Back to Journals » Cancer Management and Research » Volume 13
MRPL13 Act as a Novel Therapeutic Target and Could Promote Cell Proliferation in Non-Small Cell Lung Cancer
Authors Jing CQ, Fu R, Wang C, Li XR, Zhang W
Received 21 April 2021
Accepted for publication 1 July 2021
Published 12 July 2021 Volume 2021:13 Pages 5535—5545
DOI https://doi.org/10.2147/CMAR.S316428
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Chuanqing Jing,1,* Rong Fu,1,* Can Wang,2 Xiurong Li,3 Wei Zhang4
1The First Clinical Medical College of Shandong University of Traditional Chinese Medicine, Jinan, People’s Republic of China; 2Shandong University of Traditional Chinese Medicine, Jinan, People’s Republic of China; 3Department of Oncology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, People’s Republic of China; 4Department of Respiratory Medicine, Affiliated Hospital of Shandong University of Chinese Medicine, Jinan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Zhang
Department of Respiratory Medicine, Affiliated Hospital of Shandong University of Chinese Medicine, Jinan, People’s Republic of China
Tel +86 18805317716
Email [email protected]
Background: The latent involvement of MRPL13 in non-small cell lung cancer (NSCLC) remains unclear. This study aimed to explore the role of MRPL13 in NSCLC.
Methods: All analyses were performed in R software 4.0, SPSS version 23, and GraphPad Prism 8. The “limma” package was used to identify differentially expressed genes. Univariate and multivariate cox analyses were used to identify prognosis-related genes. A549 and H1299 lung cancer cell lines were selected for phenotypic experiments.
Results: The high level of MRPL13 was correlated with poor T classification and overall survival. In vitro experiments showed that MRPL13 was highly expressed in NSCLC tissue and cell lines. MRPL13 knockdown inhibited the proliferation of lung cancer A549 and H1299 cell lines, which was further validated by in vivo experiment. Moreover, GSEA analysis suggested that the pathway of MYC target, PI3K/AKT/mTOR/ signaling, oxidative phosphorylation, and G2/M checkpoints may be the potential pathway where MRPL13 was involved. Meanwhile, MRPL13 demonstrated a negative correlation with M1 macrophage, CD8+ T cells, and CD4+ T cells, making it an underlying immunotherapy target of NSCLC.
Conclusion: MRPL13 may promote the proliferation of NSCLC cells and serve as an independent tumor marker and an emerging therapeutic target.
Keywords: MRPL13, non-small cell lung cancer, proliferation
Introduction
Lung cancer is one of the most common malignant tumors and an extreme threat to human survival worldwide; approximately 2.1 million new cases and 1.8 million lung cancer-related deaths are recorded per year.1 According to the latest data of GLOBOCAN, lung cancer ranks first in all cancer types with 11.6% of the total cases and 18.4% of cancer deaths. Moreover, lung cancer is still the leading cause of cancer-related death throughout the world.2,3 Non-small cell lung carcinoma (NSCLC) is the most common pathological type of lung cancer and accounts for more than 80% of lung cancer cases.4,5 Despite the great advancement of NSCLC treatment, the prognosis of patients with lung tumor remains suboptimal, especially for advanced cases.6 Hence, identifying novel therapeutic targets and prognosis biomarkers related to NSCLC is of urgent need.
Mitochondrial ribosomal proteins (MRPs), encoded by nuclear genes, exist in the mitochondria of eukaryotic cells. After being transported to the mitochondria, MRPs combine with rRNA to form a mitochondrial ribosome, which translates proteins encoded by the mitochondria itself.7 Thus far, many MRPs are reported to be involved in tumorigenesis.8 Mitochondrial ribosomal protein L13 (MRPL13), a member of the MRP family, is located on chromosome 8 q24.12 and participates in the biosynthesis of mitochondrial proteins. The suppression of MRPL13 is the key to the manifestation of mitoribosome defect-mediated OXPHOS dysfunctions, which can enhance the invasiveness of hepatoma cells.9 In addition, the expression of MRPL13 in breast cancer tissues is significantly higher than that in adjacent tissues. The expression of MRPL13 is closely related to the clinicopathological factors of patients.10 Furthermore, Zhou et al revealed that the level of MRPL13 was significantly up-regulated in the worse disease-free survival group of patients with breast cancer. Hence, MRPL13 could indicate the poor prognosis of breast cancer.11 However, the role of MRPL13 in NSCLC has not been described yet.
