Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Network Pharmacology Revealed the Mechanisms of Action of Lithospermum erythrorhizon Sieb on Atopic Dermatitis
Authors Wang T, You W, Zhao L, Zhang B, Wang H
Received 5 January 2023
Accepted for publication 7 March 2023
Published 13 March 2023 Volume 2023:16 Pages 651—658
DOI https://doi.org/10.2147/CCID.S403736
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Jeffrey Weinberg
Tianyi Wang,1,* Wang You,2,* Linna Zhao,3 Bingxin Zhang,1 Hongmei Wang4
1Department of Dermatology, First Teaching Hospital of Tianjin University of TCM, National Clinical Research Center for Chinese Medicine Acupuncture and Moxibustion, Tianjin, People’s Republic of China; 2Department of Internal Medicine, Hexi Hospital of TCM, Tianjin, People’s Republic of China; 3Department of Experimental Center, First Teaching Hospital of Tianjin University of TCM, National Clinical Research Center for Chinese Medicine Acupuncture and Moxibustion, Tianjin, People’s Republic of China; 4Department of Dermatology, Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital, Tianjin, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tianyi Wang, Department of Dermatology, First teaching hospital of Tianjin University of TCM, National Clinical Research Center for Chinese Medicine Acupuncture and Moxibustion, Tianjin, 300300, People’s Republic of China, Email [email protected]
Aim: The application of network analysis algorithms promoted the development of network pharmacology. This study aimed to combine network pharmacology and signed random walk with restart (SRWR) to reveal the mechanism by which Lithospermum erythrorhizon Sieb (LES) exerts effects on atopic dermatitis (AD).
Methods: The compounds and targets of LES were retrieved from Traditional Chinese Medicine Integrated Database (TCMID) and Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), and important compounds and targets were identified by intersection analysis and protein–protein interaction (PPI) network.
Results: We found that active LES-derived compounds such as caffeic acid, Isovaleric acid, Arnebinol, and Alannan may inhibit PTGS2, HSP90AA1 and MAPK14, which are key mediators involved in PI3K-Akt pathway, vascular endothelial growth factor signaling pathway, Fc epsilon RI signaling pathway, and calcium signaling pathway.
Conclusion: The application of SRWR could identify potential targets of LES with a low false-positive rate and help elucidate the mechanism of action of traditional Chinese medicine.
Keywords: signed random walk with restart, network pharmacology, Lithospermum erythrorhizon Sieb, atopic dermatitis
Introduction
Network pharmacology is a novel method to explore the interaction between active components and potential targets of drugs.1 In addition, network pharmacology integrates the interaction into a network and analyzes the action of traditional Chinese medicine (TCM) from a systematic perspective.2 Meanwhile, some problems such as the difficulty of data integration for network pharmacology remain to be overcome. The signed random walk with restart (SRWR) has been developed to simulate the propagation of activation or inhibition effects of the drug on the signaling network.3,4
Atopic dermatitis (AD) is a chronic inflammatory skin disease that affects around 200 million people in the world. Although AD is more common in children (incidence of AD is about 20% in children and 10% in adults), approximately 1 in 4 adults with AD have adult-onset disease.5 Recent investigations have revealed the mechanisms that contribute to the pathogenesis of AD, such as the role of aryl hydrocarbon receptor.6 Lithospermum erythrorhizon Sieb (LES) has been used to treat AD. Naphthoquinone derivatives shikonin, acetylshikonin, and isobutylshikonin are widely present in the epidermis of the roots of LES.7 Wang et al showed that shikonin reduced inflammatory cell infiltration and collagen deposition by inhibiting NF-κB pathway and suppressing the expression of MMP9.8 However, the potential mechanisms of anti-inflammatory and anti-allergy activities of shikonin derivatives against AD have not been completely elucidated. Therefore, this study aimed to combine network pharmacology and signed random walk with restart (SRWR) to reveal the mechanism by which LES exerts effects on AD.
Methods
Chemical Compounds and Targets of LES
The compounds and targets of LES were retrieved from Traditional Chinese Medicine Integrated Database (TCMID)9 and Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP).10 The bioactive components were selected based on the absorption, distribution, metabolism and excretion (ADME).11 The candidate components were determined based on two parameters: (1) Pharmacokinetics GI absorption, (2) Drug-likeness (DL): the Lipinski rule of five and the Veber Ghose, Muegge and Egan rules, Compounds that meet more than two of Lipinski’s rules will be chosen.12 Eventually, 60 compounds were retrieved.
AD Related Genes
AD-related genes were screened from the DisGeNET13 and GEO (Gene Expression Omnibus) (GSE32924, GSE36842, GSE107361 and GSE130588, GSE22255).14 A total 410 differentially expressed genes were identified, including 162 up-regulated and 250 downregulated genes in AD patients.
