Back to Journals » Journal of Asthma and Allergy » Volume 17
Nasal Lavage Fluid Proteomics Reveals Potential Biomarkers of Asthma Associated with Disease Control
Authors Chen M, Ge Y, Zhang W, Wu P, Cao C
Received 24 January 2024
Accepted for publication 3 May 2024
Published 16 May 2024 Volume 2024:17 Pages 449—462
DOI https://doi.org/10.2147/JAA.S461138
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Luis Garcia-Marcos
Meiping Chen,1,2 Yijun Ge,1,3 Wen Zhang,1 Ping Wu,4 Chao Cao1
1Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital of Ningbo University, Ningbo, 315010, People’s Republic of China; 2Department of Respiratory and Critical Care Medicine, the Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, 310009, People’s Republic of China; 3Department of Respiratory and Critical Care Medicine, Ninghai First Hospital, Ningbo, 315600, People’s Republic of China; 4National Facility for Protein in Shanghai, Zhangjiang Lab, Shanghai Advanced Research Institute, CAS, Shanghai, 201210, People’s Republic of China
Correspondence: Chao Cao, The First Affiliated Hospital of Ningbo University, 59 Liuting Road, Ningbo, 315010, People’s Republic of China, Tel +86-574-87089878, Fax +86-0574-8729-1583, Email [email protected]
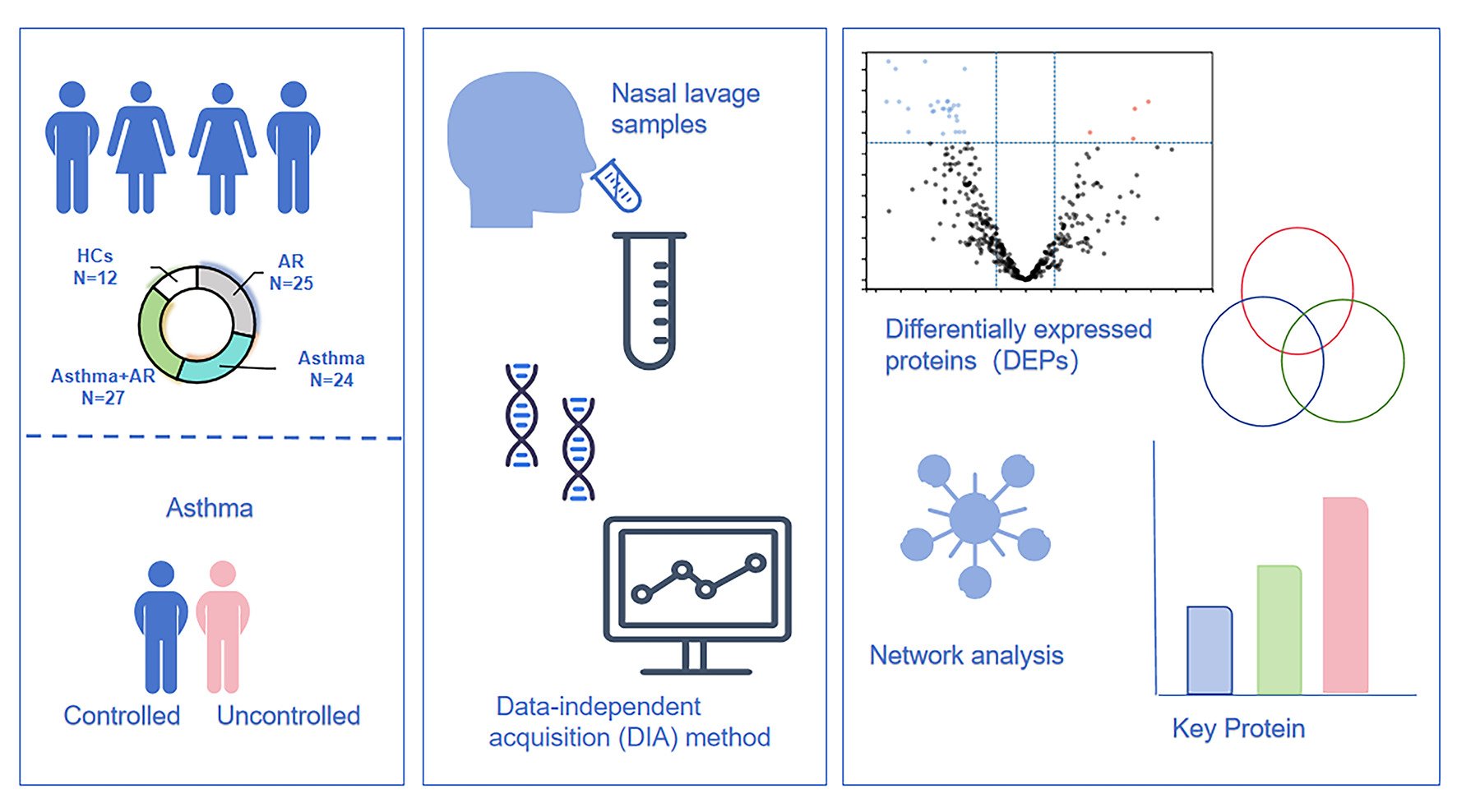
Purpose: Little research has explored the proteomic characteristics of nasal lavage fluid from asthmatic patients. This study aims to investigate whether differentially expressed proteins (DEPs) in nasal lavage fluid can serve as a biomarker to differentiate asthma patients from healthy controls (HCs) and to discern between individuals with well controlled and poorly controlled asthma.
