Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
NF1 Gene Novel Splicing Mutations in a Chinese Family with Neurofibromatosis Type 1: Case Series
Authors Wu T, Yang H, Xu L, Huang Q, He Q, Wu R, Mu YZ
Received 30 August 2022
Accepted for publication 19 October 2022
Published 31 October 2022 Volume 2022:15 Pages 2345—2351
DOI https://doi.org/10.2147/CCID.S388045
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Ting Wu,1,* Hao Yang,1,* Liuli Xu,1 Qing Huang,1 Qi He,1 Rong Wu,2 Yun-Zhu Mu1
1Department of Dermatology, the Affiliated Hospital of North Sichuan Medical College, Nanchong, People’s Republic of China; 2Pediatric department, Women’s and Children’s hospital of GaoPing District, Nanchong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yun-Zhu Mu, Department of Dermatology, the Affiliated Hospital of North Sichuan Medical College, No. 1 Maoyuan South Road, Shunqing District, Nanchong, Sichuan Province, 63700, People’s Republic of China, Tel +8615984833231, Email [email protected]
Background: Neurofibromatosis type 1 (NF1) is a common autosomal dominant genetic disorder. NF1 is a multisystemic disease and its pathogenesis involves mutations in the NF1 gene on chromosome 17q11.2 causing RAS overactivation to stimulate abnormal cell proliferation.
Purpose: To identify pathogenic mutation of the NF1 gene in a pedigree of NF1.
Patients and Methods: Collection of clinical data from one NF1 family. Peripheral blood samples were collected from the affected persons and their family members. Potential mutations of NF1 gene were screened by exome and cDNA sequencing.
Results: A splice mutation (c.4836– 10T>G) was found in exon 37 of the NF1 gene in this NFI family, and no corresponding mutation was found in healthy members of this pedigree or in the human reference genome (GRCh37/hg19).
Conclusion: Mutations of NF1 gene is a major cause of NF1. The novel splice mutation in exon 37 of NF1 gene is the underlying cause of the familial c.4836– 10T>G.
Keywords: neurofibromatosis type 1, gene, splicing mutation, sequencing
Introduction
A frequent autosomal dominant genetic condition with a frequency of 1 in 2000–4000 people is neurofibromatosis type 1.1 Café-au-lait macules (CALMs), multiple neurofibromas, axillary or inguinal freckling, iris Lisch nodules, skeleton anomalies, learning disabilities, and predisposition to malignancies are some of its clinical characteristics.2 The human NF1 gene, which spans 350 kb of DNA across 60 exons, is found on chromosome 17q11.2.3 As a negative feedback regulator of RAS signaling pathway, NF1 gene is responsible for encoding neurofibromin, which has 2818 amino acids.4,5 Under typical circumstances, neurofibromin encourages the conversion of RAS into its inactivation state, reducing cell development.6 The loss of nerve fiber protein function caused by NF1 gene mutation and functional problem results in RAS overactivation, which in turn stimulates cell proliferation and causes a variety of cancers to appear.7 NF1 has a 5–15% greater risk of malignant tumor transformation than the normal population.7 So far, >3000 different NF1 gene mutations have been identified in the human gene mutation database. In conclusion, by Exome and Sanger sequencing, we identified a novel causative NF1 mutation (c.4836–10T>G) in a Chinese family with NF1. Our research broadens the range of NF1 mutations and might help with NF1 genetic diagnostics and counseling.
Materials and Methods
Subjects
A Chinese NF1 family with autosomal dominant inheritance participated in the study. All participants provided written informed permission. The Affiliated Hospital of North Sichuan Medical College also gave its approval for this investigation. Four members (Figure 1) from the family were enrolled and performed complete dermatological and physical examination (Table 1). The National Institutes of Health’s consensus criteria were used to make the neurofibromatosis diagnosis.
 |
Table 1 Clinical Features of Affected Individuals in Family |
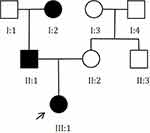 |
Figure 1 Pedigree of the Chinese family with neurofibromatosis type 1. |
DNA Extraction, RNA Extraction and cDNA Synthesis
5mL of peripheral blood was collected, placed in EDTA dipotassium anticoagulation tubes, and genomic DNA was isolated from peripheral white blood cells using the AxyPrep DNA Blood Mini Kit. RNA was isolated from the peripheral blood samples using TRIzol. First-strand cDNA was reverse-transcribed using the PrimeScript TM II 1st Strand cDNA Synthesis Kit.
