Back to Journals » Infection and Drug Resistance » Volume 15
Molecular Mechanisms Mediating Ceftazidime/Avibactam Resistance Amongst Carbapenem-Resistant Klebsiella pneumoniae Isolates from Cancer Patients
Authors El-Kady RAE, Elbaiomy MA , Elnagar RM
Received 4 August 2022
Accepted for publication 3 October 2022
Published 14 October 2022 Volume 2022:15 Pages 5929—5940
DOI https://doi.org/10.2147/IDR.S384972
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Rania Abd El-Hamid El-Kady,1,2 Mohamed Ali Elbaiomy,3 Rasha Mokhtar Elnagar1
1Department of Medical Microbiology and Immunology, Faculty of Medicine, Mansoura University, Mansoura, Egypt; 2Department of Pathological Sciences, Fakeeh College for Medical Sciences, Jeddah, Kingdom of Saudi Arabia; 3Medical Oncology Unit, Oncology Center, Mansoura University, Mansoura, Egypt
Correspondence: Rania Abd El-Hamid El-Kady, Department of Pathological Sciences, Fakeeh College for Medical Sciences, P.O. Box 2537, Jeddah, 21461, Kingdom of Saudi Arabia, Tel +966 569849897, Email [email protected]
Background: A growing body of evidence suggests that ceftazidime/avibactam (CZA) is a potential therapeutic option for carbapenem-resistant Klebsiella pneumoniae (CRKP) infections; however, resistant strains are increasingly emerged worldwide. Herein, we deemed to investigate the susceptibility profile of CRKP isolates from cancer patients to CZA and to identify the underlying resistance mechanisms.
Methods: Clinical samples were obtained from adult patients admitted to the Oncology Center of Mansoura University, Mansoura, Egypt. The antibiotic susceptibility pattern of K. pneumoniae isolates to different antibiotics was tested by the modified Kirby Bauer’s disc diffusion method. Minimum inhibitory concentrations of CZA were assessed using broth microdilution method. Screening for carbapenemase-producing strains was achieved by the modified Hodge test. Multiplex polymerase chain reactions (PCRs) were conducted for uncovering of carbapenemase-encoding genes (blaKPC, blaVIM, blaIMP, blaNDM-1, and blaOXA-48), and outer membrane porin genes (ompK35 and ompK36).
Results: A total of 12 CZA-resistant isolates were identified out of 47 CRKP isolates (25.5%). The MIC50 and MIC90 of CZA against CRKP were 1 and 64 μg/mL, respectively. Risk factors for CZA resistance included chronic kidney disease, mechanical ventilation, longer length of hospital stay, and ICU admission. The multivariate logistic regression demonstrated that longer length of hospital stay (P=0.03) was the only independent predictor for acquisition of CZA-resistant isolates. The leading mechanism for CZA resistance was sustained by blaKPC (50%), meanwhile 16.7% and 8.3% of the CZA-resistant isolates harbored blaOXA-48 and blaOXA-48/blaNDM-1, respectively. The MBL-encoding genes blaNDM-1 and blaIMP were detected in 16.7% and 8.3% of the isolates, respectively. Absence of both ompK35 and ompK36 was observed in 58.3% of the CZA-resistant isolates.
Conclusion: CZA has displayed superior in vitro activity against CRKP isolates in comparison to other antibiotics; however, thorough molecular characterization of resistant strains is highly recommended in future studies to detect and monitor the emergence of further tackling strains.