FDA Approved Drugs and Their Targets
The FDA-approved drugs for the treatment of AD were used as positive controls and their targets were retrieved from the Drugbank database.15
Construction of Signed Human Genomic Signal Network
A KEGG signaling network was established based on KEGG database. The correlation scores between genes were obtained from STRING database, and these data were then integrated to construct a network. Active or inhibitory relationships were presented as either positive or negative links.
The network is a weighted directed network with the weight of directed edges ranging from +1 to −1. The constructed network has 3892 nodes and 28,459 directed links.
Simulation of the Impact of a Drug by SRWR
SRWR allowed the restart of the walk in every step at the node with probability. SRWR algorithm was used to measure how the activation or inhibition of a seed node led to the activation or inhibition of other nodes in the human signaling network.16 The equation for the SRWR was defined as:
where r was the restart probability, W was the column-normalized adjacency matrix of the network graph, and 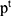 was a vector of size equal to the number of nodes in the graph where the i -element had the probability of being at node i at time step t. The initial probability vector
was a vector of size equal to the number of nodes in the graph where the i -element had the probability of being at node i at time step t. The initial probability vector 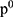 was constructed such that equal probabilities were assigned to the nodes representing the members of the disease, with the sum of the probabilities equal to 1.
was constructed such that equal probabilities were assigned to the nodes representing the members of the disease, with the sum of the probabilities equal to 1.
Results
Identification of Potential Targets of LES by Intersection Analysis
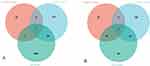
We collected two disease gene sets and one drug target set associated with AD as reference and investigated their overlap. We found that 69 of the 751 AD disease genes in DisGeNET were differentially expressed in AD patients, while the target genes for anti-AD drugs in Drugbank dataset had 11 genes in DisGeNET. Notably, the three datasets had no common genes (Figure 1A). Figure 1B shows the overlap of target genes of LES with AD disease genes and target genes for anti-AD drugs in DrugBank database, and PTGS2, NR3C1 and SLC6A4 were the overlapped genes. According to the data from TCMSP, PTGS2 was regulated by more than 10 compounds such as caffeic acid, (2R)-3-oxo-2-phenylbutanenitrile, acetylshikonin, isobutyryl shikonin, deoxyshikonin, palmitic acid, arnebinol, alpha-methyl-n-butylshikonin, alkannan, lithospermidin A, Caffeate, arnebinone, isovanillin, β-acetoxyisovalery lshikonin, lithospermidin B, shikonofuran B.
PPI Network Analysis of the Targets of LES
We constructed a compound–compound target-AD target PPI network, including 77 nodes (23 compounds, 37 intersection targets between compound targets and AD targets, and 17 AD targets) and 441 edges (Figure 2). Nine nodes with an average value of degree ≥25 could be considered as major nodes, including Interleukin-6 (IL-6), PTGS2, nuclear receptor subfamily 3, group C (NR3C1), estrogen receptor 1 (ESR1), interleukin-1β (IL-1β), androgen receptor (AR), interleukin-10 (IL-10), interleukin-4 (IL-4) and heat shock protein 90 alpha family class A member 1 (HSP90AA1).
Prediction of Genes Significantly Inhibited and Activated by LES
Anti-AD drugs from DrugBank were used to validate the effects of the drugs. There were 36 targets of DrugBank anti-AD drugs, and they were divided into three groups based on the corresponding drugs (Table 1). AD-related disease genes extracted from GEO datasets were used as controls. In Table 1, we listed the genes in common between the top genes of each compound and the three validation gene datasets. We found that most of genes in the validation sets were inhibited by some compounds, especially Acetylshikonin, Deoxyshikonin, Isobutyrylshikonin, Alkannan, Caffeic acid. These results suggest that LES exerts anti-AD effects by inhibiting the genes associated with AD.
 |
Table 1 Genes Significantly Inhibited by LES |
KEGG Pathway Enrichment Analyses
Pathway enrichment analysis showed that 107 KEGG pathways were significantly enriched with inhibited genes corresponding to LES targets (p<0.05), including all three anti-AD associated pathways. Similarly, 13 KEGG pathways were significantly enriched with anti-AD drugs (Figure 3), including metabolism pathways (Tyrosine metabolism, Tryptophan metabolism, Sphingolipid signaling pathway, Arginine and proline pathway), Cancer pathways (Small cell lung cancer, Prostate cancer, Chemical carcinogenesis), PI3K-Art pathway, Neuroactive ligand–receptor interaction, Fc epsilon RI pathway, VEGF pathway (Table 2).
