Back to Journals » Infection and Drug Resistance » Volume 13
Murine Macrophage Requires CD11b to Recognize Talaromyces marneffei
Received 5 November 2019
Accepted for publication 10 March 2020
Published 27 March 2020 Volume 2020:13 Pages 911—920
DOI https://doi.org/10.2147/IDR.S237401
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yongxuan Hu,1– 3 Sha Lu,1 Liyan Xi1,4,5
1Department of Dermatology, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Department of Dermatology and Venereology, The 3rd Affiliated Hospital of Southern Medical University, Guangzhou, People’s Republic of China; 3Guangdong Provincial Key Laboratory of Bone and Joint Degeneration Diseases, Guangzhou, People’s Republic of China; 4Dermatology Hospital of Southern Medical University, Guangzhou, People’s Republic of China; 5Department of Dermatology, Guangzhou First People’s Hospital, The Second Affiliated Hospital of South China University of Technology, Guangzhou, People’s Republic of China
Correspondence: Yongxuan Hu
Department of Dermatology and Venerology, The 3rd Affiliated Hospital of Southern Medical University, 183 West Zhongshan Road, Guangzhou 510630, People’s Republic of China
Tel/Fax +86 20 62784560
Email [email protected]
Liyan Xi
Department of Dermatology, Sun Yat-sen Memorial Hospital, Sun Yat–sen University, 107 West Yanjiang Road, Guangzhou 510120, People’s Republic of China
Tel/Fax +86 20 81332289
Email [email protected]
Introduction: Talaromyces marneffei (T. marneffei) is an emerging pathogenic fungus. Macrophage-1 antigen (Mac-1, CR3, CD11b/CD18) is an important receptor on innate immune cells and can recognize pathogens. However, the importance of CR3 in phagocytosis of T. marneffei by macrophages and their responses to T. marneffei have not been clarified.
Methods: We show that interaction of mouse peritoneal macrophages (pMacs) or RAW264.7 macrophages with T. marneffei of its conidia spores and yeast cells enhances CR3 expression on macrophages. The phagocytosis rate was determined using flow cytometry, RT-PCR and Western blotting were used to detect CD11b expression, and the levels of IFN-γ, TNF-α, IL-2, IL-4, IL-6 and IL-10 in the co-culture supernatants were determined by ELISA.
Results: Incubation of mouse macrophages with T. marneffei promoted phagocytosis of T. marneffei, which was dramatically mitigated by pretreatment with anti-CD11b antibody or knockdown of CR3 expression on macrophages. Then, interferon γ, tumor necrosis factor α, IL-4, IL-10 and IL-12 production in macrophages incubation with heat-killed T. marneffei was detected. CD11b expression on mouse macrophages was upregulated by T. marneffei. Incubation of T. marneffei promoted phagocytosis of T. marneffei by macrophages and high levels of pro-inflammatory and anti-inflammatory cytokine production by macrophages, which were mitigated and abrogated by pre-treatment with anti-CD11b or knockdown of CD11b expression.
Conclusion: These data indicated that murine macrophage requires CD11b to recognize Talaromyces marneffei and their cytokine responses to heat-killed T. marneffei in vitro.
Keywords: Talaromyces marneffei, CD11b, macrophage
Introduction
Talaromyces marneffei (T. marneffei), formerly named Penicillium marneffei (P. marneffei) is the only dimorphic member of the genus and is an emerging pathogenic fungus that can cause a fatal systemic mycosis in humans.1,2 T. marneffei can form conidia spores and yeast cells. Inhaled conidia are thought to be the infectious particles.3–5 Macrophages have various cell-surface receptors that can recognize pathogen-associated molecule pattern (PAMP) of non-opsonized pathogens by the pattern recognition receptors (PRRs).6 However, how macrophages recognize PAMP on T. marneffei is unclear.
The integrin CR3 is a heterodimer of αM (CD11b) and β2 (CD18) subunits and is one of the most versatile receptors expressed by phagocytes. CR3 can mediate adhesion, chemotaxis, and phagocytosis of innate immune cells in a complement-dependent or complement-independent manner.7–11 Previous studies have shown that the recognition of unopsonized yeast particles by CR3 depends on the binding of the yeast β-glucans to the carbohydrate-binding site located in CD11b.12,13 However, the importance of CR3 in phagocytosis of T. marneffei by macrophages has not been clarified.