Whole Exome Sequencing
The DNA was randomly broken into fragments of 150–250bp in length by Covaris ultrasonic fragmentation, and the DNA library was prepared by ligating Y-junctions on both ends of the fragments after end repair and addition of A-tails. DNA libraries pooled with different index markers were hybridized with biotin-labeled probes in liquid phase, and then all exons of the genes to be tested were captured by magnetic beads coated with streptavidin, amplified linearly by PCR and subjected to library quality control. Qualified DNA libraries were sequenced by Novaseq-6000 high-throughput sequencer with 100.00% coverage of target regions, average depth of 100× or more, and 90% of the average depth >30× loci. Primers were designed using primer blast based on high-throughput sequencing results. The candidate mutation was confirmed with Sanger sequencing using the following primers: Forward primers: 5’‐AGCCTTATTTCTCAGTGTCC‐3’; Reverse primers: 5’‐CGTGACATTTTATACACCAC‐3’. The reference sequence NM_001042492 of NF1 mRNA was used.
Sequencing Data Processing and Mutation Calling
The sequencing data were aligned to the human reference genome (GRCh37/hg19) using BWA, Samtools and Picard software. The generated bam files were locally re-matched using GATK series software to remove duplicate sequences and detect variants, and Annovar was used to annotate the vcf variant files with variants. After that, the pathogenic variations are checked for.
cDNA Sequencing
Using the father of the proband’s cDNA as a template, the NF1 gene’s coding area was amplified, and the PCR result was then directly sequenced by Sanger. The following were the amplification primer sequences: Forward primers: 5′-CAACACTTCTTGCATACCTG-3′, Reverse primers: 5′-GTTACTTGGACAGCAGTAGA-3′. The reference sequence NM_001042492 of NF1 mRNA was used.
Results
Clinical Characteristics
Figure 2 illustrates the clinical picture of three patients. At 10 days after delivery of the proband, it was discovered that the 46-days-old female kid of the prefactor had several CALMs dispersed across the extremities, back, and buttocks, ranging in size from 1 to 5 cm, with more than 6 places. II:1, a 31-year-old male patient, first saw CALMs on his back when he was 10 years old. As he aged, the spots grew larger, and over time, CALMs of various sizes, skin freckling and subcutaneous nodules spread throughout the body. After the nodule was removed in 2015, he underwent histology that was indicative of cutaneous neurofibroma. I:2, a 53-years-old female, presented with cutaneous neurofibroma at the age of 10 years, and now multiple nodules with diameters ranging from 0.3 to 5 cm were seen throughout the body, with the largest located on the left knee, several CALMs of 0.5 to 3 cm in size on the trunk, and axillary and abdominal freckles. II:2, a 29-year-old female who was examined as the mother of a prior witness were well with no features of neurofibromatosis.
Whole Exome Sequencing
The results of mutation analysis by exome sequencing are shown in Figure 3. A splice mutation (c.4836–10T>G) in exon 37 of the NF1 gene was found. The Scsnv program projected that this mutation would have a high likelihood of altering RNA splicing (0.9952) and this mutation has a frequency of 0 in ExAC(EAS). Besides, the transcriptional study of this splice mutation was novel since it had not been previously reported nor was it present in SingleNucleotide Polymorphism(dbSNP), Online Mendelian Inheritance in Man (OMIM), The Human Gene Mutation Database (HGMD), or ClinVar.
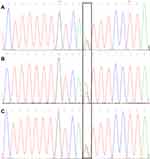 |
Figure 3 (A) the proband (III:1). (B) The proband’s father (II:1). (C) The proband’s grandmother (I:2). Rectangular box: Shown is the wildtype T allele with a small peak for the variant allele G. |
cDNA Sequencing
The results of the cDNA sequencing suggest that this mutation causes a 9bp shift forward in the splice site at the 5’ end of exon 37 (Figure 4). As a result, three amino acids (serine, phenylalanine, and leucine) are inserted in front of amino acid position 1612, adding 3 amino acids in the Sec14-PH domain of neurofibromin and lengthening the neurofibromin encoded by NF1 from 2839 amino acids to 2842 amino acids.
 |
Figure 4 (A) negative control. (B) The proband’s father (II:1). Rectangular box: 5’ end of exon 37 shifted 9bp forward to 3’ end in c.4836. |
Follow-Up
No growth of CALMs discoloration, no abnormalities in ophthalmologic examination, normal growth and development, and no abnormalities in the spine were observed in the proband after 1 month. Due to the short follow-up period, no other systemic damage or neurofibroma was observed for the time being.