 |
Table 2 Pathways Associated with Major Targets of LES |
 |
Figure 3 KEGG analysis of major targets of LES. X-axis showed the Rich factor (FDR<0.01), and Y-axis showed significantly enriched KEGG pathway of the targets. |
Notably, 3 signaling pathways were associated with cancer, indicating that LES has the potential to treat diverse cancers such as lung cancer and prostate cancer as reported previously.17–19
Phosphoinositide 3-kinase (PI3K) acts as a key signaling protein in T cell activation, proliferation, and migration. The major components of LES may inhibit IL-6 and IL-10 through PI3K/Akt pathway. Therefore, we hypothesized that PIK3-Akt pathway may mediate the effects of LES on AD. Overproduction of VEGF in AD lesions has been known as a potential marker of AD severity.20 Major compounds (Caffeic acid, Alkannan, Arnebinone) may inhibit VEGF signaling pathways through MAPK14 and PTGS2.
FcεRI is the high-affinity receptor of IgE.21 Omalizumab, cyclic peptides, and some small proteins of FcεRI signaling pathway are drug candidates against AD In this study, Caffeic acid and Alkannan have the potential to be candidate natural drugs to bind PRKCB, BTK, RAC1, and MAPK14 to inhibit FcεRI signaling pathway. Inflammation suppressing role of tacrolimus could be related to its effects of inhibiting T cell activation and blocking calcineurin signaling.22,23 ORAI1/NFAT-calcium pathway may regulate TSLP in AD.24 Combined with the PPI network, the results indicate that Arnebinol, Caffeic acid, Alkannan, Lobelanidine may inhibit TSLP through ORAI1/NFAT-calcium pathway.
Discussion
Using bioinformatics approach, we systematically investigated anti-AD mechanism of LES. We retrieved 23 active compounds from LES and predicted 99 targets. Network analysis showed that LES exerted effects on AD by regulating such targets as IL-6, PTGS2, ESR1, IL-1β, AR, IL-10, HSP90AA1 and IL-4. Analyses based on GEO database and DrugBank suggest that the active LES-derived compounds caffeic acid, Isovaleric acid, Arnebinol, Alannan may inhibit PTGS2, HSP90AA1, MAPK14, which are key mediators involved in anti-inflammation and anti-histamine pathways.
Target-pathway network analysis of potential LES targets showed that LES could modulate several signaling pathways implicated in the pathogenesis of AD, including PI3K-Akt pathway, VEGF signaling pathway, Fc epsilon RI signaling pathway and calcium signaling pathway. These pathways crosstalk to regulate inflammation, immunity, and allergy. Our results suggest that LES may exert anti-AD effects by inhibiting the proteins involved in each of the four pathways.
This study employed a computational pharmacology approach to elucidate the mechanisms of action of LES. In view of the interactions between proteins/genes, the impact of a natural drugs on targets can spread to other proteins/genes through protein–protein interaction networks or downstream signaling pathways. Several propagation-like algorithms such as SRWR have been employed to identify protein targets of drugs.16,25 However, most of these algorithms could not reflect the complete picture of drug-induced activation and/or inhibition on proteins. SRWR algorithm was used to simulate the spread of active compound-induced activation or inhibition on a group of targets on a signed human signaling network. One difficulty in the application of SRWR algorithm is the lack of a PPI network. In this study, we built a new network combining KEGG and STRING databases as a background network to conduct SRWR.
Using SRWR algorithm, we identified the top 5% genes inhibited by LES and predicted the targets of active LES compounds with the highest confidence scores. Using this strategy, we identified LES targets with a low false-positive rate. However, this study has limitations. First, this study is based on online datasets. The detailed mechanisms of action of LES on AD should be investigated by functional studies. Second, the constructed human signaling network may not include all known protein–protein interactions.
In conclusion, we constructed a signed network by combining KEGG and STRING databases and ran SRWR based on the background network. Our results show that active LES-derived compounds, such as caffeic acid, Isovaleric acid, Arnebinol, Alannan may inhibit PTGS2, HSP90AA1, MAPK14, which are key mediators involved in PI3K-Akt pathway, VEGF signaling pathway, Fc epsilon RI signaling pathway and calcium signaling pathway. These compounds may have potential application in clinical treatment of AD.
Data Sharing Statement
All data and material are available upon request.
Funding
This study was supported by Scientific research projects in key fields of Tianjin Health Commission (2022003).
Disclosure
The authors declare that there are no competing interests associated with this work.