In this study, we investigated the roles of CD11b in phagocytosis of T. marneffei by mouse macrophages and their responses to conidia spores and yeast cells of T. marneffei in vitro.
Materials and Methods
Ethical Standards
A strain of T. marneffei SUMS0152 (IFM52703) was obtained from the Fungi Research Center of Sun Yat-sen Memorial Hospital and the experimental protocol was approved by the Medical Ethics Committee of Sun Yat-sen University.14 The animal studies were reviewed and approved by the Animal Research and Care Committee of Sun Yat-sen University (SCXK Guangdong 2009–0011), and followed the guidelines for the welfare of the laboratory animals (Laboratory animal—Guideline for ethical review of animal welfare, GB/T3589-2009).
The T. marneffei experiments in this manuscript were conducted under biosafety level 3 conditions (Public experimental platform of Sun Yat-sen University).
Animals
Male BALB/c mice at 8 weeks of age were obtained from the Laboratory Animal center of Sun Yat-sen University (Guangzhou, China). To suppress the immune system, all BALB/c mice were injected intraperitoneally with 100 mg/kg Cyclophosphamide (at 0.2 mL, Sigma Aldrich, St. Louis, MO, USA) daily for consecutive 3 days. The white blood cells from their tail venous blood samples were counted at the day 1 and 4. Individual mice with reduced WBC count to ≤ 25% were considered as immunosuppressed.14
Cells
Peritoneal macrophages (pMacs) were collected from the control and immunosuppressed BALB/c mice after the third injection by peritoneal lavage as described.14 Mouse RAW264.7 macrophages were preserved in the Fungi Research Center of Sun Yat-sen Memorial Hospital.
Fungus
T. marneffei was firstly grown in Sabouraud’s dextrose agar (SDA) medium. The subsequent culture and isolation of conidia and yeast cells were carried out as described.14
Labeling of T. marneffei Yeast or Spores with Fluorescein Isothiocyanate (FITC)
T. marneffei yeast cells and conidia spores were prepared. T. marneffei yeast cells and conidia spores at 2×108/mL were labeled with 0.16 mg/mL FITC in 0.05 M carbonate-bicarbonate buffer (pH9.5) for 60 mins in the dark and washed with PBS. The efficacy of FITC-labeling reached almost 100% and the intensity of FITC-labeled yeast cells and conidia spores was 2×105, determined by flow cytometry.
Transfection
RAW264.7 cells (1×106 cells/well) were cultured in complete medium overnight and transfected with individual CD11b specific or control siRNAs (Table 1) using lipofectamine 2000 (Invitrogen, USA). The CD11b-specific siRNAs were designed using the Ambion online software (http://www.ambion.com/techlib/misc/siRNA_finder), and synthesized by GenePharma (Shanghai, China) (Table 1). After transfection for 48 hrs, the levels of CD11b expression were determined by RT-PCR and Western blot assays.
 |
Table 1 The Sequences of Primers |
Phagocytosis Assay
pMacs and RAW264.7 cells at 1×106 cells/well were cultured in FBS-free DMEM medium in 6-well plates overnight. The cells in each well were added with non-opsonized FITC-labeled T. marneffei at a ratio of 5:1 (conidia spore to macrophage) and the phagocytosis rate was determined using flow cytometry as conventional method.14
In addition, the CD11b specific or control siRNA-transfected RAW264.7 cells were tested for their phagocytosis.
RT-PCR
Macrophages were pretreated with vehicle or anti-CD11b for 1 hr and reacted with heat-killed T. marneffei at a ratio of 5:1 (conidia spores to macrophage) for 1.5 hrs. Total RNA was extracted and reversely transcribed into cDNA by conventional method.14 The primer sequences were forward 5ʹ-CAGATCAACAATGTGACCG TATGG-3ʹ and reverse 5ʹ-CATCATGTCCTTGTACTGCCGC-3ʹ for CD11b; forward 5ʹ-GAGGGAAATCGTGCGTGAC-3ʹ and reverse 5ʹ- CTGGAAGGTGGACAGTGA G-3ʹ for β-actin. The PCR reactions were operated by conventional method.14 The PCR products were resolved by gel electrophoresis on 1–5% gels and analyzed by Quantity One software (Bio-Rad, USA).
