circulating tumor cell, ATP, proteins and small molecules.58,59 5) Fluorescent probes: The fluorescence probes can greatly augment the sensitivity of detection via cell imaging (Figure 5).60 and 6) MRI: In order to improve the sensitivity of MRI, gadolinium [Gd(III)], manganese [Mn(II)] or superparamagnetic iron oxide nanoparticles (SPIONs) complexes could be used as contrast agents (Figure 6).61 In another report, monodispersed carboxylated magnetic nanocrystal (MNC, Fe3O4) with high crystallinity was conjugated with vascular endothelial growth factor receptor 2 (VEGFR2)-specific aptamers to increase the specific imaging of VEGFR2, displaying a high magnetic resonance signal and efficient VEGFR2-detecting ability.62 7) Mass cytometry: Mass cytometry is a novel technique that quantitatively analyzes cell number one by one through using biotinylated aptamers in conjunction with metal-labeled neutravidin to characterize target cells, the characteristic cells undergo rapid ionization and the ionized elemental tags are analyzed by means of time-of-flight mass spectrometry (Figure 7).63
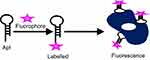 |
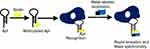
Figure 5 Aptamer labeled with fluorescent probe. Aptamer was labeled with fluorescent probe. The labeled aptamer-targeted leukemia cells and then excited fluorescence. |
Biomarkers Used for Aptamers-Mediated Diagnosis and Therapy in Leukemia
In patients with leukemia, a variety of biomarkers provide specific targets for target therapy. Such as, cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) expressed on CLL cells has been used in leukemia therapy.64 FcaRI (CD89) is a new target antigen expressed on different myeloid leukemia cell populations and mutated protein of nucleophosmin 1 (NPM1) has become an important prognostic and predictive maker.65 Collectively, accumulated findings indicate that lords of markers of leukemia are frequently applied for aptamers-mediated detection and therapy, like AP-1 aptamer targeting CD20, Sgc8 aptamer targeting protein tyrosine kinase-7 (PTK-7), anti-CD33(+)/CD34(+) aptamer targeting CD33 and CD34, K19 aptamer targeting Siglec-5 protein, CD38-specific aptamer-drug conjugates (ApDCs), CD117 aptamer-methotrexate (MTX) complexes, CD123-aptamer-mediated targeted drug train (TDT), and human OX40 (hOX40) aptamer (Table 1).
 |
Table 1 The Potential Application of Leukemic Biomarkers |
CD20 is a B-cell differentiation antigen, located only in pre-B cells and mature B cells. It can act as the diagnostic target in CLL and ALL. Haghighi M et al has selected and characterized ssDNA aptamer against CD20 via cell-SELEX, termed as AP-1, which could take place of antibodies for diagnosis and therapy of leukemia.66 PTK-7 is a significant biomarker expressed on the surface of Molt-4 cells, the specific interaction of DNA Sgc8 aptamer and PTK-7 has shown the significant potential for ALL detection.67 CD33 is expressed in more than 90% of patients with AML and is a good target for AML treatment. The functionalized gold nanoparticles (FGNs) were designed through attaching five selected antisense oligonucleotides (AOs) and one anti-CD33(+)/CD34(+) aptamer to naked gold nanoparticles (NGNs), which blocked five oncogenes expression in AML subtype 2 (AML M2) containing BAG1, MDM2, Bcl-2, BIRC5 and XIAP.68 Interestingly, AML-M2 CD33(+)/CD34(+) cells have been classified and used as targeted cells to select specific aptamers, which laid the basis for further looking for the more significant markers for diagnosis of AML-M2 leukemia.69 CD38 is widely accepted as a marker for unfavorable prognosis in leukemia. CD38-specific ApDCs provide a new avenue for targeted MM therapy with higher tissue-penetrating potential and negligible immunogenicity.70 CD44, mainly expressed in AML and B-CLL cell types, plays a key role in the proliferation, differentiation and migration of cells.71,72 The anti-CD44 aptamer has functioned as ideal tools for selective target-specific therapy when conjugated to the surface of liposome in breast cancer and lung cancer,73 which would be used in AML and B-CLL treatment by binding to CD44 on the surface of leukemic cells. CD123 (the interleukin-3 receptor α-chain) is a leukemia-associated antigen expressed on leukemia stem cells (LSCs) at high levels. CD123-aptamer-mediated TDT was synthetized to selectively deliver cytotoxic agents to CD123-expressed cells in AML.74 OX40, a TNF receptor family member, is expressed on leukemic blasts. The agonistic hOX40 aptamer has been designed to stimulate effector and memory T cell functions for leukemia treatment and even enhance the potency of dendritic cell-based tumor vaccines.75,76
Significantly, there are two important targets in AML, including CD47 and CD117. CD47 is identified as a negative prognostic marker for AML patients, anti-CD47 aptamer might also become a powerful therapeutic strategy for AML.77 CD117 is also highly expressed in AML and can be used as a myeloid immunological marker to differentially diagnose AML. The chemosynthetic CD117 aptamer was conjugated with MTX to construct Apt-MTX complex, which inhibited AML cells' growth and triggered cells' apoptosis, indicating the potential clinical value of Apt-MTX for AML targeted therapy.78
Aptamers as Diagnostic Tools in Leukemia
It is acknowledged that increasing number of preclinical applications of aptamers for leukemia diagnosis have been reported, such as Siglec-5 aptamer binding to AML, Sgc8 aptamer binding to ALL, AP-1 aptamer binding to CLL. Noteworthy, the translational application of aptamers remains to be explored in clinics (Table 2).