Keywords: CRKP, ceftazidime/avibactam, blaKPC, OmpK35, OmpK36, cancer
Introduction
Carbapenems are β-lactam antibiotics, similar to penicillins and cephalosporins, with a broad-spectrum of activity against both Gram-positive and Gram-negative bacteria. They are considered the last therapeutic option for life-threatening bacterial infections.1 The term “carbapenem” refers to the 4:5 fused ring lactam of penicillins with a double bond between C-2 and C-3 but with replacement of sulfur by carbon at C-1. They inhibit bacterial cell wall biosynthesis by interfering with transpeptidation reactions.2
In the existing era, increased incidence of carbapenem-resistant Klebsiella pneumoniae (CRKP) infections constitutes a public health concern, because of a dwindling antibiotic pipeline.3 CRKP isolates can emerge due to several causes, amongst which production of carbapenemases is the most prevalent mechanism.4 In the Ambler molecular classification scheme of β-lactamases, carbapenemases are grouped in classes A, B, and D. Both class A and D share a serine residue in the active site, whereas class B are metallo-β-lactamases (MBLs) that require zinc for catalysis.5 K. pneumoniae carbapenemases (KPCs) are the most worrisome carbapenemases of Ambler class A because of their position on self-conjugative plasmids. Although more than 20 KPC types have been identified, KPC-2 and KPC-3 are the most ubiquitous variants.6 To date, imipenemase (IMP), Verona integron-encoded MBL (VIM), and New Delhi MBL (NDM) are the three most frequent MBLs, with several variants have been described worldwide.7 On the other side, oxacillinases (OXA) – class D carbapenemases with up to 30 OXA-48-like deviations – have been discovered so far.8
Infections with CRKP are one of the leading causes of morbidity and mortality in cancer patients.9 As the currently available treatment substitutes are restricted, the discovery of novel antibiotics is of a critical priority to combat infections caused by these superbugs, especially in critically ill patients.10 A few years ago, ceftazidime/avibactam (CZA), a combination of a cephalosporin/non-β-lactam β-lactamase inhibitor, has been approved by Food and Drug Administration (FDA) for treatment of complicated intra-abdominal and urinary tract infections, as well as healthcare-associated pneumonia. Avibactam is a synthetic diazabicyclooctane with no intrinsic antibacterial activity; however, it shields ceftazidime from breakdown by Ambler class A, C, and some class D β-lactamases.11 Current evidence advocates that CZA could be a promising treatment choice for CRKP infections, but resistant strains are evolving rapidly due to multiple mechanisms including mutations in the genes encoding KPC or outer membrane porins OmpK35 and/or OmpK36.12
In literature, there is a dearth of data addressing the activity of CZA against CRKP from cancer patients. In view of that, we designed this study to a) review the susceptibility profile of CRKP isolates from adult patients diagnosed with cancer (both hematologic and solid organ) to CZA, b) determine the risk factors associated with CZA resistance, and c) explore the possible molecular mechanisms underlying the resistant phenotypes.
Materials and Methods
Study Eligibility, Design and Setting
In this prospective cohort study, we enrolled all adult patients (>18 years), with positive cultures for K. pneumoniae, who had been hospitalized in the Oncology Center of Mansoura University (OCMU), Mansoura, Egypt, between January 2020 and December 2021. The OCMU is a 500-bedded health-care facility that delivers tertiary-care for children and adults diagnosed with cancer. Samples from pediatric patients (<18 years) and those with polymicrobial infections were excluded from our cohort to adjust for the risk factors (one isolate/patient).
Sample Collection and Processing
During the study period, different clinical samples were collected from adult patients admitted to the OCMU. All samples were processed in the Microbiology Diagnostics and Infection Control unit (MDICU), Faculty of Medicine, Mansoura University, Egypt, using the standard microbiological protocols. K. pneumoniae isolates were identified to the species level based on their colony morphology on blood and MacConkey’s agar plates (Oxoid, Ltd., Basingstoke, UK), Gram staining characters, and results of biochemical reactions including Kligler iron agar (KIA) test, lysine iron agar (LIA) test, motility, indole, ornithine production (MIO) tests, and citrate utilization test. The identities of the test strains were confirmed with API 20 E (BioMérieux, Inc., Hazelwood, MO). Out of 350 collected samples, 134 yielded a positive growth for K. pneumoniae.
Antibiotic Susceptibility Testing
The susceptibility of K. pneumoniae isolates to different antibiotics was tested using the modified Kirby Bauer’s disc diffusion method on Mueller–Hinton agar plates (Oxoid Ltd., Basingstoke, UK). Antibiotic discs including sulfamethoxazole/trimethoprim (SXT; 25 μg), ampicillin/sulbactam (SAM; 20 μg), piperacillin/tazobactam (TZP; 110 μg), cefepime (FEP; 30 μg), ceftriaxone (CRO; 30 μg), ceftazidime (CAZ; 30 μg), cefotaxime (CTX; 30 μg), aztreonam (ATM; 30 μg), ciprofloxacin (CIP; 10 μg), levofloxacin (LEV; 10 μg), gentamicin (CN; 10 μg), amikacin (AK; 30 μg), imipenem (IPM; 10 μg), and meropenem (MEM; 10 μg) were purchased from Oxoid Ltd. (Basingstoke, UK), while CZA discs (50 μg) were obtained from Liofilchem®, Italy. The CRKP isolates were preliminary identified if they showed resistance (zone diameter ≤19 mm) to at least one of the used carbapenems (imipenem and meropenem). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 (Naval Medical Research Unit Three, Cairo, Egypt; NAMRU-3) were included as quality control strains. All susceptibility results were interpreted as per the recommendations of the Clinical and Laboratory Standards Institute (CLSI).13
Phenotypic Screening for the Production of Carbapenemases
The modified Hodge test (MHT) was done to determine the production of carbapenemases by K. pneumoniae isolates using E. coli ATCC 25922 as the standard strain. For each test, we also included K. pneumoniae ATCC 1705 (carbapenemase-producer) and K. pneumoniae ATCC 1706 (carbapenemase-non producer) as the positive and negative controls, respectively. Results followed the CLSI interpretive criteria,13 with strains forming cloverleaf indentations were characterized as carbapenemase producers. For subsequent molecular testing, the CRKP isolates were stored at –80°C after inoculation into brain–heart infusion (BHI) broth containing 16% glycerol.