Western Blotting
Macrophages were pretreated by RT-PCR. Protein extraction and Western blotting were manipulated by conventional method.14 The relative levels of CD11b to β-actin expression were analyzed by the densitometric scanning using the Image J software.
ELISA Assay
pMacs (1×106 cells/well) were cultured in 6-well plates overnight. The cells were pretreated with vehicle or anti-CD11b for 90 mins or RAW264.7 cells were transfected with individual siRNAs for 24 hrs. The cells were reacted in triplicate with heat-killed conidia spores of T. marneffei at a ratio of 5:1 (conidia spores to macrophage) at 37°C for 2 hrs. The levels of IFN-γ, TNF-α, IL-2, IL-4, IL-6 and IL-10 in the culture supernatants were determined by ELISA using specific kits, according to the manufacturers’ instructions (RayBiotech, USA).
Statistical Analysis
Data are present as the mean ± SD. The difference among the groups was analyzed by ANOVA and post hoc Tukey’s test using the Statistics Package for Social Science program. A P-value of <0.05 was considered statistically significant.
Results
CD11b Expression on Macrophages Is Enhanced by T. marneffei
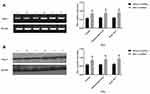
To determine the potential role of CR3 in phagocytosis of T. marneffei, we first examined whether T. marneffei modulated CR3 expression in macrophages in vitro. pMacs from control and immunosuppressed mice and mouse RAW264.7 cells were reacted with, or without, heat-killed T. marneffei at a ratio of 1:5 for 1.5 hrs. As shown in Figure 1A, there was no significant difference in the relative levels of CD11b mRNA transcripts among pMacs from healthy and immunosuppressed mice as well as in RAW264.7 cells. After reaction with T. marneffei, the relative levels of CD11b mRNA transcripts in all groups of macrophages significantly increased. A similar pattern of up-regulated CD11b protein expression was detected in the different groups of macrophages before and after T. marneffei treatment (Figure 1B).
Next, we tested whether T. marneffei spores and yeast could also enhance CD11b expression in macrophages. We found that incubation with either heat-killed or living spores or yeast also enhanced significant CD11b mRNA transcripts and protein expression in pMacs cells in vitro (Figure 2). These indicated that T. marneffei and its spores and yeast up-regulated CD11b expression in macrophages.
Murine Macrophage Requires CD11b to Recognize Talaromyces marneffei
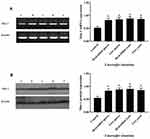
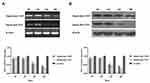
To determine the function of CD11b, RAW264.7 cells were transfected with individual CD11b specific or control siRNA for 48 hrs and the relative levels of CD11b expression were determined by RT-PCR and Western blot assays. Transfection with CD11b-specific siRNA 1369 reduced the relative levels of CD11b mRNA transcripts and protein expression and transfection with 3511 greatly diminished CD11b expression in RAW264.7 cells (Figure 3). We further tested the role of CD11b in phagocytosis of T. marneffei in vitro. RAW264.7 cells and pMacs from control and immunosuppressed mice were incubated with FITC-labeled T. marneffei. The percentages of macrophages that phagocytized T. marneffei in different groups of cells were determined by flow cytometry. The percentages of FITC+ pMac from normal and immunosuppressed BALB/c mice, and RAW264.7 cells, but the control cells without incubation with T. marneffei, were 91.63%±2.59, 89.26%±3.48, 65.75%±2.81, respectively (Figure 4). Pre-treatment with anti-CD11b reduced the percentages of FITC+ pMacs to 35.45%±3.15 (Figure 4). Similarly, transfection with CD11b-specific siRNA 3511, but not control 574, decreased the percentages of FITC+ pMacs in vitro. Together, these data clearly indicated that Murine macrophage requires CD11b to recognize Talaromyces marneffei.