 |
Table 2 The Application of Aptamers in Leukemia Diagnosis |
Profoundly, aptamer-mediated avenues can be combined with other conventional tools to achieve early diagnosis. For instance, aptamer-based network hydrogel nanostructure in combination with molecular analysis to identify the PCR products of PML/RARα presents enormous potential for early diagnosing disease.79 It is approved that leukemia-derived exosomes are composed of CD63 and nucleolin (NCL). Conjunction of anti-CD63 antibody modified magnetic beads (CD63-MBs) with NCL-specific aptamer (AS1411) is applied to sensitively diagnose leukemia through binding the exosomes.79 Furthermore, combination of novel artificial antibody with the site-enhanced multivalent aptamers (APT-CIH) also shows high capture efficiency and selectivity to leukemia cells, providing a rational basis for diagnosing leukemia and discovering more therapeutic targets.80
Some reports have shown aptamers’ potential for AML diagnosis such as Siglec-5 protein-specific aptamers JH6, JH19, and K19 selected from aptamers pools. Among them, K19 aptamer represented the highest affinity and was labeled with biotin to construct a biotin-labeled K19 aptamer, namely Siglec-5 aptamer, which effectively identified and enriched Siglec-5 protein on the surface of AML cells. Thence, Siglec-5 aptamer provided an amazing approach for the detection of AML cells.81 Importantly, the specific combination of Siglec-5 with the K19 aptamer sequence resulted in a conformational change that generated a split G-quadruplex structure recognized by the G-quadruplex-specific iridium(III) complex, which displayed an enhanced luminescent response and ultrasensitive detection.82 Another aptamer (KH1C12) also showed augmented affinity and selectivity to the target AML cell line (HL60). Moreover, KH1C12 aptamer/gold nanoparticles-poly (3,4-ethylene dioxythiophene) (PEDOT) modified GC electrode (GC/PEDOT-Au/aptamer) could precisely count concentration of HL60 cells.83 In theory, CD33 functions as the biomarker of AML, and CD33 specific aptamers have been selected and characterized to bindto CD33+/CD34-cells in patients with AML M2, which might be used for the diagnosis of leukemia.84
ALL is the most common leukemia in children. Patient’s improvement completely depends on the early of ALL. Thus, rapid, selective and specific methods are urgently in demand. Based on the specific interaction of DNA Sgc8c aptamer and PTK-7 on ALL cells,85 a growing number of sophisticated diagnostic methods have been created. In one report, biotin-modified semiconductor QDs were labeled with the Sgc8 aptamer via streptavidin, then a stronger fluorescent signal would be generated by biotin amplification interactions when Sgc8 aptamer identified CCRF-CEM cells in T-ALL,67 In another report, dual-aptamer (Sgc8c and ATP aptamers)-functionalized graphene oxide (DAFGO) complex, consisting of bispecific aptamers (Sgc8c and ATP aptamers) and graphene oxide (GO), was designed for internalization into Molt-4 T-ALL cells,86 Sgc8c aptamer was selected and characterized to target to PTK-7. FAM (fluorophore)-labeled ATP aptamer was synthetized to detect the concentration of ATP. Theoretically, in the presence of target cells, DAFGO complex internalized into Molt-4 cells through interaction of Sgc8c aptamer with PTK-7, subsequently, FAM-labeled ATP aptamer released from GO surface and then linked to ATP, leading to a strong fluorescence emission.86 Significantly, Shan et al directly connected carboxyfluorescein-labeled Sgc8 aptamer (FAM-apt) to GO to target CCRF-CEM cells, when CCRF-CEM cells were captured, Sgc8 aptamer released from GO, and quenched fluorescence was recovered rapidly and significantly.87 In addition, aptamer-modified fluorescent silica nanoparticles (FSNPs) and Cu-Au nanoparticle (Cu-Au NP)-based colorimetric or theranostic platform could also be a highly sensitive and selective method to diagnose ALL.88–90
In the further study, the combination of amplification strategy with nanoparticles has attracted immense attention. For instance, aptamer-based quartz crystal microbalance (QCM) biosensor was developed, which was based on the interaction of aminophenylboronic acid-modified gold nanoparticles (APBA-AuNPs) with silver enhancement.91 In the system, Sgc8c aptamer was immobilized on QCM to capture CCRF-CEM cells by interacting with PTK7, APBA-AuNPs were used to label the captured cells. The frequency response was amplified by silver enhancement within a short time. Lastly, the captured cells were stained and quantitatively analyzed with the fluorescence microscope.91 Another new amplification strategy also immobilized Sgc8c aptamer onto gold nanoparticles-coated magnetic FeO nanoparticles (GMNPs) to constitute Apt-GMNPs complex, the process required a nitrogen-doped graphene modified electrode. Subsequently, the hairpin structure of the aptamer would be disrupted when Sgc8c aptamer captured CCRF-CEM cells, resulting in the decrease of the electrochemical signal.92 On the other side, the combination of aptamer-conjugated magnetic beads (apt-MBs) and the magnet-QCM system was also applicable to detect and diagnose ALL. Sgc8c aptamer-conjugated MBs were used for extracting CCRF-CEM cells, and the magnet-QCM system was successfully applied for the quantitative analysis of cell. When MBs-captured CCRF-CEM cells accumulated on a quartz crystal gold electrode surface under a magnetic field, the resonant frequency would decrease.93 For minimal residual diseases (MRD) of T-ALL, a smart dual-specific detection system containing magnetic aptamer Sgc8 probe (M-sgc8 probe) and rolling cycle amplification probe (RCA-sgc8 probe) has been developed, which would be successfully utilized for diagnosing and monitoring ALL progress.94
Lately, a label-free signal-on fluorescence aptasensor based on terbium(iii)-aptamer (Tb-apt) has been designed. The aptamer sensitized the fluorescence of Tb and formed a strong fluorescent Tb-apt probe. When the probe captured the CCRF-CEM cells, the released fluorescence signal was positively associated with cell concentration. Therefore, the aptamer-sensitized terbium(iii) luminescence method was rapid, ultrasensitive, economical and highly specific, which also held great potential to diagnose other types of leukemia at the early stage.95 Excellently, the Ds-containing DNA (Ds-DNA) aptamer, 05-MB231 generated by cell-ExSELEX, could bind to CCRF-CEM cells with higher affinity, extensive specificities and biological activities, indicating that the Ds-DNA aptamers and the novel cell-ExSELEX would be broadly applied for leukemia cell imaging, biomarker discovery, and detection.26
CML is a clonal myeloproliferative disease characterized by increased neutrophils in peripheral blood and myeloid proliferation in BM. Interestingly, graphene-hemin-composite coupled with Au nanoflowers (H-RGO-Au NFs) has been demonstrated as an electrochemical aptamer biosensor, which possessed enhanced peroxidase-like properties and sensitively realized glycoprotein on the surface of K562 CML cells.96 Consistently, a new electrochemiluminescence (ECL) platform based on the complex of aptamers and carbon quantum dots (CQDs) coated ZnO nanosphere (ZnO@CQDs) could offer great promise for ultrasensitive and selective detection of K562 CML cells.97 It is accepted that the progression of CML is characterized by the generation of BCR/ABL1 fusion gene. Based upon the unique expression of BCR/ABL1 gene in CML cells, the application of BCR/ABL1-based aptamer has a great application prospect in the diagnosis, treatment and prognosis of CML.
In consideration of CLL, Haghighi M et al have selected and characterized ssDNA aptamers against CD20 via cell-SELEX, namely AP-1, AP-2 and AP-3, which could conjugate with other imaging technologies to improve diagnostic sensitivity and have the potency to take place of antibodies. AP-1 aptamer would be extensively used for diagnosis and therapy of CLL with the most stable thermodynamic property and the highest binding affinity.66 Recently, two novel truncated G-rich aptamers, R1.2 and R1.3, have been generated by “Ligand-Guided Selection” (LI-GS), both of them folded into G-quadruplex structures in the bioactive conformations, which was essential for specific and effective recognition of mIgM expressed in B cells and provided the fundamental basis for designing effective aptamer-based biosensors potentially applied for the highly sensitive and selective diagnosis of B cell lymphoma and CLL.98
Aptamers as Therapeutic Tools in Leukemia
Collectively, there are numerous preclinical applications of aptamers for leukemia treatment, for instance, CD117-specific ssDNA aptamer, ZW25 aptamer, anti-CD33(+)/CD34(+) aptamer, S30-T1 aptamer, AS-1411 aptamer, K19/Siglec-5 aptamer, RNA aptamers and peptide aptamers targeting AML, Sgc8 aptamer targeting ALL, β-arrestin 2 aptamer, NOX-A12 aptamer and F79 aptamer targeting CML. Nevertheless, only AS1411 aptamer combined with cytarabine has entered clinical trials in the treatment of patients with primary refractory or relapsed AML (Table 3).
 |
Table 3 The Application of Aptamers in Leukemia Therapy |
It is worth noting that the conjunction of aptamers with routine therapies will provide more precise and higher-quality therapeutic efficacy. Recently, increasing researches have approved aptamer-based targeted drug delivery systems prominently alleviated side effects and improved therapeutic efficacy of chemotherapy, such as CD117 ssDNA aptamer delivering MTX, S30-T1 aptamer delivering doxorubicin (Dox), Sgc8 aptamer delivering Daunorubicin (Dau) and vincristine (VCR) sulphate.