Discussion
Significant advances in understanding the etiology of NF1 have been attributed to the discovery of the NF1 gene, one of the most mutated genes in the human.8 Different types of mutations have been identified throughout the NF1 gene, including chromosomal abnormalities, insertions, point mutations, deletions, stop mutations, 3’-untranslated region mutations and splicing mutations, causing the NF1 phenotype.9 In this report, a novel splice mutation (c.4836–10T>G) was found in exon 37 of the NF1 gene in this NFI family. The splice site at the 5’ end of exon 37 is moved forward 9bp as the result of this mutation, resulting in the insertion of 3 amino acids. Three amino acids are added to the Sec14-PH structural domain of neurofibromin. The variant of the NF1 gene was predicted to be likely pathogenic according to American College of Medical Genetics and Genomics (ACMG). Up to 25.2% of NF1 patients have mutations at the NF1 splice site, and these mutations frequently result in the production of a shortened version of neurofibromin.10 In this instance, a longer neurofibromin family has developed. The GAP-related domain (GRD) and the Sec14-PH domain are two structurally identified core domains found in neurofibromin. Ras can bind to GRD, which also has a characteristic arginine finger that facilitates GTP hydrolysis in Ras’s active site.11 Sec14-PH has two subdomains: Sec14 module and pleckstrin homology (PH) module, respectively. Since the Sec14-PH domain may bind phosphoinositide lipids via its PH domain and glycerophospholipids via its Sec14 module, it is possible that lipid binding plays a direct role in neurofibromin activation or membrane recruitment.12 Additionally, it has been demonstrated that Sec14-PH interacts with endogenous opioid μ- (MORs) receptors activated G subunits to decrease NF1 RasGAP function both in vitro and in vivo.13 Therefore, the author hypothesizes that this mutation, which changed modest local changes in the Sec14-PH domain by adding three amino acids, may have affected RasGAP activity. However, before coming to any conclusion, more research is needed to investigate information on the structural features of Sec14-PH.
In contrast to individuals carrying other mutation types, such as microinsertions, microdeletions, nonsense and missense variants, NF1 patients with NF1 splice-site mutations were found to have a higher propensity to form malignant peripheral nerve sheath tumors (MPNSTs) and CNS gliomas.7 Predicting which patients will gain the most from routine clinical surveillance may be possible with its aid. Since it was established that none of the three affected family members had brain tumors, follow-up and health education (such as regular physicals, etc.) can continue for this NF1 family in future.
Unfortunately, attempts to link certain clinical characteristics of NF1 with the numerous and diverse germ line NF1 gene mutations are hard or even failing.14 Therefore, it may be challenging to carry out any surveillance choices that are based on any of these potentially curable problems in the clinic.15,16 Through a multi-institutional effort, a discovery demonstrates genotype-phenotype associations at the NF1 codons p.Met1149, p.Arg1276, and p.Lys1423. This shows that the only genotype-phenotype correlations that are clinically applicable may be established in large datasets of postpubertal individuals who have the same constitutional pathogenic variation and whose phenotype is reported in a consistent manner.17
The phenotypic variety of NF1 has previously been reported to occur within and between families, and suggested causes include genetic background differences, putative modifier variables, mosaicism in afflicted patients, environmental factors, and stochastic factors.18 NF1 phenotypes would develop as people aged.19 In the case of the proband, just CALMs are visible, whereas the afflicted family members also exhibit skin freckle-like alterations and cutaneous neurofibroma symptoms in addition to CALMs. We also hypothesize that other clinical symptoms linked to NF1 may manifest in the proband in the future. As a result, even in children with multiple (more than 6 places) CALMs, we should be on the lookout for NF1. We should also ask the child’s parents in-depth questions about any additional clinical signs of NF and, if necessary, perform whole exome sequencing to rule out any potential genetic mutations.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
This study follows the Helsinki Declaration and the research program was approved by the Affiliated Hospital of North Sichuan Medical College (approval number:2022ER339-1). Authors confirm that all family members provided informed consent for publication of the case details and images, including informed consent from the parents to cover the infant.