CD11b Is Crucial for T. marneffei Induced Cytokine Responses in Macrophages
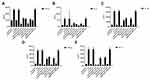
Finally, we examined the impact of CD11b on T. marneffei stimulated cytokine responses in macrophages. Different groups of macrophages were stimulated with T. marneffei and the levels of IFN-γ, TNF-α, IL-2, IL-4, IL-6 and IL-10 in the culture supernatants were determined by ELISA. There was no detectable IL-2 in different groups of macrophages (data not shown). There was very low levels of IFN-γ, TNF-α, IL-4, IL-6 and IL-10 in the supernatants of cultured control macrophages, the levels of IFN-γ, TNF-α, IL-4, IL-6 and IL-10 were significantly elevated in the supernatants of cultured macrophages that had been stimulated with T. marneffei (Figure 5). Furthermore, the levels of IFN-γ, TNF-α, IL-4, IL-6 and IL-10 in the supernatants of cultured macrophages from immunosuppressed mice were lower than from control mice regardless of T. marneffei stimulation. In addition, pre-treatment with anti-CD11b or knockdown of CD11b by its specific siRNA 3511 significantly mitigated T. marneffei-stimulated IFN-γ, TNF-α, IL-4, IL-6 and IL-10 production by macrophages. Therefore, CD11b was crucial for T. marneffei-stimulated IFN-γ, TNF-α, IL-4, IL-6 and IL-10 in macrophages in vitro.
Discussion
CR3 is one of the PRRs through which innate immune cells, such as macrophages can recognize PAMP on pathogenic microbiota. A previous study has indicated that Dectin-1 and CD3 act as major fungal receptors for beta-glucan. Dectin-1 can activate CR3 that is coordinated by the integrins to enhance phagocytosis and antifungal immunity.15 Actually, CR3 can through the I domain and the C-terminal lectin-like site on the α (CD11b) subunit, and the β (CD18) subunit to recognize and phagocytoses pathogens, such as Mycobacterium tuberculosis, Candida albicans and Histoplasma capsulatum.8,16-22 Actually, a previous study has indicated that CR3 and other pathogen recognition receptors (PRRs) in human monocytes are important for the adhesion of Penicillium marneffei, which stimulates monocyte activation and cytokine production in a PRR-dependent manner.23 In this study, we explored the importance of CD11b in phagocytosis of T. marneffei by mouse macrophages. We found that T. marneffei up-regulated CD11b expression on mouse macrophages regardless of whether macrophages were isolated from healthy and immunosuppressed mice. Similarly, either heat-killed or living spores or yeast cells from T. marneffei also significantly upregulated CD11b expression pMacs cells in vitro. These findings extended our previous findings that T. marneffei up-regulated TLR2, TLR-4 and dectin-1 expression on mouse macrophages.24 The up-regulation of CR3, TLR2, TLR4 and dectin-1 expression is usually associated with increased activation of macrophages and may reflect innate immune responses to T. marneffei by potent phagocytosis and destruction of infected T. marneffei. Srinoulprasert et al have studied the involvement of CD11b/CD18 as a phagocytic receptor relevant for T. marneffei uptake by human macrophages. The results demonstrate that various PRR such as CD11b, CD18, MR, TLR1, TLR2 and TLR4 on human monocytes participate in the initial recognition of T. marneffei conidia.23
Furthermore, we found that incubation of macrophages with T. marneffei promoted phagocytosis of T. marneffei, which was abrogated or significantly mitigated by pretreatment with anti- CD11b or knockdown of CD11b by CR3-specific siRNA on macrophages. These findings clearly indicated that CD11b was an important receptor for binding and phagocytosis of T. marneffei by mouse macrophages. Given that there are other receptors on macrophages are crucial for phagocytosis of fungus and CR3 can crosstalk with FcγRIIIB (CD16),25,26 CD14, TLR2,27 and dectin-1.28 Coccidioides posadasii,29 Aspergillus,30,31 mycobacteria,32 Sporothrix schenckii,33 Cryptococcus neoformans34 and the strong phagocytosis of T. marneffei by mouse macrophages may stem from the collaboration of multiple receptors. Heinsbroek et al35 have shown that CR3 can collaborate with dectin-1 and MR for the phagocytosis of C.albicans. The dectin-1 is considered as the fungal pattern recognition receptor to recognize structurally diverse ligands and defense fungus infection.36,37 CR3 can interact with FcγR to promote phagocytosis of fungus in a Sky-dependent manner in some types of cells.38–40 We are interested in further investigating how CR3 collaborate with other receptors such as MR for phagocytosis of T. marneffei by macrophages during the T. marneffei infection.