Ceftazidime/Avibactam Minimum Inhibitory Concentrations (MICs)
The MICs of CZA were evaluated by the reference broth microdilution (BMD) method, and results were interpreted according to the breakpoints set by the CLSI. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were employed as reference strains for quality control purposes.13
Genomic DNA Extraction from CRKP Isolates
For bacterial DNA extraction, 2–3 colonies of each CRKP strain were diluted into 500 µL distilled water. Boiling was done by incubation at 95°C for 10 minutes in a water bath followed by centrifugation for 10 minutes at 10,000 rpm. The supernatants containing bacterial DNA were then transferred into sterile Eppendorf tubes for the next polymerase chain reaction (PCR) amplifications.14
Molecular Screening for Resistance Genes by Multiplex PCR
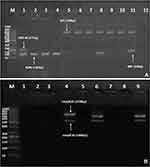
For CRKP isolates, 2 multiplex PCRs were used to detect the genes encoding for the carbapenemases (blaKPC, blaVIM, blaIMP, blaNDM-1, and blaOXA-48), and outer membrane porin genes (ompK35 and ompK36) using primer sets purchased from Sigma, Aldrich.15,16 For each amplification reaction, 50 μL final volumes were used containing 25 μL of 2X GoTaq Green Master Mix (Enzynomics, Korea), 1μL for each of the forward and reverse primers for all genes, 5 μL of the extracted DNA, and DNase free water. DNA amplifications for the carbapenemase-encoding genes were performed with the following thermal cycling conditions: 10 min at 94°C; 36 cycles (30 secs at 94°C, 40 secs at 52°C, and 50 secs at 72°C), with 5 min at 72°C for the final extension.15 Amplification conditions for the ompK35 and ompK36 included 15 min at 95°C; 40 cycles (1 min at 94°C, 1 min at 68°C, and 1 min at 72°C), with 10 min at 72 °C for the final extension.16 DNA fragments were visualized by electrophoresis in a 1.5% agarose gel stained with ethidium bromide dye. A 1000 bp ladder molecular weight marker (Lonza Rockland, Inc., USA) was used to measure the molecular weight of amplified products.
Statistical Analysis
All data were analyzed using IBM®SPSS® Statistics program version 26.0 for Windows (SPSS Inc., Chicago, IL, USA). Pearson’s Chi-Square (χ2) test was employed to analyze categorical variables. The independent samples t-tests were used to analyze the means of two independent groups. Odds ratios (OR) with 95% confidence intervals (CI) were investigated. Univariate and multivariate logistic regression analyses were done to identify the risk factors associated with the development of CRKP and CZA resistance. P-values <0.05 (2-tailed) were considered significant.
Results
Bacterial Isolates and Patients’ Characteristics
During the period of interest, a total of 134 non-duplicate (one isolate/patient), consecutive K. pneumoniae isolates were obtained from the study participants. Regarding the source, 38 (28.4%) isolates were recovered from blood samples, 34 (25.4%) from urine, 25 (18.6%) from sputum, 24 (17.9%) from wound swabs, and 13 (9.7%) from endotracheal aspirates (ETA). The average age of the infected patients was 45.56 ± 14.06 years (range, 23–81 years), of which 88 patients (65.7%) were females, whereas 46 (34.3%) were males. The most common type of hematologic malignancy was chronic lymphocytic leukemia (37.8%), followed by acute myeloid leukemia (26.7%), and non-Hodgkin’s lymphoma (17.8%). Amongst solid organ tumors, hepatocellular carcinoma, breast cancer, and nonspecific connective tissue neoplasm were the most frequent (31.5%, 25.8%, and 22.5%, respectively).