References
1. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nature Chem Biol. 2008;4:682–690. doi:10.1038/nchembio.118
2. Ruchawapol C, Fu WW, Xu HX. A review on computational approaches that support the researches on traditional Chinese medicines (TCM) against COVID-19. Phytomedicine. 2022;104:154324. doi:10.1016/j.phymed.2022.154324
3. Zhao J, Lv C, Wu Q, et al. Computational systems pharmacology reveals an antiplatelet and neuroprotective mechanism of Deng-Zhan-Xi-Xin injection in the treatment of ischemic stroke. Pharmacol Res. 2019;147:104365. doi:10.1016/j.phrs.2019.104365
4. Zhao H, Guo J, Chi Q, Fang M. Molecular mechanisms of tanshinone IIA in hepatocellular carcinoma therapy via WGCNA-based network pharmacology analysis. Biocell. 2022;46(5):1245–1259. doi:10.32604/biocell.2022.018117
5. Chovatiya R. Atopic dermatitis (eczema). JAMA. 2023;329(3):268. doi:10.1001/jama.2022.21457
6. Napolitano M, Fabbrocini G, Martora F, Picone V, Morelli P, Patruno C. Role of aryl hydrocarbon receptor activation in inflammatory chronic skin diseases. Cells. 2021;10(12):3559. doi:10.3390/cells10123559
7. Wu FY, Tang CY, Guo YM, et al. Transcriptome analysis explores genes related to shikonin biosynthesis in Lithospermeae plants and provides insights into Boraginales’ evolutionary history. Sci Rep. 2017;7:4477. doi:10.1038/s41598-017-04750-1
8. Wang TY, Zhou QL, Li M, et al. Shikonin alleviates allergic airway remodeling by inhibiting the ERK-NF-κB signaling pathway. Int Immunopharmacol. 2017;48:169–179. doi:10.1016/j.intimp.2017.05.011
9. Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013;41:D1089–1095. doi:10.1093/nar/gks1100
10. Jinlong R, Peng L, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;16:13.
11. Li B, Rui J, Ding X, Yang X. Exploring the multicomponent synergy mechanism of Banxia Xiexin decoction on irritable bowel syndrome by a systems pharmacology strategy. J Ethnopharmacol. 2019;233:158–168. doi:10.1016/j.jep.2018.12.033
12. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi:10.1016/S0169-409X(00)00129-0
13. Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET Cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960–2967. doi:10.1016/j.csbj.2021.05.015
14. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 2013;39:1005–1010. doi:10.1093/nar/gkq1184
15. Barneh F, Jafari M, Mirzaie M. Updates on drug-target network; facilitating polypharmacology and data integration by growth of DrugBank database. Brief Bioinform. 2016;17(6):1070–1080. doi:10.1093/bib/bbv094
16. Ingoo L, Hojung N. Identification of drug-target interaction by a random walk with restart method on an interactome network. BMC Bioinform. 2018;19(S8):208. doi:10.1186/s12859-018-2199-x
17. Jeung YJ, Kim HG, Ahn J, et al. Shikonin induces apoptosis of lung cancer cells via activation of FOXO3a/EGR1/SIRT1 signaling antagonized by p300. Biochim Biophys Acta. 2016;1863:2584–2593.
18. Chen Y, Zheng L, Liu J, et al. Shikonin inhibits prostate cancer cells metastasis by reducing matrix metalloproteinase-2/-9 expression via AKT/mTOR and ROS/ERK1/2 pathways. Int Immunopharmacol. 2014;21:447–455. doi:10.1016/j.intimp.2014.05.026
19. Huang H, Miao H, Wang J, et al. Advances on anticancer new drugs in China and the USA in 2020: from ongoing trial to drug approval. J Natl Cancer Center. 2021;1(4):147–152. doi:10.1016/j.jncc.2021.08.002
20. Koczy-Baron E, Jochem J, Kasperska-Zajac A. Increased plasma concentration of vascular endothelial growth factor in patients with atopic dermatitis and its relation to disease severity and platelet activation. Inflam Res. 2012;61(12):1405–1409. doi:10.1007/s00011-012-0543-6
21. Smith LD, Leatherbarrow RJ, Spivey AC. Development of small molecules to target the IgE: FcεRI protein-protein interaction in allergies. Future Med Chem. 2013;5(12):1423–1435. doi:10.4155/fmc.13.112
22. Na E, Choi M, Park I, Lim S. Effect of Black Sea bream extracts on cytokine production in lipopolysaccharide-induced inflammation. Biocell. 2020;44(2):93–199. doi:10.32604/biocell.2020.08648
23. Werfel T. Novel systemic drugs in treatment of atopic dermatitis: results from Phase II and Phase III studies published in 2017/2018. Curr Opin Allergy Clin Immunol. 2018;18(5):432–437. doi:10.1097/ACI.0000000000000477
24. Wilson SR, Thé L, Batia LM, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155(2):285–295. doi:10.1016/j.cell.2013.08.057
25. Chen Y, Wu H, Jiao A, et al. Chinese herbal prescription QYSL prevents progression of lung cancer by targeting tumor microenvironment. Oncologie. 2022;24(2):295–307. doi:10.32604/oncologie.2022.022116
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.