T. marneffei infection can activate innate immune cells, such as macrophages and the activated macrophages can secrete cytokines. In this study, we found that incubation with T. marneffei stimulated high levels of IFN-γ, TNF-α, IL-4, IL-10, and IL-12 in mouse macrophages, which also mitigated and abrogated by pre-treatment with anti-CD11b or knockdown of CD11b in macrophages. Hence, CR3 was important receptor not only for phagocytosis of T. marneffei, but also for T. marneffei-stimulated inflammatory responses in macrophages. It is possible that T. marneffei may activate mouse macrophages by enhancing the expression of CD11b and other receptors as well as cytokine production. The blocking CD11b by pre-treatment with anti-CD11b or knockdown of CD11b expression may diminish the activation of macrophages induced by T. marneffei, reducing cytokine production by macrophages. It is notable that while infection with microbial pathogens usually induces pro-inflammatory cytokine production by innate immune cells. However, we detected high levels of pro-inflammatory IFN-γ, TNF-α, and IL-12 as well as anti-inflammatory IL-4 and IL-10 produced by mouse macrophages. These data suggest that infection with T. marneffei not only induced pro-inflammatory I type macrophages, but also alternatively activated II types of macrophages, even regulatory macrophages. These may reflect homeostasis of inflammatory responses to defense T. marneffei infection and limit organ destruction.
Conclusion
In summary, our data indicated that T. marneffei upregulated CD11b expression on mouse macrophages. Incubation of T. marneffei promoted phagocytosis of T. marneffei by macrophages and high levels of pro-inflammatory and anti-inflammatory cytokine production by macrophages, which were mitigated and abrogated by pre-treatment with anti-CD11b or knockdown of CD11b expression. These indicated that murine macrophage requires CD11b to recognize Talaromyces marneffei and their cytokine responses to T. marneffei. Our findings may provide new insights into innate immune responses to T. marneffei infection.
Acknowledgment
We sincerely thank Dr. Mai Junhua for his assistance in English revision.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the grants from the Key Projects of NSFC-Guangdong Joint Fund (U0932003) and the Natural Science Foundation of Guangdong Province (2015A030310035).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249.
2. Chan JF, Lau SK, Yuen KY, Woo PC. Talaromyces (Penicillium) marneffei infection in non-HIV-infected patients. Emerg Microbes Infect. 2016;5:e19. doi:10.1038/emi.2016.18
3. Vanittanakom N, Cooper CR
4. Cooper CR, Vanittanakom N. Insights into the pathogenicity of Penicillium marneffei. Future Microbiol. 2008;3(1):43–55. doi:10.2217/17460913.3.1.43
5. Hu Y, Zhang J, Li X, et al. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia. 2013;175(1–2):57–67. doi:10.1007/s11046-012-9577-0
6. Jang JH, Shin HW, Lee JM, et al. An overview of pathogen recognition receptors for innate immunity in dental pulp. Mediators Inflamm. 2015;2015:794143. doi:10.1155/2015/794143
7. Ross GD, Vetvicka V. CR3 (CD11b, CD18): a phagocyte and NK cell membrane receptor with multiple ligand specificities and functions. Clin Exp Immunol. 1993;92(2):181–184. doi:10.1111/j.1365-2249.1993.tb03377.x
8. Le Cabec V, Carréno S, Moisand A, et al. Complement receptor 3 (CD11b/CD18) Mediates Type I and Type II phagocytosis during nonopsonic and opsonic phagocytosis, respectively. J Immunol. 2002;169(4):2003–2009. doi:10.4049/jimmunol.169.4.2003
9. Yefenof E. Complement receptor 3 (CR3): a public transducer of innate immunity signals in macrophages. Adv Exp Med Biol. 2000;479:15–25.