Results of Antibiotic Susceptibility Testing of K. pneumoniae Isolates
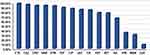
Overall, K. pneumoniae isolates demonstrated the highest in vitro susceptibility to CZA (91.04%), followed by meropenem (69.4%) and imipenem (64.9%). On the other hand, only 3.7% and 1.5% of the test isolates were susceptible to ceftazidime and cefotaxime, respectively (Figure 1). Based on the results of the MHT, 47 (35.1%) K. pneumoniae isolates were found to be carbapenem-resistant, of which 32 (68%) and 15 (32%) isolates were recovered from patients admitted to the medical and surgical wards, respectively. Sample-wise distribution of these isolates showed increased incidence from blood (51.1%), sputum (19.2%), and urine samples (14.8%). On the other side, 8.5% and 6.4% of the isolates were obtained from wound swabs and ETA, respectively. Apart from CZA, antimicrobial susceptibility testing explored high resistance rate of CRKP strains to most of the investigated antibiotics (Table 1).
 |
Table 1 Antimicrobial Susceptibility Profile of the Test CRKP Isolates |
Characteristics and Risk Factors for CZA Resistance in the Study Participants
Out of 47 obtained CRKP isolates, 12 (25.5%) were CZA-resistant, while none of the carbapenem-susceptible K. pneumoniae (CSKP) strains exhibited resistance to CZA. Of the resistant isolates, 66.7% were recovered from blood, 16.7% from urine, 8.3% from ETA, and 8.3% from sputum samples. Amongst patients identified with CZA-resistant CRKP isolates from blood samples, 5 (62.5%) were males, while 3 (37.5%) were females, with an average age of 45.71 ± 11.16 years. Of these isolates, 75% were identified from patients admitted to the medical wards compared to 25% from surgical wards. Risk factors associated with the likelihood of CZA resistance, as shown in Table 2, were chronic kidney disease (OR=11.33, 95% CI: 1.04–122.38; P= 0.018), mechanical ventilation (OR=24.29, 95% CI: 2.45–241.26; P= 0.003), longer length of hospital stay (OR=23.1, 95% CI: 3.7–144.21; P= 0.001), and ICU admission (OR=6.76, 95% CI: 1.59–28.72; P= 0.006). In the multivariate logistic regression analysis, longer length of hospital stay was the only identified predictor for CZA resistance (Table 3). Importantly, no outbreaks of CZA-resistant CRKP isolates were detected during the study period.
 |
Table 2 Risk Factors and Outcome of Ceftazidime/Avibactam Resistance Amongst the Study Cohort |
 |
Table 3 Multivariate Analysis for Predictors of Ceftazidime/Avibactam Resistance Amongst the Study Cohort |
Results of MICs, MIC50, and MIC90 of CZA Against CRKP Isolates
The MICs of CZA inhibiting CRKP isolates ranged from 0.25 to 128 µg/mL. The MIC50 and MIC90 values of CZA were 1 and 64 µg/mL, respectively (Table 4). Among the CZA-sensitive isolates, the MICs ranged from 0.25 to 4 µg/mL compared to 16–128 µg/mL in the CZA-resistant isolates (P= 0.0001).
 |
Table 4 Phenotypic and Genotypic Characterization of the Investigated CRKP Isolates (N= 47) |
Distribution of Carbapenem Resistance and Outer Membrane Porin Genes in CRKP Isolates
Amongst our 47 CRKP isolates, blaKPC was the most common carbapenemase-encoding gene (42.6%), followed by blaOXA-48 (23.4%), blaNDM-1 (10.6%), and blaIMP (2.1%). None of the test CRKP isolates harbored blaVIM. About 12.7% and 4.3% of the isolates coharbored blaOXA-48/blaNDM-1 and blaKPC/blaNDM-1, respectively. Also, 4.3% of the isolates displayed coexistence of blaKPC/blaIMP. Half (50%) of CZA-resistant CRKP strains harbored blaKPC, whereas 16.7% and 8.3% carried blaOXA-48 and blaOXA-48/blaNDM-1, respectively. The MBL-encoding genes blaNDM-1 and blaIMP were identified in 16.7% and 8.3% of the CZA-resistant isolates, respectively. Regarding omp genes, both ompK35 and ompK36 were not observed in 10 out of the total CRKP isolates (21.3%). Out of 12 CZA-resistant CRKP strains, 7 (58.3%) showed negative results for both porin genes, compared to 3 of the CZA-susceptible CRKP isolates (8.6%). Results of molecular testing are shown in Figure 2A, B and Table 4.
Discussion
In cancer patients, acquisition of CRKP infections signals an escalating threat to the health-care providers owing to the declining antimicrobial armamentarium. Therefore, regular detection and combating of these infectious agents are of utmost importance to avoid subsequent detrimental effects. In this context, we aimed to explore the in vitro activity of the rather new antibiotic CZA against the recovered CRKP isolates from adult patients suffering from cancer, as well as the molecular mechanisms driving the emergence of CZA resistance.