Of note, the polyvalent aptamer system composed of multiple aptamer units and physically inserted chemotherapy agents, namely “Poly-Aptamer-Drug”, naturally presents enhanced binding affinity (~ 40 fold greater) and cellular internalization efficiency than monovalent counterpart in targeting and killing leukemia cells due to multivalent effects.99,100 For further monitoring anti-leukemia drug effect, the complex of localized surface plasmon resonance (LSPR) and gold nanorods (AuNR) functionalized with aptamers has been effectively used in preclinical stage through sensing the cytochrome-c released from apoptotic leukemia cells.101
The current treatment strategies for AML might induce relapse and immunological rejection. Thus, promising therapeutic approaches for AML with fewer side effects are urgently needed. CD117, a transmembrane receptor highly expressed on AML cells, is beneficial for detecting MRD, which indicates that the CD117 may be a potential therapeutic biomarker for AML.102 Recently, CD117-specific ssDNA aptamer was produced, and the Apt-MTX complex was formulated by conjugating aptamer with MTX. Importantly, Apt-MTX efficiently inhibited the growth of primary AML cells and had no side effects on off-target normal marrow cells.78 Another AML marker, CD123-aptamer (ZW25)-mediated TDT was created to selectively deliver drugs to target cells via binding to CD123, which demonstrated potential applications for effectively delivering therapeutic drugs to CD123-expressed cells.74 Similarly, CD33 is also the significant marker of AML, in the further study, FGNs were designed by attaching five selected antisense AOs and one anti-CD33(+)/CD34(+) aptamer to NGNs, which hindered AML-M2 development through blocking five important oncogenes expression, including BAG1, MDM2, Bcl-2, BIRC5 and XIAP.68 Moreover, the optimized CD33-targeting aptamer S30-T1 could highly recognize the C2 domain of the CD33 antigen in vitro and in vivo and specifically arrest the cell cycle at the G2 phase to inhibit CD33 positive AML HL-60 cell proliferation via delivering Dox to target cells.103 In consideration of Siglec-5 protein, three new aptamers (JH6, JH19, and K19) were selected and characterized, and K19 had the highest affinity and specificity. The biotin-labeled K19 aptamer was used to enrich and identify Siglec-5 protein, which acted as a prospective strategy for AML targeted therapy.81 Additionally, we have found AS-1411, a guanosine-rich oligonucleotide aptamer, was designed for specifically binding to NCL, demonstrating obvious inhibitive activity against AML cell lines. Combination of AS-1411 with cytarabine showed the superiority to cytarabine alone.104
Further, a recent research has reported RNA aptamers were created to link with the Runt domain (RD) of the AML1 protein with high affinity. AMLI (RUNX1) is a transcription factor isolated from a chromosomal breakpoint and its genetic variations are frequently observed in human leukemia, leading to abnormal hemocytopoiesis and immunodeficiency.105 The repair of AML1 function is dependent on RNA aptamers targeting to RD, providing new insights into treatment of AML1-related diseases.106,107 Besides, some aptamers were designed for treating AML via controlling cell cycle and inhibiting malignant cell proliferation. For instance, the peptide aptamers were selected to prohibit translationally controlled tumor protein (TCTP) expression in Molt-4 cells in AML, which down-regulated the expressions of cell-cycle-related downstream proteins, suggesting potential promise for AML therapy.108
Clinically, the treatment of ALL is also confronted with great challenges. Dau has been attractive in the treatment of ALL but with limitations due to its cardiotoxicity. To reduce the cardiotoxicity, Sgc8 Apt-Dau conjugate, a simple and efficient complex was constructed to specifically deliver Dau to ALL T cells through the interaction between Sgc8 and PTK7 with negligible toxicity.109,110 In order to further enhance the specificity and sensitivity of treatment. The Apt-Dau complex was connected to AuNPs which has widely been used to deliver therapeutic drugs with a large surface to volume ratio, simple synthesis and low toxicity.111 The release of Dau from the complex depended on the pH levels. In pH 7.4, about 18% of Dau was released from Apt-Dau-AuNPs complex, while switching to pH 5.5, the release of drug from the complex was up to 75% under the same condition. Subsequently, flow cytometry analysis indicated the internalization efficiency of Dau was dramatically increased in comparison with Dau- and Apt-Dau conjugate-treated Molt-4 cells. Hence, Apt-Dau-AuNPs complex has been explored as a specific delivery system to internalize agents into target cells with reduced cytotoxic effects.110
In addition to Dau and DOX, VCR is widely used in hemato-oncology, especially ALL. The VCR-loaded and Sgc8 aptamer-conjugated liposomal drug delivery system (Sgc8/VCR-Lipo) significantly enhanced the therapeutic effects of VCR against ALL.112 Amazingly, the novel N-heterocyclic carbene (NHC)-gold(I) complexes and aptamer-guided DNA tetrahedral nanostructure (s-TDN) could also be utilized to transport drugs when conjugated with Sgc8c aptamer.113,114 As we know, NCL is a 9-O-acetylated sialoglycoprotein presenting on the surface of pre-B ALL, NCL-recognition aptamer AS1411 has shown evident therapeutic effects.115 Strikingly, the conjugation of 5-MB231 Ds-DNA aptamer with personalized medicine held the potency to augment anti-ALL activity.26
In terms of CML, regulators of signaling pathways and oncogenes might be ideal therapeutic targets. It has been reported that β-arrestin 2 was critical for the onset and maintenance of both the chronic and blast crisis stages of CML (bcCML). The specific β-arrestin 2-binding aptamers were constructed, including β–arr2A1, β–arr2A2 and β–arr2A3. β–arr2A3 had the highest binding affinity. In order to selectively deliver β-arrestin 2 aptamer to leukemia cells, β–arr2A3 was linked to the NCL aptamer through complementary base-pair annealing. The NCL aptamer-β-arr2A3 chimera internalized into cells and delivered the β-arr2A3 aptamer to its intracellular target, which effectively inhibited the activation of the Hh/Smo and Wnt/Fz (Wingless/Frizzled) signaling pathways, and thereby suppressed CML progression.116 Besides, there is an RNA aptamer targeting oncogene-induced Leukemia. NOX-A12 is an RNA oligonucleotide in L-configuration spiegelmer aptamer, which could block stromal-cell derived factor-1 (SDF-1) or chemokine (C-X-C motif) ligand-12 (CXCL-12)-induced BCR-ABL1 expression through preventing SDF-1 from reacting with its receptors, chemokine (C-X-C motif) receptor 4 (CXCR4) and CXCR7.117,118 Moreover, the cooperation of NOX-A12 with nilotinib (ABL-kinase inhibitor) could suppress CML development, indicating a cooperative effect of two inhibitors for diminishing or eradicating residual CML cells. The finding also suggested that NOX-A12 had the ability to improve the efficacy of tyrosine kinase inhibitors (TKI) against oncogene-driven leukemia.118
It is identified that RAD52 has 2 DNA binding domains I and II (DNA I and DNA II) that bind to amino acid residues F79 and K102, respectively, which plays a critical role in BCR-ABL1-mediated CML cells growth. Of note, F79 aptamer could abrogate DNA binding activity of RAD52, leading to homologous recombination repair (HRR) inhibition and CML elimination through inducing synthetic lethality in BRCA-deficient CML cells. Theoretically, targeting RAD52 by F79 aptamer might enhance the therapeutic effect of oncogenes-induced leukemia including B-ALL, T-ALL and PML-RAR-positive acute promyelocytic leukemia (APL).119,120
Prospective