With rapid technological development, massive data generated from next-generation sequencing are commonly used in biomedical research. Bioinformatics is a powerful tool for processing such data and deriving useful information. This study aimed to identify novel differentially expressed genes that are associated with NSCLC prognosis and might be therapeutic targets. In this study, we first identified MRPL13 as a candidate gene through a series of bioinformatics analysis. The high level of MPRL13 was associated with poor clinical-pathological features and prognosis. In vitro experiment showed that MRPL13 was highly expressed in NSCLC cell lines and promoted tumor proliferation, which was further validated by in vivo experiment. Overall, our data demonstrated that MRPL13 has the potential to be a valuable target of NSCLC treatment.
Materials and Methods
Data Acquisition and Preprocessing
Bulk transcriptome data as well as clinical and survival information of NSCLC were obtained from the TCGA database (TCGA-LUAD and TCGA-LUSC), a public database that integrates the comprehensive data of 33 types of cancer. In detail, the whole expression profile of 1037 NSCLC samples and 107 paracancerous tissues were downloaded in the “FPKM” form and then converted into the “TPM” form for better comparability with chip data. The clinical data were “bcr-xml” file. Chip data were acquired from the GEO database, including GSE18842, GSE21933, GSE116959, and GSE43458. The detailed information of the included databases is shown in Table 1. Prior to the analysis, all data were preprocessed as follows: 1. gene annotation; 2. background correction; 3. normalization and removing batch effects; and 4. supplementing missing values.
 |
Table 1 The Detailed Information of Databases Included in the Analysis |
Differentially Expressed Genes (DEGs) Identification and Prognosis Analysis
DEGs between tumor and normal tissues were analyzed using “limma” package in R software with the threshold of |logFC (fold-change)|>1 and P-value <0.05. Intersection analysis was conducted to identify common DEGs among TCGA, GSE18842, and GSE21933. Univariate and multivariate cox analyses were performed to screen genes associated with prognosis, with the threshold set as P-value < 0.05. Kaplan-Meier survival curves were used to visualize patients in different groups.
Immunohistochemistry (IHC)
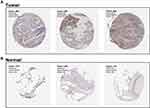
The IHC images of MRPL13 in NSCLC tissue were obtained from The Human Protein Atlas database, an open-accessed project that maps all the human proteins in cells, tissues, and organs. The antibody of MRPL13 in lung cancer and normal lung tissues was HPA060899.
Gene Set Variation Analysis (GSVA)
GSEA analysis was conducted to explore the underlying biological pathway of MRPL13 in tumor development. The GSVA package in R software was used to perform GSVA analysis with “ssGSEA” method. The gene set was the “Hallmark” signature.
Immune Infiltration Analysis
The tumor immune microenvironment was quantitatively evaluated using the “CIBERSOT” algorithm developed by Newman et al, a powerful tool that utilizes expression profiles to estimate the abundance of member cell types in a mixed cell population. The parameters of CIBERSORT were set as follows: the number of permutations was “100”; the kappa was “999”; the q-value was “0.25”.
Cell Line and qPCR Analysis
Normal lung epithelial cell line BEAS-2B and four NSCLC cell lines, namely, A549, H23, H1299, and H520 were purchased from iCell (Shanghai, China). All the five cell lines were incubated in a 5% CO2 humidified incubator at 37 °C. The total RNA of all cell lines was isolated using Trizol (Invitrogen, Carlsbad, CA, USA). cDNA was reversely transcribed from the total RNA by using a reverse transcription kit (Promega) and PrimeScript RT Master Mix (Takara, Japan). The target gene was measured with SYBR Green assay in the 96-well plates following the manufacturer’s instructions (Applied Biosystems, Carlsbad, CA, USA). The primers used were as follows: MRPL13, forward: 5′-ACATAAACCTGTGTACCATGCAC-3′; MRPL13, reverse: 5′- GGTAGCCAGTATGCGAAGAGT-3′; GAPDH, forward: 5′-AC CACAGTCCATGCCATCAC-3′; and GAPDH, reverse: 5′-TCCACCACCCTG TTGCTGTA-3′.