Patients and Methods: We enrolled patients with allergic rhinitis (AR), asthma, or both conditions, and HCs in this study. We recorded patients’ demographic and medical history data and administered asthma quality of life questionnaire (AQLQ) and asthma control questionnaire (ACQ). Nasal fluid samples were collected, followed by protein measurements, and proteomic analysis utilizing the data-independent acquisition (DIA) method.
Results: Twenty-four with asthma, 27 with combined asthma+ AR, 25 with AR, and 12 HCs were enrolled. Four proteins, superoxide dismutase 2 (SOD2), serpin B7 (SERPINB7), kallikrein-13 (KLK13), and bleomycin hydrolase (BLMH) were significantly upregulated in nasal lavage fluid samples of asthma without AR, compared to HCs (Fold change ≥ 2.0, false-discovery rate [FDR] < 0.05). Conversely, 56 proteins including secretoglobin family 2A member 1 (SCGB2A1) were significantly downregulated (fold change ≥ 2.0, FDR < 0.05). Furthermore, 96.49% of DEPs including peptidase inhibitor 3 (PI3) and C-X-C motif chemokine 17 (CXCL17) were upregulated in poorly controlled asthma patients without AR relative those with well- or partly controlled asthma (fold change ≥ 1.5, FDR < 0.05). Search tool for the retrieval of interacting genes/proteins (STRING) analysis showed that PI3, with 18 connections, may be pivotal in asthma control.
Conclusion: The study revealed significant alteration in the nasal lavage proteome in asthma without AR patients. Moreover, our results indicated a potential association between the expression of proteome in the upper airway and the level of asthma control. Specifically, PI3 appears to be a key role in the regulation of asthma without AR.
Keywords: allergy, proteomic analysis, nasal lavage fluid, asthma control
Graphical Abstract:
Introduction
Asthma is a chronic inflammatory disorder of the airway, which affected approximately 1–18% of the population, with over 339 million individuals impacted, and poses a major public health concern.1 Characterized by hyperreactivity of the airway, inflammation, and reversible airflow limitation.2 The diagnosis and management of asthma largely rely on respiratory symptoms, clinical examination, and lung function test. However, the substantial heterogeneity and complexity of the condition pose challenges in the assessment.
In recent years, high-throughput sequencing technology has revolutionized the diagnosis and assessment of respiratory diseases.3–5 Proteomic techniques have facilitated the quantitative analysis of thousands of proteins and have been utilized to identify disease-specific proteins through molecular and protein pathway analyses.6 Weitoft et al research identified 150 proteins in asthmatic subjects that significantly changed following respiratory challenge with inhaled allergens.7 Notably, coagulation factor XIII, which is impacted in macrophage polarization and associated with airflow limitation,8,9 was among these proteins. Isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomic analysis has further revealed seven proteins, such as alpha1-microglobulin bikunin [AMBP], insulin-like growth factor-binding protein complex acid labile subunit [IGFALS], kininogen-1 [KNG1], and others, which were differentially expressed in asthma patients compared to healthy controls (HCs).10 Sputum proteomics has unveiled former smokers in severe asthma exhibited higher expression of C-X-C motif chemokine ligand 8 (CXCL8) and neutrophil elastase.11
Despite these advances, the majority of studies have concentrated on biological specimen samples like sputum and serum, with limited exploration of nasal lavage fluid proteomic profile in asthma. Furthermore, the utility of nasal lavage fluid proteomics in evaluating asthma control has not been well established.
Allergic rhinitis (AR) presents with nasal obstruction, itching, congestion, rhinorrhoea, and sneezing, and often co-presents with asthma.12 Given this co-morbidity, we hypothesized that differentially expressed proteins (DEPs) in nasal lavage fluid could serve as biomarkers to differentiate asthmatic adults from HCs and distinguish between well controlled and poorly controlled asthma.
Materials and Methods
Patient Characteristics
Adults diagnosed with asthma, AR, or both conditions were recruited from a random sample of the outpatient department for respiratory and critical care medicine at the First Affiliated Hospital of Ningbo University in Ningbo, China, between August 2020 and February 2022. Additionally, 12 volunteers without the history of allergic rhinitis and asthma were recruited from the community to serve as HCs. Asthma was defined according to the Global Initiative for Asthma guidelines.13 AR and combined asthma + AR were diagnosed based on symptom presentation (sneezing, itching, rhinorrhoea, nasal obstruction) and physical examination using the Allergic Rhinitis and its Impact on Asthma guidelines.14 The study exclusion criteria were as follows: pregnancy, lactation, other respiratory disorders, antibiotic therapy, cancer, glucocorticoids administered orally or intravenously within the past four weeks, and patients with chronic rhinosinusitis (CRS) in asthma group.
We collected demographic data, such as sex, age, height, weight, body mass index (BMI), and cigarette smoking history. Forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), maximal mid-expiratory flow (MMEF75/25), and peak expiratory flow (PEF) were used as the primary parameters of lung function. Patients’ medical histories, including their history of allergies, were also obtained. All subjects completed the total nasal symptom score (TNSS), asthma quality of life questionnaire (AQLQ),15 and asthma control questionnaire (ACQ).16 Additionally, the level of asthma control was evaluated with the Juniper ACQ and used to categorize the patients into two groups: well- or partly controlled (average ACQ score <1.5) and poorly controlled asthma (average ACQ score >1.5).17
Nasal Lavage Fluid Sample Collection
Nasal lavage fluid samples were obtained by injecting 10 mL of sterile saline solution bilaterally into the nasal cavity.18 Samples were centrifuged at 3000 rpm for 10 min at 4°C, and the supernatants were stored at −80°C until analysis.