Recently, rapid, ultrasensitive, and highly specific diagnostic methods for leukemia have been emerging. Based on aptamer–target interaction, a functional DNA nanodevice has been built for recognizing target cells by aptamer-integrated DNA nanoassembly, which presented the potential applications for point-of-care leukemia diagnosis with excellent sensitivity and selectivity.121 Besides conventional biomarkers, some promising markers have already emerged, like miRNA, exosomes and so on. Herein, leukemia-derived exosomes containing CD63 and NCL have also been identified as nanosized biomarkers highly sensitively detected by a fluorescent biosensing platform, namely a dual-signal amplification. The process required anti-CD63 antibody modified magnetic bead conjugates (MB-CD63) and a DNA primer comprising an NCL-recognition AS1411 aptamer.122 Meanwhile, an aptamer biosensor provided a novel strategy for label-free detection of leukemia-derived miR-16 with high sensitivity due to the polymerase cyclic amplification and the aggregation of the illuminator. Significantly, the neoteric technique would offer a promising strategy for early diagnosis of fusion genes.123
Generally, aptamers or aptamer/antibody bispecific system (AAbs) are often designed to target to biomarkers on the surface of leukemia cells or to influence the intracellular targets. However, there are only a few reports about aptamers directly against fusion genes. A novel research has developed an enzyme-free and label-free surface plasmon resonance (SPR) biosensor applied for ultrasensitive detection of fusion genes, which required the DNA self-assembly aptamer-based network hydrogel nanostructure via streptavidin (SA) encapsulation, and the SA aptamer, as a capture aptamer, dramatically amplified the SPR signal by strongly linking to SA. Furthermore, the sophisticated biosensing method has been applied to capture the PML/RARα, a gene marker of M4E0, displaying excellent feasible potency for detecting fusion genes and monitoring leukemia progress.79 Besides, there are other traditional fusion genes such as AML1-MTG8 and CBFβ-MYH11, typical symbols of M2b and M3, respectively, based on which we speculate that it is available to extract fusion genes from leukemia samples to directly construct ssDNA library or indirectly formulate RNA library, which provides qualifications for selecting and characterizing fusion genes-specific nucleic acid aptamers. Nevertheless, we may face inevitable challenges due to the insufficient sample and uncertain length of the randomized sequence. In the future, efforts should be focused on how to design and optimize nucleic acid aptamers to directly target and prohibit fusion genes' expressions and further control leukemia development. Interestingly, aptamers can also be utilized to discover more gene targets related to leukemia.
Conclusion
Even though monoclonal antibody-based complexes still dominated in leukemia diagnosis and therapy, aptamers and SELEX technology have brought a revolution to biomedical fields. Compared with monoclonal antibody therapy, aptamers can be used to directly kill leukemia cells or deliver agents to targets with faster internalization, higher selectivity and affinity, lower toxicity, lower immunogenicity, longer half-life and more stable properties at a vast range of variable environment. What’s more, after being conjugated with imaging and other advanced technologies, such as nanoparticles, microfluidic devices, QBs probes, MB, fluorescent probes, MRI and mass cytometry, the sensitivity and specificity of aptamers are significantly improved in diagnosis and treatment. However, aptamers require chemical modifications to significantly overcome nuclease degradation, rapid renal excretion and deficient binding affinity due to the nucleic acid characteristics, which result in increased cost and time procedure. In addition, SELEX is an expensive and tedious process. All of these problems bring great challenges to the clinical application of aptamers. Here, future researches should try to make aptamers’ selection and modifications procedures faster, cheaper and simpler.
In summary, nucleic acid aptamers have attracted great attention and achieved tremendous progress in increasingly diverse fields. It is recognized that nucleic acid aptamers are powerful diagnostic and therapeutic tools for leukemia, which also can conjunct with conventional strategies. But numerous clinical applications of aptamers are still confronted with immense challenges including the automation of SELEX process, the increase of biostability and bioavailability and so on. Herein, the aptamer-based strategies actually hold the potential to be excellent alternatives to traditional approaches for diagnosis and treatment of leukemia, and the application of aptamers in detecting fusion genes is urgent to be explored.
Abbreviations
AAbs, aptamer/antibody bispecific system; BM, bone marrow; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CPG, controlled pore glass; APBA-AuNPs, aminophenylboronic acid-modified gold nanoparticles; Ara-C, arabinoside; Apt, aptamer; Apt-AuNP, aptamer–gold nanoparticle; Apt-MNP, aptamer–magnetic nanoparticle; ATX, autotoxin; AuNPs, gold nanoparticles; Bn-Du, 5-(N-benzylcarboxamide)-2-deoxyuridine; CD, cluster of differentiation; CLL, chronic lymphoblastic leukemia; CML, chronic myeloid leukemia; CTLA-4, cytotoxic T lymphocyte-associated antigen 4; DAFGO, dual-aptamer-functionalized graphene oxide; Dau, daunorubicin; Dox, doxorubicin; FGNs, functionalized gold nanoparticles; GO, graphene oxide; MNC, monodispersed carboxylated magnetic nanocrystal; NMR, nuclear magnetic resonance; VEGFR2, vascular endothelial growth factor receptor 2; MNPs, magnetic nanoparticles; MRD, minimal residual disease; ApDCs, aptamer-drug conjugates; MTX, methotrexate; NGNs, naked gold nanoparticles; NPM1, nucleophosmin 1; ECL, electrochemiluminescence; QCM, quartz crystal microbalance; QDs, quantum dots; RD, runt domain; SELEX, systemic evolution of ligands by exponential enrichment; ExSELEX, genetic alphabet Expansion for SELEX; SOMAmers, slow off-rate modified aptamers; SPIONs, super paramagnetic iron oxide nanoparticles; TCR, T cell receptor; NCL, nucleolin; PTK-7, protein tyrosine kinase-7; AML M2, AML subtype 2; AOs, antisense oligonucleotides; siRNA, small interfering RNA; RNAi, RNA interference; DAG, dialkylglycerol; MB, molecular beacon; MRI, magnetic resonance imaging; SPR, surface plasmon resonance; SA, streptavidin; APL, acute promyelocytic leukemia; VCR, vincristine sulphate; TCTP, translationally controlled tumor protein; Aunano-PEDOT/GC, gold nanoparticles-poly(3,4-ethylene dioxythiophene) modified GC electrode; LSCs, leukemia stem cells; UB, unnatural base; UBPs, UB pairs; Px, 2-nitro-4-propynylpyrrole; Ds, 7-(2-thienyl)imidazo[4,5-b]pyridine; Ds-DNA, Ds-containing DNA; GMNPs, gold nanoparticles-coated magnetic FeO nanoparticles; FSNPs, fluorescent silica nanoparticles; Cu-Au NP, Cu-Au nanoparticle; FAM, fluorophore; LI-GS, Ligand-Guided Selection; LSPR, localized surface plasmon resonance; NHC, N-heterocyclic carbine; TKI, tyrosine kinase inhibitors; HRR, homologous recombination repair; H-RGO-Au NFs, graphene-hemin-composite coupled with Au nanoflower; ZnO@CQDs, carbon quantum dots (CQDs) coated ZnO nanosphere; AuNR, gold nanorods; bcCML, blast crisis stages of CML; SDF-1, stromal-cell derived factor-1; CXCL-12, chemokine (C-X-C motif) ligand-12; CXCR4, chemokine (C-X-C motif) receptor 4.
Acknowledgments
We thank the members of the Clinical Laboratory of Hunan Cancer Hospital and Xiangya Medical School of Central South University for contributions and Dr. Dong Z from the Hormel Institute of University of Minnesota for helpful discussion.