Western Blot Analysis
Total proteins were extracted using the Total Protein Extraction Kit (Beyotime, P0013; Beijing, China). Protein concentration was determined with a BCA reagent kit (Pierce™ BCA Protein Assay Kit). SDS-PAGE gels were run with 8–12% gradient and transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were incubated with the diluted primary antibodies (anti-MRPL13, anti-caspase-3, anti-GAPDH, anti-Bax, and anti-Bcl-2) at 4 °C overnight. The second antibodies were incubated with the membranes for 2 h at RT (anti-rabbit or anti-mouse IgG (H+L), CST, USA).
Gene Transfection and RNAi
Control siRNA and MRPL13 siRNA were purchased from GenePharma. A549 and H1299 cell lines were transfected with control siRNA and siRNA-MRPL13 by using Lipofectamine 2000 (Invitrogen). The target sequence of shRNA MRPL13 was 5′-GGGATCCAGTGGCAATTGT-3′.
CCK8 Assay
CCK8 assays was performed using a CCK8 Kit (Dojindo, Shanghai, China). The cells were resuspended and cultivated in 96-well plates at a concentration of 1×104 cells per well. All the plates were incubated at 37 °C for 1 h. ELISA plate reader (BioRad 680 ELISA Reader) was used to record absorbance at OD 450 nm.
Colony Formation Assay
Cell proliferation ability was assessed using colony formation assay. Cells were inoculated into 6-well plates with 250 cells/mL and cultured for 14 days. The medium was changed every 2–3 days. The number of cell was conducted after 14 days.
Flow Cytometry Assay
Flow cytometry was performed for cell apoptosis detection (FACSCalibur) with Annexin V/propidium iodide (PI) staining. Cells were washed with phosphate-buffered saline (PBS) and resuspended in 1× binding buffer. FlowJo software was used for data processing.
In vivo Tumor Xenografts
Cells were trypsinized with 0.25% trypsin (Invitrogen) and resuspended in PBS. Afterward, 0.4 mL of the cell suspension was inoculated into the armpit of 40-week-old male BALB/c nude mice. The mice were randomly divided into control and shRNA groups. On day 20, all the mice were sacrificed, and tumors were isolated.
Statistical Analysis
All the data were analyzed using R software 4.0, SPSS version 23 and GraphPad Prism 8. P-value < 0.05 was regarded statistically significant. All the experiments were repeated three times. Data were presented with mean ± standard Deviation (SD). For the continuous variables following the normal distributions, an independent t-test was used to compare the statistical difference between two groups. Mann–Whitney U-test was used to compare the continuous variables that were not normally distributed.
Results
Identification of DEGs
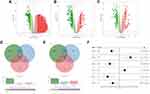
Genes that are differentially expressed in tumor and corresponding paracancerous tissues in TCGA, GSE18842, and GSE21933 datasets were analyzed using the “limma” package with the threshold of |logFC| > 1 and P-value < 0.05 (Figure 1A–C). Forty-three common upregulated genes and 483 down-regulated genes were identified (Figure 1D and E).
Screening of Prognosis-Related Genes and Clinical Correlation
We then correlated the gene expression data of the 916 DEGs with survival outcomes. Univariate and multivariate cox analyses were performed to identify prognosis-related genes (Table S1 and Figure 1F). The genes IGSF10, MRPL13, MMP14, FLRT3, RAD51, and DAAM2 were finally identified. MRPL13 has not been reported in NSCLC and was therefore selected for further analysis. Based on the public data of TCGA, GSE116959, and GSE43458, MRPL13 has higher expression level in tumor tissues (Figure 2A–D). Meanwhile, Kaplan-Meier survival curve indicated poor prognosis in the patients highly expressing MRPL13 (Figure 2E, P<0.05). The MRPL13 level was significantly higher in T3-4 than in T1-2 classification but was not significantly associated with N and M classification (Figure 2F–H).