Protein Measurements in Nasal Lavage Fluid
Nasal lavage samples (1.5 mL) were freeze-dried under a vacuum (Labconco, USA). The samples were then dissolved in 100 µL ddH2O and incubated with 20% acetone overnight at −20°C. After removing the supernatant, the protein was extracted by centrifugation at 12,000 rpm for 15 min and dried under a vacuum. Disulphide reduction was conducted with 5 mM Tris(2-carboxyethyl) phosphine hydrochloride (Thermo Scientific, USA) at 25°C for 30 min, followed by alkylation with 0.25 M iodoacetamide (Sigma, USA) at 25°C for 30 min. The protein was digested overnight at 37°C at a trypsin: protein ratio of 50:1. The peptide mixture was then desalted on a C18 column (Thermo Scientific) and resuspended in an aqueous solution of 0.1% formaldehyde (FA). The peptide concentration was then determined.
Nasal Lavage Fluid Proteomic Analysis: Data-Independent Acquisition (DIA) Pipeline
Peptides were resuspended in 30 µL FA, followed by a separate 9 µL from each sample. Next, mixed 1 µL indexed retention time (iRT), and 1 µL was taken for detection. DIA runs were acquired using an Easy-nLC1200 mass spectrometer (Thermo Fisher Scientific, USA) interfaced with an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific, USA). The full mass spectrometer settings were as follows: scan range = 300–1500 m/z, resolution = 60,000, AGC = 4 × 105. The subsequent mass spectrometer settings were as follows: resolution = 15,000, AGC = 5 × 104. Data were analysed with Spectronaut software (Biognosys AG, Switzerland) using the Direct-DIA method. Finally, protein quantification was performed, and a false-discovery rate (FDR) of 1% was used for intensity and identification.
Bioinformatics Analysis
The UniProt database (https://www.uniprot.org) was employed to perform functional annotation of proteins in nasal lavage fluid samples.19 Volcano plot analyses of DEPs (log scale) were conducted at NetworkAnalyst 3.0 software (https://www.networkanalyst.ca)20 and Sangerbox 3.0 (http://sangerbox.com). A Venny plot was generated using Venny 2.0 software (https://bioinfogp.cnb.csic.es/tools/venny/index.htm).21 Proteins with a fold change ≥1.5 or ≤0.5 and a Benjamini–Hochberg FDR <0.05 were considered differentially expressed. Protein–protein interaction (PPI) network analysis with a confidence score >0.4 was performed using the search tool for the retrieval of interacting genes/proteins (STRING) database (https://cn.string-db.org/) and visualized using Cytoscape software version 3.9.1. Gene Ontology (GO) enrichment and Reactome pathway analyses were carried out for DEPs using the STRING database.
Statistics Analysis
Data were processed using SPSS (version 13.0; SPSS Inc., Chicago, IL, USA). Subjects’ baseline data were presented as mean values (standard deviation) for normally distributed data and as medians (interquartile range, IQR) for skewed data. Normally distributed values were analysed with Student’s t-tests, and non-normally distributed data were analysed with Mann–Whitney U-tests for two-group comparisons. The Kruskal–Wallis test was used for comparisons of three or more groups. Categorical variables were compared using the chi-squared test or Fisher’s exact test. The signal intensity corresponds to the protein expression levels in nasal lavage samples, and the analysis of protein expression was performed using a Mann–Whitney U or Kruskal–Wallis tests. Pearson’s correlation coefficient was computed to assess correlations between protein expression and categorical variables (ie, sex, BMI, smoking status, and lung function parameters). Receiver operating characteristic (ROC) analysis was conducted to assess performance of the proteins in asthma, and to calculate the area under curve (AUC) and 95% confidence intervals (CI). The sensitivity and specificity of proteins were determined by standard techniques. P-values < 0.05 indicated statistical significance. Pearson correlation heat maps and scatter plots were generated with GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA).
Ethical Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the ethics committee of the First Affiliated Hospital of Ningbo University (approval 2020-R145). All subjects provided written informed consent before participating in the study.
Results
Participants’ Baseline Characteristics
This analysis included a total of 88 adults into four groups: 24 with asthma, 27 with combined asthma + AR, 25 with AR, and 12 HCs. As shown in Table 1, there were no significant differences in sex, age, BMI, or smoking status between groups. Lung function parameters, ACQ, and AQLQ questionnaires were assessed. Furthermore, participants were separated into two groups – well- or partly controlled asthma and poorly controlled asthma according to ACQ score.