In the present study, phenotypic screening for carbapenemases by the MHT proved that 47 out of 134 K. pneumoniae isolates (35.1%) were carbapenemase-producers, consistent with the results from a prior Egyptian study,17 but lower than that reported from another recent Egyptian literature.18 The high pattern of carbapenem resistance amongst our isolates could be ascribed to the selective pressure stemming from inappropriate empiric use of carbapenems in our health-care facility, merged with lack of regular molecular characterization of bacterial isolates. Disappointingly, our recovered CRKP isolates showed dramatically increased resistance threshold to the other commonly used antibiotics including β-lactams, trimethoprim/sulfamethoxazole, aminoglycosides, and fluoroquinolones (Table 1). This highlights the importance of continued vigilance and combating of the rising resistant strains to avoid additional propagation.
The antimicrobial susceptibility testing confirmed that 8.96% of K. pneumoniae (Figure 1) and 25.5% of the CRKP isolates (Table 1) investigated in this study were resistant to CZA. This result suggests a striking rise in CZA resistance when the strains are carbapenem-resistant, compatible with a previous study.19 Our relatively high rate is alarming, since the obtained isolates were collected from patients without prior exposure to CZA-based treatment regimens. In agreement with our finding, a recent literature indicated 21% resistance of CRKP to CZA,20 while other researchers described 8.2% resistance from a Chinese tertiary hospital.21 Moreover, results from a retrospective observational Spanish study reported resistance following CZA treatment in 6 out of 47 (12.7%) patients.22 Heterogeneity across diverse studies could be justified by variations in the study design, geographic region, sample size, as well as study cohort.
It is worth-mentioning that CZA-resistant strains have been developed at low rates prior to the clinical application of this antibiotic in 2015. This should be taken into account, since the indiscriminate use of CZA in health-care settings may favor the spread of strains with baseline resistance to CZA and the advent of further resistance.23 Notably, the overall studies about the susceptibility profile to CZA from cancer patients are rare; however, per a previous multicenter study involving 31 patients with hematologic malignancies, patients who received CZA therapy had a significantly higher remission rate after 14 days of antibiotic treatment compared to patients who received other antibiotic regimens.24 In addition, an elsewhere study involving a cohort of three patients with leukemia concluded that the use of CZA could improve the prognosis of patients infected with multidrug-resistant bacteria, including KPC-K. pneumoniae.25
At present, only a handful of studies have depicted the risk factors for acquisition of CZA-resistant CRKP infections. In our work, no significant differences were observed between patients infected with CZA-susceptible CRKP versus those infected with CZA-resistant strains in terms of the associated comorbidity except for chronic kidney disease, where renal patients were 11.33 times more likely to develop infection with CZA-resistant strains (Table 2). A recent study identified renal replacement therapy as an independent predictor for the incidence of CZA-resistant CRKP infections.26 These findings reflect the urgent need for CZA dosage adjustment in case of renal insufficiency to reach optimum microbiological and clinical response. In contrast to our results, an earlier study by Shields et al denoted that pneumonia was an independent factor for CZA resistance amongst a panel of carbapenem-resistant Enterobacteriaceae; however, the contributing mechanism was not entirely understood.27
In the current study, exposure to invasive procedures was another risk factor for CZA resistance, with patients submitted to mechanical ventilation were 24.29 times more likely to develop resistance (Table 2). A recently published, retrospective, observational study revealed that undergoing mechanical ventilation was significantly associated with clinical failure to CZA.28 Likewise, a univariate analysis by Liu and his colleagues demonstrated that mechanically ventilated patients were more probable to develop infections with CZA-resistant CRKP strains.26 No doubt that patient’s intubation interferes with the natural defense mechanisms, causing opportunistic bacteria to adhere to the inner wall of the endotracheal tube, where they initiate a biofilm formation that acts as an armor from the host’s immunity as well as antibiotic effect.29
Amongst our cohort, admission to the ICU was correlated with the occurrence of CZA-resistant strains (OR=6.76, 95% CI: 1.59–28.72; P=0.006). Cancer patients in ICUs are critically ill and vulnerable to different healthcare-associated infections. As a consequence, the irrational consumption of wide-spectrum antibiotics leads to a growing surge in the incidence of antibiotic-resistant bugs.30 Outstandingly, patients infected with CZA-resistant strains in our study showed a higher 30-day mortality rate in comparison to those infected with susceptible isolates (Table 2). However, we could not trace this difference only to the in vitro CZA resistance without testing other confounders, as in vivo models, that may be associated with this observation. In our multivariate regression analysis, the only factor independently associated with CZA resistance was longer length of hospital stay (OR=0.10, 95% CI: 0.01–0.86; P=0.03), possibly due to repeated invasive manipulations and/or antibiotic regimens (Table 3).