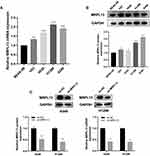
MRPL13 is Upregulated in NSCLC Tissue and Cell Lines
The representative IHC images illustrating MRPL13 protein levels are shown in Figure 3A and B. The results indicated high protein level in NSCLC tissues. The MPRL13 mRNA level increased in NSCLC cell lines, as determined by qPCR analysis (Figure 4A). In parallel, Western blot assay showed a higher protein level of MRPL13 in cancer cell lines (Figure 4B). A549 and H1299 cell lines were transfected with siRNA due to their highest mRNA and protein level of MRPL13. Western blot and qPCR assay revealed the successful knockdown of MRPL13 (Figure 4C).
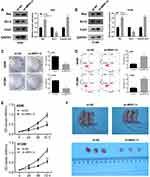
MRPL13 Promotes Proliferation of NSCLC Cells
Considering that the Bax, Bcl2 and cleaved-Cas3 were the vital protein involved in the apoptosis process, we therefore assessed their protein level in control and MRPL13 knockdown cells, which could measure cell proliferation capacity. The Western blot assay indicated that the MRPL13 knockdown decreased the Bcl-2 protein level but increased the levels of Bax and cleaved-Cas3, indicating that MRPL13 might promote the cell proliferation and inhibit apoptosis of lung cancer cells (Figure 5A and B). Colony formation assay revealed that the knockdown of MRPL13 remarkably decreased the number of colony cells (Figure 5C). In addition, a high apoptosis rate was observed in the sh-MRPL13 group (Figure 5D). The promotion effect of MRPL13 on NSCLC cells was validated by CCK8 assay (Figure 5E). In vivo experiments suggested that mice inoculated with sh-MRPL13 cells developed smaller tumors (Figure 5F).
Pathway Enrichment and Immune Infiltration Analyses
GSVA analysis was performed on patients with high and low MRPL13 in TCGA to explore the potential biological pathway. As shown in Figure 6, in the high MRPL13 phenotype, the pathway of MYC target, mTORC1 signaling, DNA repair, oxidative phosphorylation, E2F target, PI3K/mTOR signaling, and others, were enriched. Increasing lines of evidence has revealed that the recruitment of tumor cells to immune cells could affect tumor progression. Thus, we explored the potential correlation between MRPL13 and multiple immune cells. All the patients were divided into low and high MRPL13 groups. Based on the transcriptional profiling data, the tumor microenvironment was assessed using the “CIBERSOT” algorithm. The result demonstrated that MRP13 was negatively correlated with M1 macrophage, CD8+ T cells, CD4+ memory activated T cells, follicular helper T cells, gamma delta T cells, resting mast cells, monocyte, native B cells, neutrophil, yet positively correlated with memory B cells, CD4+ memory resting T cells, Treg, activated mast cells (Figure 7).
 |
Figure 6 GSVA analysis in high and low MRPL13 group. Abbreviation: GSVA, gene set variation analysis. |
 |
Figure 7 Immune infiltration analysis in high and low MRPL13 group. Notes: *P <0.05, **P<0.01, ***P <0.001. |
Discussion
As the most dominant pathological subtype of lung cancer, NSCLC accounts for approximately 85% of patients with lung cancer and has the largest number of cancer-related deaths worldwide.12 Despite great advances in NSCLC treatment, the 5-year survival rate of patients with lung cancer is still less than 20% in most countries.13 In this regard, novel therapeutic and prognostic targets of NSCLC should be identified. In the study, we first identified DEGs between NSCLC tumors and adjacent normal tissues. MAPL13 has gained research interest because of its tight association with prognosis. Next, we found the high expression level of MAPL3 in NSCLC tissue and cancer cell lines. The knockdown of MRPL13 inhibited the proliferation of NSCLC cancer cells.
To our knowledge, this study is the first to explore the role of MRPL13 in lung cancer. As a mitochondrial coding gene, MRPL13 was previously reported in breast cancer and liver cancer. For example, Zuo et al found that MRPL13 is a poor prognostic biomarker and associated with the immune infiltration status of breast cancer.10 Considering the pivotal role of MRPL13 in the mitochondrion, MRPL13 may also be involved in cell metabolism. Lee et al revealed that the low expression of MRPL13 could result in ribosomal defect, subsequently attenuating mitochondrial oxidative phosphorylation capacity. This phenomenon leads to increased glycolysis and cell invasiveness.9 As one of the core components of cells, the mitochondrion plays a central role in tumor progression, and defects may be the underlying reason for tumorigenesis.14 We observed the abnormal activation of MRPL13 in NSCLC, which may be a promising therapeutic target.