 |
Table 1 Demographic Data of Study Participants |
Comparison of Differentially Expressed Proteins
Protein and peptide concentrations of nasal lavage fluid was showed in eTable 1. Then, a total of 620 proteins were identified in the nasal lavage fluid via DIA analysis. To further distinguish the profiles between patients with asthma, those with AR, and HCs, we specifically analysed the up- and downregulated proteins in AR and asthma patients compared to HCs. The volcano plot revealed four upregulated proteins. The resulting volcano plot highlighted significant differences in protein expression, with four proteins found to be upregulated in AR group compared to HCs: decay-accelerating factor ([DAF], also known as CD55), alkaline phosphatase (ALPL), Aminopeptidase N (ANPEP), and immunoglobulin heavy variable 3–49 (IGHV3-49). Additionally, 28 proteins were identified as downregulated in nasal lavage fluid of the AR group (fold change ≥2.0 or ≤0.5, FDR <0.05, Figure 1A). Notably, ANPEP, also known as CD13, plays a critical role in various biological processes, including peptide cleavage, modulation of host–virus interactions, promotion of angiogenesis, and prevention of cholesterol crystallization (Table 2).22
 |
Table 2 The List of Proteins Upregulated in Nasal Lavage Fluid Between AR, Asthma, and Asthma +AR as Compared with HCs |
In our analysis of nasal lavage fluid samples from patients with asthma without AR, we identified four proteins that were significantly upregulated compared to HCs. Superoxide dismutase 2 (SOD2), serpin B7 (SERPINB7), kallikrein-13 (KLK13), and bleomycin hydrolase (BLMH) (Fold change ≥2.0, FDR <0.05) showing the most significant enrichment. Concurrently, 56 proteins were found to be downregulated, with the most significant suppression observed in secretoglobin family 2A member 1 (SCGB2A1), keratin, type II cytoskeletal 78 (KRT78), secretoglobin family 1D member 1 (SCGB1D1), phosphoglucomutase-1 (PGM1), destrin (DSTN), drebrin-like protein (DBNL), and phosphoglucomutase-2 (PGM2) being the most significantly suppressed in isolated asthma compared to HCs (fold change ≤0.5, FDR <0.05, Figure 1B). Notably, SOD2 and ANPEP also exhibited increased expression in combined asthma + AR groups when compared to HCs (Figure 1C, fold change ≥2.0, FDR <0.05). A Venn diagram was employed to depict the number of shared or unique DEPs between each allergic disease group and HCs in nasal lavage fluid proteome. It revealed that 27 proteins were specific to asthma without AR, including upregulated SERPINB7, KLK13, and BLMH, while 31 proteins were found to overlap between asthma without AR and combined asthma +AR group, compared to HCs. Of these, SOD2 is recognized for its crucial role in catalysing the dismutation of superoxide anion radicals16 (Figure 1D).
To further evaluate the DEPs, scatter plots were utilized to assess the signal intensity levels across all four groups. Nasal lavage levels of CD55 were significantly downregulated in both asthma (P< 0.001), and combined asthma + AR (P< 0.001) compared to the AR group (Figure 2A). CD55 is known to inhibit the activation of complement factor (complement C3 [C3], C4), thereby preventing complement-mediated tissue damage (Table 2).15 Consistently, C3 levels were decreased in both the asthma, and combined asthma + AR groups compared to the AR group (Figure 2B). Moreover, the expression of C5, C7, ANPEP, ALPL was downregulated in asthma without AR compared to those with AR (Figure 2C–F). However, a significant higher relative expression of SOD2, SERPINB7, KLK13 was observed in asthma without AR compared to those with AR (Figure 2G–I).
To explore their potential as biomarker for asthma diagnosis, we subjected three upregulated proteins, SOD2, SERPINB7, and KLK13 found in the upper respiratory tract of asthma without AR, to ROC analysis. The AUC value showed 0.686 (95% CI, 0.552–0.820, p = 0.015) for SOD2, 0.755 (95% CI, 0.633–0.876, p = 0.001) for SERPINB7, and 0.792 (95% CI, 0.677–0.906, p < 0.001) for KLK13. When these three proteins were combined for analysis, the results yielded an AUC of 0.794 (95% CI, 0.681–0.907, p < 0.001), offering a sensitivity of 66.7% and a specificity of 86.5% for the asthma diagnosis (eFigure 1).
Nasal Lavage Fluid Proteomic Analysis in Asthma Patients in Association with Asthma Control Level
The asthma statuses and baseline characteristics of the participants are presented in eTable 2. There were no significant differences in terms of sex, age, or BMI between the groups with well- or partly controlled asthma (n = 15) and those with poorly controlled asthma (n = 9). However, higher proportion of smokers among poorly controlled asthma group (33.33%) compared to the well- or partially controlled group (6.67%, P = 0.031). The poorly controlled asthma group exhibited markedly lower value for FEV1 (mean: 51.12% vs 89.52%, P < 0.001), FEV1/FVC (57.90% vs 83.26%, P < 0.001), PEF (51.09% vs 94.12%, P < 0.001), and MMEF75/25 (20.73% vs 73.67%, P < 0.001). These findings indicated more severe pulmonary impairment in patients with poorly controlled asthma than in those with well- or partly controlled asthma, aligning with clinical data (eTable 2).
We identified 57 DEPs by volcano plot. The majority of those proteins (n = 55, 96.49%), including C-X-C motif chemokine 17 (CXCL17), transforming growth factor-beta-induced protein ig-h3 (TGFBI), basic salivary proline-rich protein 2 (PRB2), plasma kallikrein (KLKB1), ras-related protein Rab-1A (RAB1A), phospholipase A2, membrane associated (PLA2G2A), basic salivary proline-rich protein 4 (PRB4), dermcidin (DCD), glutaminyl-peptide cyclotransferase (QPCT), neuroserpin (SERPINI1), and secretoglobin family 1D member 2 (SCGB1D2), were found to be increased in poorly controlled asthma patients compared to well- or partly controlled asthma patients, while two proteins (glycogen phosphorylase, liver form [PYGL] and hemoglobin subunit beta [HBB]), were decreased (fold change ≥1.5 or ≤0.66, P < 0.05, Figure 3A). Pearson correlation coefficient analysis revealed significant inverse relationship between the expression levels of PRB2 and Ly6/PLAUR domain-containing protein 3 (LYPD3) with key lung function parameters such as FEV1, FEV1/FVC, PEF, and MMEF75/25, in asthmatics. Additionally, peptidase inhibitor 3 (PI3) showed a positive correlation with WAP four-disulfide core domain protein 2 (WFDC2), beta-defensin 1 (DEFB1), and midkine (MDK) expression, whereas CXCL17 levels correlated positively with antileukoproteinase (SLPI), lysozyme C (LYZ), and MDK expression (Figure 3B).