Author Contributions
All authors contributed towards data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi:10.1182/blood-2016-03-643544
2. Keegan A, Charest K, Schmidt R, et al. Flow cytometric minimal residual disease assessment of peripheral blood in acute lymphoblastic leukaemia patients has potential for early detection of relapsed extramedullary disease. J Clin Pathol. 2018;71(7):653–658. doi:10.1136/jclinpath-2017-204828
3. Dorge P, Meissner B, Zimmermann M, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98(3):428–432. doi:10.3324/haematol.2011.056135
4. Marcinkowska-Swojak M, Handschuh L, Wojciechowski P, et al. Simultaneous detection of mutations and copy number variation of NPM1 in the acute myeloid leukemia using multiplex ligation-dependent probe amplification. Mutat Res. 2016;786:14–26. doi:10.1016/j.mrfmmm.2016.02.001
5. Maino E, Bonifacio M, Scattolin AM, Bassan R. Immunotherapy approaches to treat adult acute lymphoblastic leukemia. Expert Rev Hematol. 2016;9(6):563–577. doi:10.1586/17474086.2016.1170593
6. Palanca-Wessels MC, Czuczman M, Salles G, et al. Safety and activity of the anti-CD79B antibody-drug conjugate polatuzumab vedotin in relapsed or refractory B-cell non-Hodgkin lymphoma and chronic lymphocytic leukaemia: a phase 1 study. Lancet Oncol. 2015;16(6):704–715. doi:10.1016/S1470-2045(15)70128-2
7. Beckwith KA, Frissora FW, Stefanovski MR, et al. The CD37-targeted antibody-drug conjugate IMGN529 is highly active against human CLL and in a novel CD37 transgenic murine leukemia model. Leukemia. 2014;28(7):1501–1510. doi:10.1038/leu.2014.32
8. Guo J, Luan X, Cong Z, et al. The potential for clinical translation of antibody-targeted nanoparticles in the treatment of acute myeloid leukaemia. J Control Release. 2018;286:154–166. doi:10.1016/j.jconrel.2018.07.024
9. Dickinson H, Lukasser M, Mayer G, Huttenhofer A. Cell-SELEX: in vitro selection of synthetic small specific ligands. Methods Mol Biol. 2015;1296:213–224.
10. Yazdian-Robati R, Arab A, Ramezani M, Abnous K, Taghdisi SM. Application of aptamers in treatment and diagnosis of leukemia. Int J Pharm. 2017;529(1–2):44–54. doi:10.1016/j.ijpharm.2017.06.058
11. Mallikaratchy P, Tang Z, Kwame S, et al. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Mol Cell Proteomics. 2007;6(12):2230–2238. doi:10.1074/mcp.M700026-MCP200
12. Bouvet P. Identification of nucleic acid high affinity binding sequences of proteins by SELEX. Methods Mol Biol. 2015;1334:333–343.
13. Park KS. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens Bioelectron. 2018;102:179–188. doi:10.1016/j.bios.2017.11.028
14. Wang P, Yang Y, Hong H, et al. Aptamers as therapeutics in cardiovascular diseases. Curr Med Chem. 2011;18(27):4169–4174. doi:10.2174/092986711797189673
15. Bouvier-Muller A, Duconge F. Nucleic acid aptamers for neurodegenerative diseases. Biochimie. 2018;145:73–83. doi:10.1016/j.biochi.2017.10.026
16. Li W, Lan X. Aptamer oligonucleotides: novel potential therapeutic agents in autoimmune disease. Nucleic Acid Ther. 2015;25(4):173–179. doi:10.1089/nat.2014.0529
17. Ashrafuzzaman M. Aptamers as both drugs and drug-carriers. Biomed Res Int. 2014;2014:697923. doi:10.1155/2014/697923
18. Marangoni K, Neves AF, Rocha RM, et al. Prostate-specific RNA aptamer: promising nucleic acid antibody-like cancer detection. Sci Rep. 2015;5:12090. doi:10.1038/srep12090
19. Ruiz Ciancio D, Vargas MR, Thiel WH, et al. Aptamers as diagnostic tools in cancer. Pharmaceuticals (Basel). 2018;11:3. doi:10.3390/ph11030086
20. Schneider V, Zhang L, Bullinger L, et al. Leukemic stem cells of acute myeloid leukemia patients carrying NPM1 mutation are candidates for targeted immunotherapy. Leukemia. 2014;28(8):1759–1762. doi:10.1038/leu.2014.116
21. Ni S, Yao H, Wang L, et al. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int J Mol Sci. 2017;18:8. doi:10.3390/ijms18081683
22. Chen M, Yu Y, Jiang F, et al. Development of cell-SELEX technology and its application in cancer diagnosis and therapy. Int J Mol Sci. 2016;17:12. doi:10.3390/ijms17122079
23. Sun H, Zhu X, Lu PY, et al. Oligonucleotide aptamers: new tools for targeted cancer therapy. Mol Ther Nucleic Acids. 2014;3:e182. doi:10.1038/mtna.2014.32
24. Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv. 2015;33(6 Pt 2):1141–1161. doi:10.1016/j.biotechadv.2015.02.008
25. Drabik A, Ner-Kluza J, Mielczarek P, et al. Advances in the study of aptamer-protein target identification using the chromatographic approach. J Proteome Res. 2018;17(6):2174–2181. doi:10.1021/acs.jproteome.8b00122
26. Futami K, Kimoto M, Lim YWS, Hirao I. Genetic alphabet expansion provides versatile specificities and activities of unnatural-base DNA aptamers targeting cancer cells. Mol Ther Nucleic Acids. 2019;14:158–170. doi:10.1016/j.omtn.2018.11.011
27. Stovall GM, Bedenbaugh RS, Singh S, et al. In vitro selection using modified or unnatural nucleotides. Curr Protoc Nucleic Acid Chem. 2014;56:96 1–33.
28. Friedman AD, Kim D, Liu R. Highly stable aptamers selected from a 2ʹ-fully modified fGmH RNA library for targeting biomaterials. Biomaterials. 2015;36:110–123. doi:10.1016/j.biomaterials.2014.08.046
29. Campbell MA, Wengel J. Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem Soc Rev. 2011;40(12):5680–5689. doi:10.1039/c1cs15048k
30. Gupta S, Hirota M, Waugh SM, et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J Biol Chem. 2014;289(12):8706–8719. doi:10.1074/jbc.M113.532580
31. Arnold AE, Malek-Adamian E, Le PU, et al. Antibody-antisense oligonucleotide conjugate downregulates a key gene in glioblastoma stem cells. Mol Ther Nucleic Acids. 2018;11:518–527. doi:10.1016/j.omtn.2018.04.004
32. Maio G, Enweronye O, Zumrut HE, et al. Systematic optimization and modification of a DNA aptamer with 2ʹ-O-methyl RNA analogues. ChemistrySelect. 2017;2(7):2335–2340. doi:10.1002/slct.201700359
33. Lapa SA, Chudinov AV, Timofeev EN. The toolbox for modified aptamers. Mol Biotechnol. 2016;58(2):79–92. doi:10.1007/s12033-015-9907-9
34. Fujino T, Suzuki T, Okada K, et al. Chimeric RNA oligonucleotides incorporating triazole-linked trinucleotides: synthesis and function as mRNA in cell-free translation reactions. J Org Chem. 2016;81(19):8967–8976. doi:10.1021/acs.joc.6b01618
35. Gao S, Zheng X, Jiao B, Wang L. Post-SELEX optimization of aptamers. Anal Bioanal Chem. 2016;408(17):4567–4573. doi:10.1007/s00216-016-9556-2
36. Palframan MJ, Alharthy RD, Powalowska PK, Hayes CJ. Synthesis of triazole-linked morpholino oligonucleotides via Cu(I) catalysed cycloaddition. Org Biomol Chem. 2016;14(11):3112–3119. doi:10.1039/C6OB00007J
37. Flur S, Micura R. Chemical synthesis of RNA with site-specific methylphosphonate modifications. Methods. 2016;107:79–88. doi:10.1016/j.ymeth.2016.03.024
38. Abeydeera ND, Egli M, Cox N, et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016;44(17):8052–8064. doi:10.1093/nar/gkw725