Several resistance mechanisms to CZA have been proposed, thus far, including mutations within the blaKPC associated with lost or reduced function of KPC enzyme, hyperexpression of blaKPC, and modifications in the OMPs.31 In an attempt to uncover the underlying molecular mechanisms of resistance to CZA, all of the collected CRKP isolates were further subjected to multiplex PCRs (Figure 2 and Table 4). Our investigations demonstrated that blaKPC was the most predominant genotype identified in 42.6% of the CRKP isolates, while none of the examined isolates harbored blaVIM. Our finding corroborates a previous study conducted at our institution, where blaKPC was the most prevalent variant.32 In contrast, another Egyptian study reported blaKPC in 26.8% of the CRKP isolates, meanwhile blaNDM was the most frequent genotype.33
Data from other countries also showed considerable involvement of blaKPC in carbapenem resistance in K. pneumoniae.34 Since the first detection of KPCs in the USA in 1996, they spread globally and turned to be the most common carbapenemases in various countries.35 Despite the preponderance of OXA-48 and NDM type enzymes in Saudi Arabia, a recent report indicated the emergence of the first KPC-producing K. pneumoniae isolate in Riyadh, from a urine sample, which demands rigorous interventions to prevent further dissemination.36 It is notable that blaKPC was a major mechanism of resistance amongst our CZA-resistant CRKP isolates (50%), possibly due to the high rate of this gene in the overall CRKP isolates. In favor of our result, an Italian study attributed the emergence of CZA resistance to blaKPC expression in 57.1% of the analyzed K. pneumoniae isolates.37 In addition, Liao and others reported the development of CZA resistance in a CRKP clinical isolate secondary to blaKPC-78; a new blaKPC-2 variant induced by point mutation following CZA therapy.38
Although blaKPC was the leading carbapenemase-encoding gene, we also observed blaOXA-48 in 23.4% of our CRKP isolates, with 16.7% of CZA resistance was associated with the presence of this gene. Unfortunately, within a few years of being recognized in a Turkish patient, data involving dissemination of blaOXA-48 into Europe, the Mediterranean, and the Middle East have been published.39 Out of 12 CZA-resistant CRKP isolates in our study, 8.3% coharbored blaOXA-48/blaNDM-1. An analogous Indian study observed 51% susceptibility to CZA amongst CRKP isolates, with 52% and 27% of K. pneumoniae strains produced OXA-48 like and NDM/OXA-48 like carbapenemases, respectively.40 In the present work, the MBL-encoding genes blaNDM-1 and blaIMP were identified in 16.7% and 8.3% of CZA-resistant strains, respectively, lower than that reported formerly.41 Importantly, CZA retained activity against most of our OXA-48- and KPC-producing isolates (81.8% and 70%, respectively) as shown in Table 4. Also, 60% activity was identified against NDM-1 carbapenemase, which contradicts literature describing poor activity of CZA against MBL-producing organisms.42 This discrepancy could be allocated to other undetermined mechanisms that need future validation.
The OmpK35 and OmpK36 facilitate the diffusion of avibactam throughout the outer membrane of K. pneumoniae.20 In our study, 7 out of 12 CZA-resistant isolates (58.3%) yielded negative multiplex PCR results for ompK35 and ompK36. Intriguingly, we also observed absence of both genes in 3 of the susceptible strains (8.6%) as per Table 4. Existing literature suggests that alteration of OmpK35 and OmpK36 porins is not an initial pathway for development of CZA resistance; however, loss of porins associated with enhanced blaKPC expression are possible underlying mechanisms for CZA-resistant phenotypes, which endorses our results.43 Similar conclusions were also drawn by earlier authors.44
What is more, the present work proved that the mean MICs of CZA in ompK35- and ompK36-negative isolates were significantly higher compared to positive strains (P= 0.0001 for each gene). In keeping with our findings, a comparable study observed elevated MICs of CZA amongst a panel of carbapenem-resistant Enterobacterales secondary to Ompk36 L3 alterations and production of β-lactamases.45 Also, a former study by Shen and his colleagues reported that mending of OmpK35 resulted into more reduction of the MIC of ceftazidime as opposed to OmpK36, but without a considerable decrease in the MIC of avibactam.46
Our study has some limitations to be addressed. First, we excluded children from our cohort, therefore future studies including pediatric patients should be planned. Second, detailed molecular analyses of porin genes in relation to point mutations, deletions, and insertions, and phenotypic analysis of porin loss, were not done, which highlights the need for further complementing studies.