GO term enrichment analysis indicated that the following biological processes such as: antibacterial humoral response, cytolysis, peptide cross-linking, cornification, antimicrobial humoral response, and defense response to bacteria were significantly enriched in samples from patients with poorly controlled asthma (eTable 3 and eFigure 2). Reactome pathway analysis further revealed that pathway related to antimicrobial peptide (PI3, LYZ, DCD, and DEFB1), formation of the cornified envelope (PI3, caspase-14 [CASP14], keratin, type I cytoskeletal 16 [KRT16]. Cornifin-B [SPRR1B], filaggrin [FLG], keratin, type II cytoskeletal 6A [KRT6A], desmocollin-1 [DSC1], and desmocollin-2 [DSC2]), neutrophil degranulation (SLP, LYZ, metalloproteinase inhibitor 2 [TIMP2], haptoglobin [HP], HBB, CD59 glycoprotein [CD59], DSC1, calmodulin-like protein 5 [CALML5], and PYGL), and innate immune system (SLPI, PI3, LYZ, TIMP2, DCD, DEFB1, HBB, HP, CD59, DSC1, CALML5, and PYGL) were upregulated in poorly controlled asthma patients compared to those with well- or partly controlled asthma patients (eTable 4 and eFigure 3).
To explore the interrelationship among DEPs, we conducted STRING PPI network analysis, focusing on Betweenness Centrality, Closeness Centrality, and Degree Centrality. This analysis highlighted PI3 as a central node with 18 connections, suggesting its pivotal role for asthma control (Figure 3C). The top 10 proteins ranked by “Betweenness” in poorly controlled compared to well- or partly controlled asthma were PI3, Zinc-alpha-2-glycoprotein (AZGP1), SLPI, LYZ, semenogelin-2 (SEMG2), corneodesmosin (CDSN), prolactin-inducible protein (PIP), DEFB1, HP, and WFDC2 (eTable 5 and Figure 3C). Furthermore, patient with poorly controlled asthma showed significant upregulation of PI3, SLPI, LYZ, SEMG2, and DEFB1 compared to those with well- or partly controlled asthma (Figure 3D).
Nasal Lavage Fluid Proteomic Analysis in Combined Asthma + AR Patients in Association with Asthma Control Level
In our analysis of patients with combined asthma + AR, baseline characteristics were found to be similar between the two groups categorized by different asthma status. Consistent with disease progression, poorly lung function was observed in patients with poorly controlled compared to those with well- or partly controlled combined asthma + AR patients (eTable 6).
We identified 19 DEPs, all of which exhibited downregulation in the poorly controlled group with a fold change ≤0.66. (P < 0.05, Figure 4A). Notably, a robust positive association was observed between the levels of nitric oxide synthase, inducible (NOS2) and mucin-4 (MUC4) (r = 0.732, P < 0.001, Figure 4B). Additionally, a moderate positive association was detected between MMP10 and MUC16 (r = 0.476, P < 0.001, Figure 4B). The median abundances of stromelysin-2 [MMP10], SCGB1D2, NOS2, insulin-like growth factor-binding protein 2 (IGFBP2), MUC4, and MUC16 were significantly lower in the poorly controlled group when compared to well- or partly controlled groups (Figure 4C). A trend toward lower protein S100-A12 (S100A12) and SEMG2 abundance was also observed in the poorly controlled group, although this did not reach statistical significance (Figure 4C). Furthermore, STRING PPI network analysis showed that 19 nodes connected via three edges (eFigure 4).
Discussion
This study employed proteomic analyses to investigate DEPs in nasal lavage fluid samples from asthmatic patients, both with and without AR. SOD2 was consistently upregulated in both asthmatic groups compared to HCs. In contrast, the majority of proteins, including SCGB2A1 and SCGB1D1 were observed to be downregulated in asthma patients without AR compared to HCs. Notably, the expression levels of SOD2, SERPINB7, KLK13 were found to be upregulated in asthma without AR compared to those with only AR. The combination of SOD2, SERPINB7, and KLK13 demonstrated strong diagnostic potential for asthma, with an AUC of 0.794 (95% CI, 0.681–0.907, p < 0.001). Furthermore, the study identified potential biomarkers indicate asthma control levels. A total of 55 DEPs were upregulated in poorly controlled asthma patients compared to these with well- or partly controlled. The STRING PPI network analysis demonstrated that the PI3, which with 18 connections, may play a key role in asthma control.
The study results indicated a decrease in the expression of SCGB2A1 and SCGB1D1 in the upper airway of patients with asthma without AR. This observation is consistent with previous investigation, which also reported the downregulation of SCGB2A1 in the nasal mucosa of patients in CRS, and demonstrated the varied responses of secretoglobins (SCGBs) genes to inflammatory cytokines.23 Given that the SCGBs family generally produced by the secretory epithelial and plays a critical role in the innate immune and anti-inflammatory processes.24 Therefore, the decreased expression of SCGB2A1 and SCGB1D1 may contribute to the pathophysiology of airway diseases. However, there were limited available data for SCGBs family, and further research is needed to elucidate the complex interplay between the innate and adaptive immune responses and their antagonistic regulation.
The aim of this study was to develop protein biomarkers in the upper airway tract to assist in the diagnosis and evaluation of asthma. Bioinformatics analysis showed a significant increase in SOD2, SERPINB7, and KLK13 in asthma without AR. The ROC analysis of the combination of these three proteins yielded AUCs of 0.794. With regard to SOD2, it belongs to the iron/manganese superoxide dismutase family and catalyses the dismutation of superoxide anion radicals.25 Overexpression of SOD2 was found in human bronchial epithelial (16-HBE) cells following exposed to monoaromatic hydrocarbons and polycyclic aromatic hydrocarbons, leading to increased levels of thymic stromal lymphopoietin (TSLP), IL-25 and IL-33, posing a potential risk of asthma and exacerbation.26 Moreover, a clinical study demonstrated that blood sample SOD2 levels are associated with disease exacerbation and prolonged hospital admission duration in patients with asthma and chronic obstructive pulmonary disease.27 Our data showed increased expression of SOD2 in nasal lavage fluid samples from asthma even without AR. The non-invasive biofluid may contribute to understanding pathological mechanism, definition novel phenotypes, and even aiding in therapeutic assessment.