39. Volk DE, Lokesh GLR. Development of Phosphorothioate DNA and DNA Thioaptamers. Biomedicines. 2017;5:3.
40. Hoellenriegel J, Zboralski D, Maasch C, et al. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123(7):1032–1039. doi:10.1182/blood-2013-03-493924
41. Schwoebel F, van Eijk LT, Zboralski D, et al. The effects of the anti-hepcidin Spiegelmer NOX-H94 on inflammation-induced anemia in cynomolgus monkeys. Blood. 2013;121(12):2311–2315. doi:10.1182/blood-2012-09-456756
42. Lee CH, Lee SH, Kim JH, et al. Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol Ther Nucleic Acids. 2015;4:e254. doi:10.1038/mtna.2015.30
43. Ninomiya K, Yamashita T, Kawabata S, Shimizu N. Targeted and ultrasound-triggered drug delivery using liposomes co-modified with cancer cell-targeting aptamers and a thermosensitive polymer. Ultrason Sonochem. 2014;21(4):1482–1488. doi:10.1016/j.ultsonch.2013.12.023
44. Taghdisi SM, Danesh NM, Sarreshtehdar Emrani A, et al. Targeted delivery of epirubicin to cancer cells by PEGylated A10 aptamer. J Drug Target. 2013;21(8):739–744. doi:10.3109/1061186X.2013.812095
45. Xing H, Li J, Xu W, et al. The effects of spacer length and composition on aptamer-mediated cell-specific targeting with nanoscale PEGylated liposomal doxorubicin. Chembiochem. 2016;17(12):1111–1117. doi:10.1002/cbic.201600092
46. Liu Y, Ouyang Q, Li H, et al. Turn-on fluoresence sensor for Hg(2+) in food based on FRET between aptamers-functionalized upconversion nanoparticles and gold nanoparticles. J Agric Food Chem. 2018;66(24):6188–6195. doi:10.1021/acs.jafc.8b00546
47. Lee KY, Kang H, Ryu SH, et al. Bioimaging of nucleolin aptamer-containing 5-(N-benzylcarboxyamide)-2ʹ-deoxyuridine more capable of specific binding to targets in cancer cells. J Biomed Biotechnol. 2010;2010:168306. doi:10.1155/2010/168306
48. Miller LM, Keune WJ, Castagna D, et al. Structure-activity relationships of small molecule autotaxin inhibitors with a discrete binding mode. J Med Chem. 2017;60(2):722–748. doi:10.1021/acs.jmedchem.6b01597
49. Wolter AC, Duchardt-Ferner E, Nasiri AH, et al. NMR resonance assignments for the class II GTP binding RNA aptamer in complex with GTP. Biomol NMR Assign. 2016;10(1):101–105. doi:10.1007/s12104-015-9646-7
50. Souard F, Perrier S, Noel V, et al. Optimization of experimental parameters to explore small-ligand/aptamer interactions through use of (1) H NMR spectroscopy and molecular modeling. Chemistry. 2015;21(44):15740–15748. doi:10.1002/chem.201501527
51. Hausmann J, Keune WJ, Hipgrave Ederveen AL, et al. Structural snapshots of the catalytic cycle of the phosphodiesterase autotaxin. J Struct Biol. 2016;195(2):199–206. doi:10.1016/j.jsb.2016.06.002
52. Davies DR, Gelinas AD, Zhang C, et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A. 2012;109(49):19971–19976. doi:10.1073/pnas.1213933109
53. Kimoto M, Yamashige R, Matsunaga K, Yokoyama S, Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat Biotechnol. 2013;31(5):453–457. doi:10.1038/nbt.2556
54. Jo H, Ban C. Aptamer-nanoparticle complexes as powerful diagnostic and therapeutic tools. Exp Mol Med. 2016;48:e230. doi:10.1038/emm.2016.44
55. Reinholt SJ, Craighead HG. Microfluidic device for aptamer-based cancer cell capture and genetic mutation detection. Anal Chem. 2018;90(4):2601–2608. doi:10.1021/acs.analchem.7b04120
56. Cao J, Zhao XP, Younis MR, et al. Ultrasensitive capture, detection, and release of circulating tumor cells using a nanochannel-ion channel hybrid coupled with electrochemical detection technique. Anal Chem. 2017;89(20):10957–10964. doi:10.1021/acs.analchem.7b02765
57. Qian ZS, Shan XY, Chai LJ, Chen JR, Feng H. A fluorescent nanosensor based on graphene quantum dots-aptamer probe and graphene oxide platform for detection of lead (II) ion. Biosens Bioelectron. 2015;68:225–231. doi:10.1016/j.bios.2014.12.057
58. Park Y, Nim-Anussornkul D, Vilaivan T, Morii T, Kim BH. Facile conversion of ATP-binding RNA aptamer to quencher-free molecular aptamer beacon. Bioorg Med Chem Lett. 2018;28(2):77–80. doi:10.1016/j.bmcl.2017.12.008
59. Hwang JY, Kim ST, Han HS, Kim K, Han JS. Optical aptamer probes of fluorescent imaging to rapid monitoring of circulating tumor cell. Sensors (Basel). 2016;16:11. doi:10.3390/s16111909
60. Li H, Hu H, Zhao Y, et al. Multifunctional aptamer-silver conjugates as theragnostic agents for specific cancer cell therapy and fluorescence-enhanced cell imaging. Anal Chem. 2015;87(7):3736–3745. doi:10.1021/ac504230j
61. Zhou W, Zhou Y, Wu J, et al. Aptamer-nanoparticle bioconjugates enhance intracellular delivery of vinorelbine to breast cancer cells. J Drug Target. 2014;22(1):57–66. doi:10.3109/1061186X.2013.839683
62. Kim B, Yang J, Hwang M, et al. Aptamer-modified magnetic nanoprobe for molecular MR imaging of VEGFR2 on angiogenic vasculature. Nanoscale Res Lett. 2013;8(1):399. doi:10.1186/1556-276X-8-399
63. Mironov GG, Bouzekri A, Watson J, et al. Aptamer-facilitated mass cytometry. Anal Bioanal Chem. 2018;410(13):3047–3051. doi:10.1007/s00216-018-1011-0
64. Ciszak L, Frydecka I, Wolowiec D, Szteblich A, Kosmaczewska A. Patients with chronic lymphocytic leukaemia (CLL) differ in the pattern of CTLA-4 expression on CLL cells: the possible implications for immunotherapy with CTLA-4 blocking antibody. Tumour Biol. 2016;37(3):4143–4157. doi:10.1007/s13277-015-4217-1
65. Mladenov R, Hristodorov D, Cremer C, et al. The Fc-alpha receptor is a new target antigen for immunotherapy of myeloid leukemia. Int J Cancer. 2015;137(11):2729–2738. doi:10.1002/ijc.29628
66. Haghighi M, Khanahmad H, Palizban A. Selection and characterization of single-stranded DNA aptamers binding human B-cell surface protein CD20 by cell-SELEX. Molecules. 2018;23(4):715. doi:10.3390/molecules23040715
67. Yu Y, Duan S, He J, et al. Highly sensitive detection of leukemia cells based on aptamer and quantum dots. Oncol Rep. 2016;36(2):886–892. doi:10.3892/or.2016.4866
68. Zaimy MA, Jebali A, Bazrafshan B, et al. Coinhibition of overexpressed genes in acute myeloid leukemia subtype M2 by gold nanoparticles functionalized with five antisense oligonucleotides and one anti-CD33(+)/CD34(+) aptamer. Cancer Gene Ther. 2016;23(9):315–320. doi:10.1038/cgt.2016.33
69. Zhang SQ, Wang GP, Zhu P, et al. [Screening and structure analysis of nucleic acid aptamers binding to surface of CD33(+)/CD34(+) cells from patients with acute myeloid leukemia subtype M(2)]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011;19(3):561–565.