10. Tandon R, Sha’afi RI, Thrall RS. Neutrophil beta2-integrin upregulation is blocked by a p38 MAP kinase inhibitor. Biochem Biophys Res Commun. 2000;270(3):858–862.
11. Cox D, Dale BM, Kashiwada M, et al. A regulatory role for Src homology 2 Domain–Containing Inositol 5′-Phosphatase (Ship) in Phagocytosis Mediated by Fcγ Receptors and Complement Receptor 3 (αMβ2; Cd11b/Cd18). J Exp Med. 2001;193(1):61–71. doi:10.1084/jem.193.1.61
12. Thornton BP, Vĕtvicka V, Pitman M, et al. Analysis of the sugar specificity and molecular location of the beta-glucan-binding lectin site of complement receptor type 3 (CD11b/CD18). J Immunol. 1996;156(3):1235–1246.
13. Xia Y, Ross GD. Generation of recombinant fragments of CD11b expressing the functional beta-glucan-binding lectin site of CR3 (CD11b/CD18). J Immunol. 1999;162(12):7285–7293.
14. Lu S, Hu Y, Lu C, et al. Development of in vitro macrophage system to evaluate phagocytosis and intracellular fate of Penicillium marneffei conidia. Mycopathologia. 2013;176(1–2):11–22. doi:10.1007/s11046-013-9650-3
15. Li X, Utomo A, Cullere X, et al. The β-glucan receptor Dectin-1 activates the integrin Mac-1 in neutrophils via Vav protein signaling to promote Candida albicans clearance. Cell Host Microbe. 2011;10(6):603–615. doi:10.1016/j.chom.2011.10.009
16. Ehlers MR. CR3: a general purpose adhesion-recognition receptor essential for innate immunity. Microbes Infect. 2000;2(3):289–294. doi:10.1016/S1286-4579(00)00299-9
17. van Bruggen R, Drewniak A, Jansen M, et al. Complement receptor 3, not Dectin-1, is the major receptor on human neutrophils for beta-glucan-bearing particles. Mol Immunol. 2009;47(2–3):575–581. doi:10.1016/j.molimm.2009.09.018
18. Zerria K, Jerbi E, Hammami S, et al. Recombinant integrin CD11b A-domain blocks polymorphonuclear cells recruitment and protects against skeletal muscle inflammatory injury in the rat. Immunology. 2006;119(4):431–440. doi:10.1111/imm.2006.119.issue-4
19. Obregón-Henao A, Henao-Tamayo M, Orme IM, Ordway DJ. Gr1(int)CD11b+ myeloid-derived suppressor cells in Mycobacterium tuberculosis infection. PLoS One. 2015;8(11):e80669. doi:10.1371/journal.pone.0080669
20. Paulovičová E, Bujdáková H, Chupáčová J, et al. Humoral immune responses to Candida albicans complement receptor 3-related protein in the atopic subjects with vulvovaginal candidiasis. Novel sensitive marker for Candida infection. FEMS Yeast Res. 2015;15(2):fou001. doi:10.1093/femsyr/fou001
21. Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170(1):487–494. doi:10.4049/jimmunol.170.1.487
22. Lin JS, Huang JH, Hung LY, et al. Distinct roles of complement receptor 3, Dectin-1, and sialic acids in murine macrophage interaction with Histoplasma yeast. J Leukoc Biol. 2010;88(1):95–106. doi:10.1189/jlb.1109717
23. Srinoulprasert Y, Pongtanalert P, Chawengkirttikul R. Engagement of Penicillium marneffei conidia with multiple pattern recognition receptors on human monocytes. Microbiol Immunol. 2009;53(3):162–172. doi:10.1111/j.1348-0421.2008.00102.x
24. Zhao WJ, Liyan X, Ma L. [Effect of Penicillium marneffei on TLR-2, TLR-4, and Dectin-1 expression and TNF-alpha production in macrophage]. Nan Fang Yi Ke Da Xue Xue Bao. 2008;28(1):37–40. Chinese.