SERPINs are serine protease inhibitors associated with the fibrinolytic system and have been identified as implicated in mucous production, potentially involved in the development of asthma.28,29 SERPINB7 is essential for stabilizing epidermal strength and integrity.30 Indeed, a genome-wide meta-analysis of 796,661 individuals, including 22,474 with atopic dermatitis, indicated that SERPINB7 mutations may play a role in the development of atopic dermatitis.30 Furthermore, upregulation of SERPINB7 in this context could potentially increase the risk of asthma and other allergic diseases by impacting nasal epithelial cell barrier function and lymphocyte-mediated immunity disorder.31 The present study observed that SERPINB7 was markedly elevated in asthma patients in nasal lavage fluid. The findings suggested that asthma may cause damage to the epithelial barriers in the upper airway tract, even without AR.32 Furthermore, human tissues produce kallikreins, which are secreted serine proteases (KLK1-KLK15). Multiple KLKs were elevated in patients with atopic dermatitis, and the expression of KLK7 in serum was correlated with blood levels of eosinophils.33 It is worth noted that our study is the first to observe an upregulated KLK13 in the nasal lavage fluid of asthma without AR. Furthermore, the combination SOD2, SERPINB7, and KLK13 may serve as a potential biomarker helpful in determining some immune patterns or endotypes for the asthma. However, due to the small sample size of this study and its early stage of research, further study is necessary to evaluate the practicality of using protein biomarkers for diagnosing asthma.34
Proteins associated with immune responses were enriched in the upper airways of poorly controlled asthma patients. It was noteworthy that PI3 is highly expressed in uncontrolled asthma. A previous study has demonstrated the PI3 associated with Th17 inflammatory processes, suggesting that increased PI3 expression in asthma may promote Th17 responses and prevent effective asthma control.35 Furthermore, a study adapted to sputum macrophage transcriptomic analysis indicated that individuals with neutrophilic asthma had a higher level of PI3 compared to those without neutrophilic.36 Both the neutrophilic asthma and Th17 are inflammatory phenotypes associated with worse clinical condition and poorly asthma control. Our work was in line with the previous study findings. However, our study provided direct evidence of decreased PI3 expression in poorly controlled asthma through minimal invasive sampling of the upper airway tract. CXCL17, a myeloid cell-attracting chemokine, modulates immune responses associated with pulmonary fibrosis, asthma, and infectious diseases.37 Higher expression of CXCL17 in bronchial brushing samples was found in type 2 asthma patients compared to HCs.38 Not surprisingly, our results showed CXCL17 also elevated in poorly controlled asthma in upper airway tract. Given that CXCL17 suppressed the expression of microRNA 221–3p by decreasing the levels of eosinophils-associated molecules, such as (C-C motif) ligand (CCL) 24, CCL26, and periostin (POSTIN) in vitro.38 The inverse correlation was also observed between CXCL17 and microRNA 221–3p. Nonetheless, it is possible for apply CXCL17 for determining endotypes and evaluating asthma.
There were several limitations to this study. First, only a small number of HCs were enrolled. Second, we did not utilize other assays, such as flow cytometry or enzyme-linked immunosorbent assay (ELISA) analysis, to investigate the expression of cytokines associated with the inflammatory response. Third, the absence of lower airway samples hindered our ability to demonstrate a potential correlation between uncontrolled inflammation in the bronchi and its manifestation in the upper airways. Moreover, we did not categorize asthma into T2 and non-T2 variants, resulting in a failure to identify the DEPs associated with Th type. However, nasal lavage fluid proteomics reveals potential biomarkers of asthma associated with the degree of asthma control. Less invasive testing may provide new utility for the diagnosis and monitoring of asthma.
Conclusion
In sum, this study observed altered proteomic in the upper airway tract in asthma patients without upper airway symptoms. Bioinformatic analyses indicated that the upregulated of SOD2, SERPINB7, KLK13, and BLMH in asthma. Moreover, the combination of SOD2, SERPINB7, and KLK13 could potentially serve as a potential biomarker for asthma diagnosing. Furthermore, this work demonstrated the expression of proteome like PI3 in the upper airway tract may play a key role in asthma control.