70. Wen J, Tao W, Hao S, Iyer SP, Zu Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia. 2016;30(4):987–991. doi:10.1038/leu.2015.216
71. D’Arena G, Calapai G, Deaglio S. Anti-CD44 mAb for the treatment of B-cell chronic lymphocytic leukemia and other hematological malignancies: evaluation of WO2013063498. Expert Opin Ther Pat. 2014;24(7):821–828. doi:10.1517/13543776.2014.915942
72. Chen P, Huang H, Wu J, et al. Bone marrow stromal cells protect acute myeloid leukemia cells from anti-CD44 therapy partly through regulating PI3K/Akt-p27(Kip1) axis. Mol Carcinog. 2015;54(12):1678–1685. doi:10.1002/mc.22239
73. Alshaer W, Hillaireau H, Vergnaud J, Ismail S, Fattal E. Functionalizing liposomes with anti-CD44 aptamer for selective targeting of cancer cells. Bioconjug Chem. 2015;26(7):1307–1313. doi:10.1021/bc5004313
74. Wu H, Wang M, Dai B, et al. Novel CD123-aptamer-originated targeted drug trains for selectively delivering cytotoxic agent to tumor cells in acute myeloid leukemia theranostics. Drug Deliv. 2017;24(1):1216–1229. doi:10.1080/10717544.2017.1367976
75. Dollins CM, Nair S, Boczkowski D, et al. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem Biol. 2008;15(7):675–682. doi:10.1016/j.chembiol.2008.05.016
76. Pratico ED, Sullenger BA, Nair SK. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013;23(1):35–43. doi:10.1089/nat.2012.0388
77. Wang Y, Yin C, Feng L, Wang C, Sheng G. Ara-C and anti-CD47 antibody combination therapy eliminates acute monocytic leukemia THP-1 cells in vivo and in vitro. Genet Mol Res. 2015;14(2):5630–5641. doi:10.4238/2015.May.25.15
78. Zhao N, Pei SN, Qi J, et al. Oligonucleotide aptamer-drug conjugates for targeted therapy of acute myeloid leukemia. Biomaterials. 2015;67:42–51. doi:10.1016/j.biomaterials.2015.07.025
79. Guo B, Wen B, Cheng W, et al. An enzyme-free and label-free surface plasmon resonance biosensor for ultrasensitive detection of fusion gene based on DNA self-assembly hydrogel with streptavidin encapsulation. Biosens Bioelectron. 2018;112:120–126. doi:10.1016/j.bios.2018.04.027
80. Liu L, Yang K, Gao H, et al. Artificial antibody with site-enhanced multivalent aptamers for specific capture of circulating tumor cells. Anal Chem. 2019;91(4):2591–2594. doi:10.1021/acs.analchem.8b05259
81. Yang M, Jiang G, Li W, et al. Developing aptamer probes for acute myelogenous leukemia detection and surface protein biomarker discovery. J Hematol Oncol. 2014;7:5. doi:10.1186/1756-8722-7-5
82. Lin S, Lu L, Kang TS, et al. Interaction of an Iridium(III) complex with G-Quadruplex DNA and its application in luminescent switch-on detection of siglec-5. Anal Chem. 2016;88(20):10290–10295. doi:10.1021/acs.analchem.6b03128
83. Amouzadeh Tabrizi M, Shamsipur M, Saber R, Sarkar S. Isolation of HL-60 cancer cells from the human serum sample using MnO2-PEI/Ni/Au/aptamer as a novel nanomotor and electrochemical determination of thereof by aptamer/gold nanoparticles-poly(3,4-ethylene dioxythiophene) modified GC electrode. Biosens Bioelectron. 2018;110:141–146. doi:10.1016/j.bios.2018.03.034
84. Zhu P, Wang G, Zhang S, et al. [In vitro selection of single strand deoxyribonucleic acid aptamers binding to cells from patients with acute myeloblastic leukemia]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2012;37(8):771–776. doi:10.3969/j.issn.1672-7347.2012.08.003
85. Leitner M, Poturnayova A, Lamprecht C, et al. Characterization of the specific interaction between the DNA aptamer sgc8c and protein tyrosine kinase-7 receptors at the surface of T-cells by biosensing AFM. Anal Bioanal Chem. 2017;409(11):2767–2776. doi:10.1007/s00216-017-0238-5
86. Bahreyni A, Yazdian-Robati R, Ramezani M, et al. Identification and imaging of leukemia cells using dual-aptamer-functionalized graphene oxide complex. J Biomater Appl. 2017;32(1):74–81. doi:10.1177/0885328217712111
87. Tan J, Lai Z, Zhong L, et al. A graphene oxide-based fluorescent aptasensor for the turn-on detection of CCRF-CEM. Nanoscale Res Lett. 2018;13(1):66. doi:10.1186/s11671-017-2403-3
88. Ye X, Shi H, He X, et al. Cu-Au alloy nanostructures coated with aptamers: a simple, stable and highly effective platform for in vivo cancer theranostics. Nanoscale. 2016;8(4):2260–2267. doi:10.1039/C5NR07017A
89. Tan J, Yang N, Hu Z, et al. Aptamer-functionalized fluorescent silica nanoparticles for highly sensitive detection of leukemia cells. Nanoscale Res Lett. 2016;11(1):298. doi:10.1186/s11671-016-1512-8
90. Ye X, Shi H, He X, et al. Iodide-responsive Cu-Au nanoparticle-based colorimetric platform for ultrasensitive detection of target cancer cells. Anal Chem. 2015;87(14):7141–7147. doi:10.1021/acs.analchem.5b00943
91. Shan W, Pan Y, Fang H, et al. An aptamer-based quartz crystal microbalance biosensor for sensitive and selective detection of leukemia cells using silver-enhanced gold nanoparticle label. Talanta. 2014;126:130–135. doi:10.1016/j.talanta.2014.03.056
92. Khoshfetrat SM, Mehrgardi MA. Amplified detection of leukemia cancer cells using an aptamer-conjugated gold-coated magnetic nanoparticles on a nitrogen-doped graphene modified electrode. Bioelectrochemistry. 2017;114:24–32. doi:10.1016/j.bioelechem.2016.12.001
93. Pan Y, Guo M, Nie Z, et al. Selective collection and detection of leukemia cells on a magnet-quartz crystal microbalance system using aptamer-conjugated magnetic beads. Biosens Bioelectron. 2010;25(7):1609–1614. doi:10.1016/j.bios.2009.11.022
94. Li X, Zhou B, Zhao Z, et al. A smart detection system based on specific magnetic and rolling cycle amplification signal-amplified dual-aptamers to accurately monitor minimal residual diseases in patients with T-ALL. J Biomed Nanotechnol. 2016;12(12):2151–2160. doi:10.1166/jbn.2016.2302
95. Wu S, Yang N, Zhong L, et al. A novel label-free terbium(iii)-aptamer based aptasensor for ultrasensitive and highly specific detection of acute lymphoma leukemia cells. Analyst. 2019;144(12):3843–3852. doi:10.1039/C8AN02342E