Consent for Publication
All authors have read and approved the final manuscript for submission.
Acknowledgments
We thank the patients and their family members for participating in our study.
Funding
Basic research project of the Affiliated Hospital of North Sichuan Medical College, Grant/Award Number:2022JC007.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jordan JT, Plotkin SR. Neurofibromatoses. Hematol Oncol Clin North Am. 2022;36(1):253–267. doi:10.1016/j.hoc.2021.08.010
2. Legius E, Messiaen L, Wolkenstein P, et al. Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet Med. 2021;23(8):1506–1513. doi:10.1038/s41436-021-01170-5
3. Fountain JW, Wallace MR, Brereton AM, et al. Physical mapping of the von Recklinghausen neurofibromatosis region on chromosome17. Am J Hum Genet. 1989;44(1):58–67.
4. Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi:10.1038/nrc3911
5. Hinman MN, Sharma A, Luo G, Lou H. Neurofibromatosis type 1 alternative splicing is a key regulator of Ras signaling in neurons. Mol Cell Biol. 2014;34(12):2188–2197. doi:10.1128/MCB.00019-14
6. Martin GA, Viskoohil D, Bollag G, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63(4):843–849. doi:10.1016/0092-8674(90)90150-D
7. Alkindy A, Chuzhanova N, Kini U, Cooper DN, Upadhyaya M. Genotype-phenotype associations in neurofibromatosis type 1 (NF1): an increased risk of tumor complications in patients with NF1 splice-site mutations? Hum Genomics. 2012;6(1):12. doi:10.1186/1479-7364-6-12
8. Koczkowska M, Chen Y, Callens T, et al. Genotype-phenotype correlation in NF1: evidence for a more severe phenotype associated with missense mutations affecting NF1 codons 844-848. Am J Hum Genet. 2018;102(1):69–87. doi:10.1016/j.ajhg.2017.12.001
9. Messiaen LM, Callens T, Mortier G, et al. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15(6):541–555. doi:10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N
10. Xu W, Yang X, Hu X, Li S. Fifty-four novel mutations in the NF1 gene and integrated analyses of the mutations that modulate splicing. Int J Mol Med. 2014;34(1):53–60. doi:10.3892/ijmm.2014.1756
11. Rabara D, Tran TH, Dharmaiah S, et al. KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis. Proc Natl Acad Sci U S A. 2019;116(44):22122–22131. doi:10.1073/pnas.1908353116
12. Chaker-Margot M, Werten S, Dunzendorfer-Matt T, et al. Structural basis of activation of the tumor suppressor protein neurofibromin. Mol Cell. 2022;82(7):1288–1296. doi:10.1016/j.molcel.2022.03.011
13. Xie K, Colgan LA, Dao MT, et al. NF1 Is a Direct G Protein Effector Essential for Opioid Signaling to Ras in the Striatum. Curr Biol. 2016;26(22):2992–3003. doi:10.1016/j.cub.2016.09.010
14. Castle B, Baser M, Huson SM, Cooper DN, Upadhyaya M. Evaluation of genotype-phenotype correlations in neurofibromatosis type 1. J Med Genet. 2003;40(10):e109. doi:10.1136/jmg.40.10.e109
15. Upadhyaya M. Neurofibromatosis type 1: diagnosis and recent advances. Exp Opin Med Diagn. 2010;4(4):307–322. doi:10.1517/17530059.2010.494660
16. Ferner RE, Huson SM, Thomas N, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44(2):81–88. doi:10.1136/jmg.2006.045906
17. Koczkowska M, Callens T, Chen Y, et al. Clinical spectrum of individuals with pathogenic NF1 missense variants affecting p.Met1149, p.Arg1276, and p.Lys1423: genotype-phenotype study in neurofibromatosis type 1. Hum Mutat. 2020;41(1):299–315. doi:10.1002/humu.23929
18. Easton DF, Ponder MA, Huson SM, Ponder BA. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): evidence for modifying genes. Am J Hum Genet. 1993;53(2):305–513.
19. Banerjee S, Lei D, Liang S, et al. Novel phenotypes of NF1 patients from unrelated Chinese families with tibial pseudarthrosis and anemia. Oncotarget. 2017;8(24):39695–39702. doi:10.18632/oncotarget.13932
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