We subsequently performed a pathway enrichment analysis of MPRL13 to explore the biological pathway. MRPL13 was associated with several cancer pathways, including MYC target, PI3K/AKT/mTOR/ signaling, oxidative phosphorylation, and G2/M checkpoints. The MYC oncogene promotes various cancers and is strongly associated with tumor metabolism.15 Enhanced MYC could upregulate L-lactate dehydrogenase A chain (LDHA) expression, a key molecule participating in glycolysis, thereby promoting tumor development.16,17 Additionally, overexpressed MYC leads to a lower ATP level in the cells and activates AMP‐activated kinase (AMPK) to induce metabolic stresses.18 Our analysis and previous study reported the association between MRPL13 and tumor metabolism.9 Therefore, MRPL13 may be involved in the complex interplay of MYC on cancers. The PI3L/AKT/mTOR signaling pathway was regarded as the principal modulator of cancer.19 Zhang et al revealed that the acquisition of epithelial-mesenchymal transition (EMT) and cancer stem cells (CSS) were regulated by the PI3K/AKT/mTOR signaling pathway in prostate cancer.20 Maric et al found that GPNMB could facilitate tumor growth and metastasis by enhancing the PI3K/AKT/mTOR pathway signaling and β-catenin activity.21 The G2/M checkpoint is an essential link in cell proliferation and mitosis.22 Su et al demonstrated that diallyl disulfide (DADS) could induce G2/M checkpoint arrest through the Cdc25C/cyclin B1 pathway mediated by Chk1 phosphorylation.23 Chen et al found that CtIP could promote G2/M phase arrest and enhance the cell sensitivity to Eto through the ATR/Chk1/CDC25C pathway.24
Our result also found that MRPL13 significantly decreased the component of M1 macrophages, CD8+ T cells, and CD4+ T cells in the NSCLC tumor microenvironment. Yuan et al suggested that M1 macrophages could suppress tumor development by inducing apoptosis, whereas M2a/M2c macrophages can promote lung cancer invasion and xenograft tumor growth.25 CD8+ T cells, an important type of immune cells, could produce cytotoxic molecules, such as perforin and granzymes, to kill tumor cells.26 Activated CD8+ T cells participate in oxidative phosphorylation, glycolysis, and glutaminolysis, which are important for acquiring effector functions.27 Zander et al demonstrated that CD4+ T cells might enhance the kill effect of CD8+ T cells on tumor cells with the help of IL21.28
This study has some limitations. First, the clinical information of the samples included in our analysis is limited, which would bring potential bias for the final result. Thus, detailed clinical information is needed to reduce bias in subsequent studies. Second, data obtained from TCGA were predominantly originated from Western countries. Our conclusion may not apply to Asian countries. Subsequent studies should validate these results in large Asian populations. Third, the mechanisms by which MRPL13 promoted the proliferation of lung cancer need to be further investigated.
Conclusion
We first identified MRPL13 as a prognosis-related gene in NSCLC. The qPCR and Western blot results showed that MRPL13 is highly expressed in A549 and H1299 cell lines than in normal BEAS-2B cell line. In vitro and in vivo experiments showed MRPL13 could promote proliferation in NSCLC, which has not been reported previously. The underlying interaction between MRPL13 with multiple immune cells makes it a potential target for NSCLC immunotherapy.
Funding
This study was supported by Taishan character construction project special fund support project (ts201712096).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Brody H. Lung cancer. Nature. 2014;513(7517):S1. doi:10.1038/513S1a
2. Henley SJ, Ward EM, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2020;126(10):2225–2249. doi:10.1002/cncr.32802
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
4. Lancet T. Lung cancer: some progress, but still a lot more to do. Lancet. 2019;394(10212):1880. doi:10.1016/S0140-6736(19)32795-3