96. Liu J, Cui M, Niu L, Zhou H, Zhang S. Enhanced peroxidase-like properties of graphene-hemin-composite decorated with Au nanoflowers as electrochemical aptamer biosensor for the detection of K562 leukemia cancer cells. Chemistry. 2016;22(50):18001–18008. doi:10.1002/chem.201604354
97. Zhang M, Liu H, Chen L, et al. A disposable electrochemiluminescence device for ultrasensitive monitoring of K562 leukemia cells based on aptamers and ZnO@carbon quantum dots. Biosens Bioelectron. 2013;49:79–85. doi:10.1016/j.bios.2013.05.003
98. Moccia F, Platella C, Musumeci D, et al. The role of G-quadruplex structures of LIGS-generated aptamers R1.2 and R1.3 in IgM specific recognition. Int J Biol Macromol. 2019;133:839–849. doi:10.1016/j.ijbiomac.2019.04.141
99. Han X, Jiang Y, Li S, et al. Multivalent aptamer-modified tetrahedral DNA nanocage demonstrates high selectivity and safety for anti-tumor therapy. Nanoscale. 2018;11(1):339–347. doi:10.1039/C8NR05546G
100. Zhang Z, Ali MM, Eckert MA, et al. A polyvalent aptamer system for targeted drug delivery. Biomaterials. 2013;34(37):9728–9735. doi:10.1016/j.biomaterials.2013.08.079
101. Loo JF, Lau PM, Kong SK, Ho HP. An assay using localized surface plasmon resonance and gold nanorods functionalized with aptamers to sense the cytochrome-c released from apoptotic cancer cells for anti-cancer drug effect determination. Micromachines (Basel). 2017;8:11.
102. Scolnik MP, Morilla R, de Bracco MM, Catovsky D, Matutes E. CD34 and CD117 are overexpressed in AML and may be valuable to detect minimal residual disease. Leuk Res. 2002;26(7):615–619. doi:10.1016/S0145-2126(01)00182-5
103. Yang C, Wang Y, Ge MH, et al. Rapid identification of specific DNA aptamers precisely targeting CD33 positive leukemia cells through a paired cell-based approach. Biomater Sci. 2019;7(3):938–950. doi:10.1039/C8BM01393D
104. Mongelard F, Bouvet P. AS-1411, a guanosine-rich oligonucleotide aptamer targeting nucleolin for the potential treatment of cancer, including acute myeloid leukemia. Curr Opin Mol Ther. 2010;12(1):107–114.
105. Amano R, Takada K, Tanaka Y, et al. Kinetic and thermodynamic analyses of interaction between a high-affinity RNA aptamer and its target protein. Biochemistry. 2016;55(45):6221–6229. doi:10.1021/acs.biochem.6b00748
106. Fukunaga J, Nomura Y, Tanaka Y, et al. The Runt domain of AML1 (RUNX1) binds a sequence-conserved RNA motif that mimics a DNA element. RNA. 2013;19(7):927–936. doi:10.1261/rna.037879.112
107. Takada K, Amano R, Nomura Y, et al. Characterisation of an aptamer against the Runt domain of AML1 (RUNX1) by NMR and mutational analyses. FEBS Open Bio. 2018;8(2):264–270. doi:10.1002/feb4.2018.8.issue-2
108. Kadioglu O, Efferth T. Peptide aptamer identified by molecular docking targeting translationally controlled tumor protein in leukemia cells. Invest New Drugs. 2016;34(4):515–521. doi:10.1007/s10637-016-0339-6
109. Taghdisi SM, Abnous K, Mosaffa F, Behravan J. Targeted delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer. J Drug Target. 2010;18(4):277–281. doi:10.3109/10611860903434050
110. Danesh NM, Lavaee P, Ramezani M, Abnous K, Taghdisi SM. Targeted and controlled release delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer-modified gold nanoparticles. Int J Pharm. 2015;489(1–2):311–317. doi:10.1016/j.ijpharm.2015.04.072
111. Cabuzu D, Cirja A, Puiu R, Grumezescu AM. Biomedical applications of gold nanoparticles. Curr Top Med Chem. 2015;15(16):1605–1613. doi:10.2174/1568026615666150414144750
112. Duan S, Yu Y, Lai C, et al. Vincristine-loaded and sgc8-modified liposome as a potential targeted drug delivery system for treating acute lymphoblastic leukemia. J Biomed Nanotechnol. 2018;14(5):910–921. doi:10.1166/jbn.2018.2530
113. Liu M, Ma W, Li Q, et al. Aptamer-targeted DNA nanostructures with doxorubicin to treat protein tyrosine kinase 7-positive tumours. Cell Prolif. 2019;52(1):e12511. doi:10.1111/cpr.2019.52.issue-1
114. Niu W, Chen X, Tan W, Veige AS. N-Heterocyclic Carbene-Gold(I) complexes conjugated to a leukemia-specific DNA aptamer for targeted drug delivery. Angew Chem Int Ed Engl. 2016;55(31):8889–8893. doi:10.1002/anie.201602702
115. Joo EJ, Wasik BR, Parrish C, et al. Pre-B acute lymphoblastic leukemia expresses cell surface nucleolin as a 9-O-acetylated sialoglycoprotein. Sci Rep. 2018;8(1):17174. doi:10.1038/s41598-018-33873-2
116. Kotula JW, Sun J, Li M, et al. Targeted disruption of beta-arrestin 2-mediated signaling pathways by aptamer chimeras leads to inhibition of leukemic cell growth. PLoS One. 2014;9(4):e93441. doi:10.1371/journal.pone.0093441
117. Kalatskaya I, Berchiche YA, Gravel S, et al. AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol Pharmacol. 2009;75(5):1240–1247. doi:10.1124/mol.108.053389
118. Weisberg EL, Sattler M, Azab AK, et al. Inhibition of SDF-1-induced migration of oncogene-driven myeloid leukemia by the L-RNA aptamer (Spiegelmer), NOX-A12, and potentiation of tyrosine kinase inhibition. Oncotarget. 2017;8(66):109973–109984. doi:10.18632/oncotarget.v8i66
119. Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122(7):1293–1304. doi:10.1182/blood-2013-05-501072
120. Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32(30):3552–3558. doi:10.1038/onc.2012.391
121. Norouzi A, Ravan H, Mohammadi A, et al. Aptamer-integrated DNA nanoassembly: a simple and sensitive DNA framework to detect cancer cells. Anal Chim Acta. 2018;1017:26–33. doi:10.1016/j.aca.2018.02.037
122. Huang L, Wang DB, Singh N, et al. A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale. 2018;10(43):20289–20295. doi:10.1039/C8NR07720G
123. Zhang M, Zhou F, Zhou D, et al. An aptamer biosensor for leukemia marker mRNA detection based on polymerase-assisted signal amplification and aggregation of illuminator. Anal Bioanal Chem. 2019;411(1):139–146. doi:10.1007/s00216-018-1424-9
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.