Abbreviations
ACQ, asthma control questionnaire; ALPL, alkaline phosphatase; AMBP, alpha1-microglobulin bikunin; ANPEP, aminopeptidase N; AQLQ, asthma quality of life questionnaire; AR, allergic rhinitis; AUC, area under curve; AZGP1, Zinc-alpha-2-glycoprotein; BLMH, bleomycin hydrolase; BMI, body mass index; C3, complement C3; CALML5, calmodulin-like protein 5; CASP14, caspase-14; CCL, C-C motif ligand; CD59, CD59 glycoprotein; CDSN, corneodesmosin; C1R, complement C1r subcomponent; CI, confidence intervals; CXCL17, C-X-C motif chemokine 17; CXCL8, C-X-C motif chemokine ligand 8; CD55 (DAF), decay-accelerating factor; DBNL, drebrin-like protein; DCD, dermcidin; DEFB1, beta-defensin 1; DEPs, differentially expressed proteins; DSC1, desmocollin-1; DC, dendritic cell; DIA, data-independent acquisition; DSC2, desmocollin-2; DSTN, destrin; ELISA, enzyme-linked immunosorbent assay; FDR, false-discovery rate; FEV1, forced expiratory volume in 1 second; FLG, filaggrin; FVC, forced vital capacity; GO, Gene Ontology; HBB, hemoglobin subunit beta; HCs, healthy controls; HDAC2, histone deacetylase 2; HP, haptoglobin; IGFALS, insulin-like growth factor-binding protein complex acid labile subunit; IGFBP2, insulin-like growth factor-binding protein 2; IGHV3-49, immunoglobulin heavy variable 3-49; IL, interleukin; iRT, indexed retention time; iTRAQ, isobaric tags for relative and absolute quantitation; KLK13, kallikrein-13; KLKB1, plasma kallikrein; KNG1, kininogen-1; KRT16, keratin, type I cytoskeletal 16; KRT6A, keratin, type II cytoskeletal 6A; KRT78, keratin, type II cytoskeletal 78; LYZ, lysozyme C; MDK, midkine; MMEF75/25, maximal mid-expiratory flow; MMP10, stromelysin-2; MUC4, mucin-4; NOS2, nitric oxide synthase, inducible; PEF, peak expiratory flow; PGM1, phosphoglucomutase-1; PGM2, phosphoglucomutase-2; PI3, peptidase inhibitor 3; PIP, prolactin-inducible protein; PLA2G2A, phospholipase A2, membrane associated; POSTIN, periostin; PPI, protein–protein interaction; PRB2, basic salivary proline-rich protein 2; PRB4, basic salivary proline-rich protein 4; PROS1, protein S; PYGL, glycogen phosphorylase, liver form; QPCT, glutaminyl-peptide cyclotransferase; RAB1A, ras-related protein Rab-1A; ROC, receiver operating characteristic; S100A12, protein S100-A12; SCGB1A1, uteroglobin; SCGB1D1, secretoglobin family 1D member 1; SCGB1D2, secretoglobin family 1D member 2; SCGB2A1, secretoglobin family 2A member 1; SEMG2, semenogelin-2; SERPINA1, alpha-1-antitrypsin; SERPINB3, serpin B3; SERPINB7, serpin B7; SERPINI1, neuroserpin; SLPI, antileukoproteinase; SOD2, superoxide dismutase 2; SPRR1B, cornifin-B; STRING, search tool for the retrieval of interacting genes/proteins; TGFBI, transforming growth factor-beta-induced protein ig-h3; Th, T-helper type; TIMP2, metalloproteinase inhibitor 2; TNSS, total nasal symptom score; TSLP, thymic stromal lymphopoietin; WFDC2, WAP four-disulfide core domain protein 2.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the ethics committee of the First Affiliated Hospital of Ningbo University (approval 2020-R145). All participants agreed and signed informed consents before enrolment.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was supported by the National Natural Science Foundation of China (Grant number [82170016]).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chowdhury NU, Guntur VP, Newcomb DC, Wechsler ME. Sex and gender in asthma. Eur Respir Rev. 2021;30(162):210067. doi:10.1183/16000617.0067-2021
2. Papi A, Brightling C, Pedersen SE, Reddel HK. Asthma. Lancet. 2018;391(10122):783–800. doi:10.1016/S0140-6736(17)33311-1
3. Diao Z, Han D, Zhang R, Li J. Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections. J Adv Res. 2022;38:201–212. doi:10.1016/j.jare.2021.09.012
4. Du M, Garcia JGN, Christie JD, et al. Integrative omics provide biological and clinical insights into acute respiratory distress syndrome. Intensive Care Med. 2021;47(7):761–771. doi:10.1007/s00134-021-06410-5
5. Radzikowska U, Baerenfaller K, Cornejo-Garcia JA, et al. Omics technologies in allergy and asthma research: an EAACI position paper. Allergy. 2022;77:2888–2908. doi:10.1111/all.15412
6. Vizuet-de-Rueda JC, Montero-Vargas JM, Galván-Morales M, Porras-Gutiérrez-de-Velasco R, Teran LM. Current insights on the impact of proteomics in respiratory allergies. Int J Mol Sci. 2022;23(10):5703. doi:10.3390/ijms23105703
7. Weitoft M, Kadefors M, Stenberg H, et al. Plasma proteome changes linked to late phase response after inhaled allergen challenge in asthmatics. Respir Res. 2022;23(1):50. doi:10.1186/s12931-022-01968-0
8. Jamil MA, Singh S, El-Maarri O, Oldenburg J, Biswas A. Exploring diverse coagulation factor xiii subunit expression datasets: a bioinformatic analysis. Int J Mol Sci. 2022;23(9):4725. doi:10.3390/ijms23094725
9. Esnault S, Kelly EA, Sorkness RL, Evans MD, Busse WW, Jarjour NN. Airway factor XIII associates with type 2 inflammation and airway obstruction in asthmatic patients. J Allergy Clin Immunol. 2016;137(3):767–773.e766. doi:10.1016/j.jaci.2015.05.053
10. Nieto-Fontarigo JJ, González-Barcala FJ, Andrade-Bulos LJ, et al. iTRAQ-based proteomic analysis reveals potential serum biomarkers of allergic and nonallergic asthma. Allergy. 2020;75(12):3171–3183. doi:10.1111/all.14406