5. Slatore C, Sockrider M. Lung cancer prevention. Am J Respir Crit Care Med. 2014;190(10):P7–P8.
6. DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271.
7. Greber BJ, Bieri P, Leibundgut M, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348(6232):303–308. doi:10.1126/science.aaa3872
8. Li Z, Akar S, Yarkan H, et al. Genome-wide association study in Turkish and Iranian populations identify rare familial Mediterranean fever gene (MEFV) polymorphisms associated with ankylosing spondylitis. PLoS Genet. 2019;15(4):e1008038. doi:10.1371/journal.pgen.1008038
9. Lee YK, Lim JJ, Jeoun UW, et al. Lactate-mediated mitoribosomal defects impair mitochondrial oxidative phosphorylation and promote hepatoma cell invasiveness. J Biol Chem. 2017;292(49):20208–20217. doi:10.1074/jbc.M117.809012
10. Tao Z, Suo H, Zhang L, et al. MRPL13 is a prognostic cancer biomarker and correlates with immune infiltrates in breast cancer. Onco Targets Ther. 2020;13:12255–12268. doi:10.2147/OTT.S263998
11. Zhou X, Xiao C, Han T, et al. Prognostic biomarkers related to breast cancer recurrence identified based on logit model analysis. World J Surg Oncol. 2020;18(1):254. doi:10.1186/s12957-020-02026-z
12. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
13. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546. doi:10.1038/nrc3775
14. Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61(5):667–676. doi:10.1016/j.molcel.2016.02.011
15. Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi:10.1016/j.cell.2012.03.003
16. Shim H, Chun YS, Lewis BC, Dang CV. A unique glucose-dependent apoptotic pathway induced by c-Myc. Proc Natl Acad Sci U S A. 1998;95(4):1511–1516. doi:10.1073/pnas.95.4.1511
17. Wong WJ, Qiu B, Nakazawa MS, Qing G, Simon MC. MYC degradation under low O2 tension promotes survival by evading hypoxia-induced cell death. Mol Cell Biol. 2013;33(17):3494–3504. doi:10.1128/MCB.00853-12
18. von Eyss B, Jaenicke LA, Kortlever RM, et al. A MYC-driven change in mitochondrial dynamics limits YAP/TAZ function in mammary epithelial cells and breast cancer. Cancer Cell. 2015;28(6):743–757. doi:10.1016/j.ccell.2015.10.013
19. Xia P, Xu XY. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res. 2015;5(5):1602–1609.
20. Chang L, Graham PH, Hao J, et al. Acquisition of epithelial-mesenchymal transition and cancer stem cell phenotypes is associated with activation of the PI3K/Akt/mTOR pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4(10):e875. doi:10.1038/cddis.2013.407
21. Maric G, Annis MG, MacDonald PA, et al. GPNMB augments Wnt-1 mediated breast tumor initiation and growth by enhancing PI3K/AKT/mTOR pathway signaling and β-catenin activity. Oncogene. 2019;38(26):5294–5307. doi:10.1038/s41388-019-0793-7
22. Akaike Y, Chibazakura T. Aberrant activation of cyclin A-CDK induces G2/M-phase checkpoint in human cells. Cell Cycle. 2020;19(1):84–96. doi:10.1080/15384101.2019.1693119
23. Bo S, Hui H, Li W, et al. Chk1, but not Chk2, is responsible for G2/M phase arrest induced by diallyl disulfide in human gastric cancer BGC823 cells. Food Chem Toxicol. 2014;68:61–70. doi:10.1016/j.fct.2014.03.007
24. Chen H, Shan J, Chen D, et al. CtIP promotes G2/M arrest in etoposide-treated HCT116 cells in a p53-independent manner. J Cell Physiol. 2019;234(7):11871–11881. doi:10.1002/jcp.27824
25. Yuan A, Hsiao YJ, Chen HY, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. 2015;5:14273. doi:10.1038/srep14273
26. Tsukumo SI, Yasutomo K. Regulation of CD8(+) T cells and antitumor immunity by notch signaling. Front Immunol. 2018;9:101. doi:10.3389/fimmu.2018.00101
27. Zhang L, Romero P. Metabolic control of CD8(+) T cell fate decisions and antitumor immunity. Trends Mol Med. 2018;24(1):30–48. doi:10.1016/j.molmed.2017.11.005
28. Zander R, Schauder D, Xin G, et al. CD4(+) T cell help is required for the formation of a cytolytic CD8(+) T cell subset that protects against chronic infection and cancer. Immunity. 2019;51(6):1028–1042.e1024. doi:10.1016/j.immuni.2019.10.009
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.