25. Krauss JC, Xue W, Mayo-Bond L, et al. Reconstitution of antibody-dependent phagocytosis in fibroblasts expressing Fc-receptor IIIB and the complement receptor type 3. J Immunol. 1994;153(4):1769–1777.
26. Peyron P, Bordier C, N’Diaye EN, et al. Nonopsonic phagocytosis of Mycobacterium kansasii by human neutrophils depends on cholesterol and is mediated by CR3 associated with glycosylphosphatidylinositol- anchored proteins. J Immunol. 2000;165(9):5186–5191. doi:10.4049/jimmunol.165.9.5186
27. Sendide K, Reiner NE, Lee JS, et al. Cross-talk between CD14 and complement receptor 3 promotes phagocytosis of mycobacteria: regulation by phosphatidylinositol 3-kinase and cytohesin-1. J Immunol. 2005;174(7):4210–4219. doi:10.4049/jimmunol.174.7.4210
28. Willcocks S, Offord V, Seyfert HM, et al. Species-specific PAMP recognition by TLR2 and evidence for species-restricted interaction with Dectin-1. J Leukoc Biol. 2013;94(3):449–458. doi:10.1189/jlb.0812390
29. Viriyakosol S, Fierer J, Brown GD, et al. Innate immunity to the pathogenic fungus Coccidioides posadasii is dependent on Toll-like receptor 2 and Dectin-1. Infect Immun. 2005;73(3):1553–1560. doi:10.1128/IAI.73.3.1553-1560.2005
30. Luther K, Torosantucci A, Brakhage AA, et al. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the Dectin-1-glucan receptor and Toll-like receptor 2. Cell Microbiol. 2007;9(2):368–381. doi:10.1111/j.1462-5822.2006.00796.x
31. Cho SY, Kwon EY, Choi SM, et al. Immunomodulatory effect of mesenchymal stem cells on the immune response of macrophages stimulated by Aspergillus fumigatus conidia. Med Mycol. 2016;54(4):377–383. doi:10.1093/mmy/myv110
32. Yadav M, Schorey JS. The β-glucan receptor Dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108(9):3168–3175. doi:10.1182/blood-2006-05-024406
33. Alegranci P, de Abreu Ribeiro LC, Ferreira LS, et al. The predominance of alternatively activated macrophages following challenge with cell wall peptide-polysaccharide after prior infection with Sporothrix schenckii. Mycopathologia. 2013;176(1–2):57–65. doi:10.1007/s11046-013-9663-y
34. Stukes S, Coelho C, Rivera J, et al. The membrane phospholipid binding protein annexin A2 promotes phagocytosis and nonlytic exocytosis of Cryptococcus neoformans and impacts survival in fungal infection. J Immunol. 2016;197(4):1252–1261. doi:10.4049/jimmunol.1501855
35. Heinsbroek SE, Taylor PR, Martinez FO, et al. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 2008;4(11):e1000218. doi:10.1371/journal.ppat.1000218
36. Huysamen C, Brown GD. The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol Lett. 2009;290(2):121–128. doi:10.1111/j.1574-6968.2008.01418.x
37. Johnson EE, Srikanth CV, Sandgren A, et al. Siderocalin inhibits the intracellular replication of Mycobacterium tuberculosis in macrophages. FEMS Immunol Med Microbiol. 2010;58(1):138–145. doi:10.1111/j.1574-695X.2009.00622.x
38. Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106(7):2543–2550. doi:10.1182/blood-2005-03-1239
39. Huang JH, Lin CY, Wu SY, et al. CR3 and Dectin-1 Collaborate in Macrophage Cytokine Response through Association on Lipid Rafts and Activation of Syk-JNK-AP-1 Pathway. PLoS Pathog. 2015;11(7):e1004985. doi:10.1371/journal.ppat.1004985
40. Tohyama Y, Yamamura H. Protein tyrosine kinase, syk: a key player in phagocytic cells. J Biochem. 2009;145(3):267–273. doi:10.1093/jb/mvp001
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.