11. Takahashi K, Pavlidis S, Ng Kee Kwong F, et al. Sputum proteomics and airway cell transcripts of current and ex-smokers with severe asthma in U-BIOPRED: an exploratory analysis. Eur Respir J. 2018;51(5):1702173. doi:10.1183/13993003.02173-2017
12. Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet. 2011;378(9809):2112–2122. doi:10.1016/S0140-6736(11)60130-X
13. Reddel HK, Bacharier LB, Bateman ED, et al. Global initiative for asthma strategy 2021: executive summary and rationale for key changes. Eur Respir J. 2022;59(1):2102730. doi:10.1183/13993003.02730-2021
14. Brożek JL, Bousquet J, Agache I, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol. 2017;140(4):950–958. doi:10.1016/j.jaci.2017.03.050
15. Ozen A, Comrie WA, Ardy RC, et al. CD55 deficiency, early-onset protein-losing enteropathy, and thrombosis. N Engl J Med. 2017;377(1):52–61. doi:10.1056/NEJMoa1615887
16. Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med. 2009;47(4):344–356. doi:10.1016/j.freeradbiomed.2009.05.018
17. Juniper EF, O’Byrne PM, Guyatt GH, Ferrie PJ, King DR. Development and validation of a questionnaire to measure asthma control. Eur Respir J. 1999;14(4):902–907. doi:10.1034/j.1399-3003.1999.14d29.x
18. Eifan AO, Orban NT, Jacobson MR, Durham SR. Severe persistent allergic rhinitis. inflammation but no histologic features of structural upper airway remodeling. Am J Respir Crit Care Med. 2015;192(12):1431–1439. doi:10.1164/rccm.201502-0339OC
19. Bateman A, Martin M-J, Orchard S. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–d489. doi:10.1093/nar/gkaa1100
20. Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–w241. doi:10.1093/nar/gkz240
21. Oliveros JC. (2007-2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. Available from: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
22. Dong X, An B, Salvucci Kierstead L, Storkus WJ, Amoscato AA, Salter RD. Modification of the amino terminus of a class II epitope confers resistance to degradation by CD13 on dendritic cells and enhances presentation to T cells. J Immunol. 2000;164(1):129–135. doi:10.4049/jimmunol.164.1.129
23. Lu X, Wang N, Long XB, You XJ, Cui YH, Liu Z. The cytokine-driven regulation of secretoglobins in normal human upper airway and their expression, particularly that of uteroglobin-related protein 1, in chronic rhinosinusitis. Respir Res. 2011;12(1):28. doi:10.1186/1465-9921-12-28
24. Mootz M, Jakwerth CA, Schmidt-Weber CB, Zissler UM. Secretoglobins in the big picture of immunoregulation in airway diseases. Allergy. 2022;77(3):767–777. doi:10.1111/all.15033
25. MacMillan-Crow LA, Thompson JA. Tyrosine modifications and inactivation of active site manganese superoxide dismutase mutant (Y34F) by peroxynitrite. Arch Biochem Biophys. 1999;366(1):82–88. doi:10.1006/abbi.1999.1202
26. Shen Q, Yu H, Liu Y, Li G, An T. Combined exposure of MAHs and PAHs enhanced amino acid and lipid metabolism disruption in epithelium leading asthma risk. Environ Pollut. 2024;343:123261. doi:10.1016/j.envpol.2023.123261
27. Canova C, Dunster C, Kelly FJ, et al. PM10-induced hospital admissions for asthma and chronic obstructive pulmonary disease: the modifying effect of individual characteristics. Epidemiology. 2012;23(4):607–615. doi:10.1097/EDE.0b013e3182572563
28. Bai J, Zhong JY, Liao W, et al. iTRAQ‑based proteomic analysis reveals potential regulatory networks in dust mite‑related asthma treated with subcutaneous allergen immunotherapy. Mol Med Rep. 2020;22(5):3607–3620. doi:10.3892/mmr.2020.11472
29. Sivaprasad U, Askew DJ, Ericksen MB, et al. A nonredundant role for mouse Serpinb3a in the induction of mucus production in asthma. J Allergy Clin Immunol. 2011;127(1):
30. Sliz E, Huilaja L, Pasanen A, et al. Uniting biobank resources reveals novel genetic pathways modulating susceptibility for atopic dermatitis. J Allergy Clin Immunol. 2022;149(3):1105–1112.e1109. doi:10.1016/j.jaci.2021.07.043
31. Ferreira MA, Vonk JM, Baurecht H, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752–1757. doi:10.1038/ng.3985
32. Bousquet J, Melén E, Haahtela T, et al. Rhinitis associated with asthma is distinct from rhinitis alone: the ARIA-MeDALL hypothesis. Allergy. 2023;78(5):1169–1203. doi:10.1111/all.15679
33. Komatsu N, Saijoh K, Kuk C, et al. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp Dermatol. 2007;16(6):513–519. doi:10.1111/j.1600-0625.2007.00562.x
34. Habib N, Pasha MA, Tang DD. Current understanding of asthma pathogenesis and biomarkers. Cells. 2022;11(17):2764. doi:10.3390/cells11172764
35. Choy DF, Hart KM, Borthwick LA, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. 2015;7(301):301ra129. doi:10.1126/scitranslmed.aab3142
36. Fricker M, Qin L, Sánchez-Ovando S, et al. An altered sputum macrophage transcriptome contributes to the neutrophilic asthma endotype. Allergy. 2022;77(4):1204–1215. doi:10.1111/all.15087
37. Choreño-Parra JA, Thirunavukkarasu S, Zúñiga J, Khader SA. The protective and pathogenic roles of CXCL17 in human health and disease: potential in respiratory medicine. Cytokine Growth Factor Rev. 2020;53:53–62. doi:10.1016/j.cytogfr.2020.04.004
38. Zhang K, Liang Y, Feng Y, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol. 2018;315(2):L253–l264. doi:10.1152/ajplung.00567.2017
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