Conclusion
Our study illustrated that CZA has an excellent in vitro activity against CRKP isolates from cancer patients. Despite CZA drug has not been introduced into our institution for in vivo usage until now, considerable rate of resistant mutants expressing multiple virulence genes have been emerged. This troublesome finding emphasizes the urgent need to cautious implementation of stringent infection control measures to preclude additional recirculation of mutant strains in the hospital environment. Moreover, our results underscore the importance of molecular characterization of resistant strains to prevent clonal expansion and propagation that could jeopardize current/or future antibiotics.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
The present study was conducted in accordance with the Declaration of Helsinki. The study design was granted approval by the Institutional Review Board (IRB) of the Faculty of Medicine, Mansoura University, Egypt (Decision no. R.19.05.502.R1). Written informed consents were taken from all contributors in the study. Patients’ data privacy and confidentiality were respected in all stages of the study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in relation to this work.
References
1. Lee Y, Bradley N. Overview and insights into carbapenem allergy. Pharmacy. 2019;7(3):110. doi:10.3390/pharmacy7030110
2. Elshamy AA, Aboshanab KM. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Future Sci OA. 2020;6(3):FSO438. doi:10.2144/fsoa-2019-0098
3. Wyres K, Holt K. Regional differences in carbapenem-resistant Klebsiella pneumoniae. Lancet Infect Dis. 2022;22(3):309–310. doi:10.1016/S1473-3099(21)00425-4
4. Zhang WX, Chen HY, Chen C, et al. Resistance phenotype and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Shanghai. Microb Drug Resist. 2021;27(10):1312–1318. doi:10.1089/mdr.2020.0390
5. Codjoe FS, Donkor ES. Carbapenem resistance: a Review. Med Sci. 2017;6(1):1.
6. Hammoudi Halat D, Ayoub Moubareck C. The current burden of carbapenemases: review of significant properties and dissemination among gram-negative bacteria. Antibiotics. 2020;9(4):186. doi:10.3390/antibiotics9040186
7. Mojica MF, Rossi MA, Vila AJ, Bonomo RA. The urgent need for metallo-β-lactamase inhibitors: an unattended global threat. Lancet Infect Dis. 2022;22(1):e28–e34. doi:10.1016/S1473-3099(20)30868-9
8. Dabos L, Oueslati S, Bernabeu S, Bonnin RA, Dortet L, Naas T. To be or not to be an OXA-48 carbapenemase. Microorganisms. 2022;10(2):258. doi:10.3390/microorganisms10020258
9. Di Domenico EG, Cavallo I, Sivori F, et al. Biofilm production by carbapenem-resistant Klebsiella pneumoniae significantly increases the risk of death in oncological patients. Front Cell Infect Microbiol. 2020;10(10):561741. doi:10.3389/fcimb.2020.561741
10. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
11. Criscuolo M, Trecarichi EM. Ceftazidime/avibactam and ceftolozane/tazobactam for multidrug-resistant gram negatives in patients with hematological malignancies: current experiences. Antibiotics. 2020;9(2):58. doi:10.3390/antibiotics9020058
12. Cavallini S, Unali I, Bertoncelli A, Cecchetto R, Mazzariol A. Ceftazidime/avibactam resistance is associated with different mechanisms in KPC-producing Klebsiella pneumoniae strains. Acta Microbiol Immunol Hung. 2021;2021:1626.
13. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
14. Bina M, Pournajaf A, Mirkalantari S, Talebi M, Irajian G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae isolated from the clinical samples by the phenotypic and genotypic methods. Iran J Pathol. 2015;10(3):199–205.
15. Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother. 2012;67(4):906–909. doi:10.1093/jac/dkr563
16. Wassef M, Abdelhaleim M, AbdulRahman E, Ghaith D. The role of OmpK35, OmpK36 porins, and production of β-lactamases on imipenem susceptibility in Klebsiella pneumoniae clinical isolates, Cairo, Egypt. Microb Drug Resist. 2015;21(6):577–580. doi:10.1089/mdr.2014.0226
17. Metwally L, Gomaa N, Attallah M, Kamel N. High prevalence of Klebsiella pneumoniae carbapenemase–mediated resistance in K. pneumoniae isolates from Egypt. East Mediterr Health J. 2013;19(11):947–952. doi:10.26719/2013.19.11.947
18. Kotb S, Lyman M, Ismail G. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare-associated Infections Surveillance Data, 2011-2017. Antimicrob Resist Infect Control. 2020;9(1):2. doi:10.1186/s13756-019-0639-7
19. Senchyna F, Gaur RL, Sandlund J, et al. Diversity of resistance mechanisms in carbapenem-resistant Enterobacteriaceae at a health care system in Northern California, from 2013 to 2016. Diagn Microbiol Infect Dis. 2019;93(3):250–257. doi:10.1016/j.diagmicrobio.2018.10.004
20. Wang Y, Wang J, Wang R, Cai Y. Resistance to ceftazidime-avibactam and underlying mechanisms. J Glob Antimicrob Resist. 2020;22:18–27. doi:10.1016/j.jgar.2019.12.009
21. Li D, Liao W, Huang HH, et al. Emergence of hypervirulent ceftazidime/avibactam-resistant Klebsiella pneumoniae isolates in a Chinese tertiary hospital. Infect Drug Resist. 2020;3(13):2673–2680. doi:10.2147/IDR.S257477
22. Castón JJ, Gallo M, García M, et al. Ceftazidime–avibactam in the treatment of infections caused by KPC-producing Klebsiella pneumoniae: factors associated with clinical efficacy in a single-center cohort. Int J Antimicrob Agents. 2020;56(3):106075. doi:10.1016/j.ijantimicag.2020.106075
23. Di Bella S, Giacobbe DR, Maraolo AE, et al. Resistance to ceftazidime/avibactam in infections and colonisations by KPC-producing Enterobacterales: a systematic review of observational clinical studies. J Glob Antimicrob Resist. 2021;25:268–281. doi:10.1016/j.jgar.2021.04.001
24. Castón JJ, Lacort-Peralta I, Martín-Dávila P, et al. Clinical efficacy of ceftazidime/avibactam versus other active agents for the treatment of bacteremia due to carbapenemase–producing Enterobacteriaceae in hematologic patients. Int J Infect Dis. 2017;59:118–123. doi:10.1016/j.ijid.2017.03.021
25. Metafuni E, Criscuolo M, Spanu T, Sica S. Ceftazidime–avibactam for gram–negative multidrug–resistant bacteria in hematological patients: a single–center experience. Ann Hematol. 2019;98(6):1495–1497. doi:10.1007/s00277-018-3535-y
26. Liu X, Chu Y, Yue H, Huang X, Zhou G. Risk factors for and clinical outcomes of ceftazidime–avibactam–resistant carbapenem–resistant Klebsiella pneumoniae nosocomial infections: a single–center retrospective study. Infection. 2022. doi:10.1007/s15010-022-01781-3
27. Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. Pneumonia and renal replacement therapy are risk factors for ceftazidime–avibactam treatment failures and resistance among patients with carbapenem–resistant Enterobacteriaceae infections. Antimicrob Agents Chemother. 2018;62(5):e02497–17. doi:10.1128/AAC.02497-17
28. Di Pietrantonio M, Brescini L, Candi J, et al. Ceftazidime–avibactam for the treatment of multidrug–resistant pathogens: a retrospective, single center study. Antibiotics. 2022;11(3):321. doi:10.3390/antibiotics11030321
29. Baidya S, Sharma S, Mishra SK, Kattel HP, Parajuli K, Sherchand JB. Biofilm formation by pathogens causing ventilator–associated pneumonia at intensive care units in a tertiary care hospital: an armor for refuge. Biomed Res Int. 2021;28(2021):8817700.
30. Garg VK, Mishra S, Gupta N, et al. Microbial and antibiotic susceptibility profile among isolates of clinical samples of cancer patients admitted in the intensive care unit at regional tertiary care cancer center: a retrospective observational study. Indian J Crit Care Med. 2019;23(2):67–72. doi:10.5005/jp-journals-10071-23119
31. Li X, Ke H, Wu W, Tu Y, Zhou H, Yu Y. Molecular mechanisms driving the in vivo development of kpc-71-mediated resistance to ceftazidime-avibactam during treatment of carbapenem-resistant Klebsiella pneumoniae infections. mSphere. 2021;6(6):e0085921. doi:10.1128/mSphere.00859-21
32. Moemen D, Masallat DT. Prevalence and characterization of carbapenem–resistant Klebsiella pneumoniae isolated from intensive care units of Mansoura University hospitals. Egypt J Basic Appl Sci. 2017;4:37–41.
33. Abdulall AK, Tawfick MM, El Manakhly AR, El Kholy A. Carbapenem-resistant Gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur J Clin Microbiol Infect Dis. 2018;37(9):1647–1652. doi:10.1007/s10096-018-3294-7
34. Chatzidimitriou M, Chatzivasileiou P, Sakellariou G, et al. Ceftazidime/avibactam and eravacycline susceptibility of carbapenem-resistant Klebsiella pneumoniae in two Greek tertiary teaching hospitals. Acta Microbiol Immunol Hung. 2021;68(2):65–72. doi:10.1556/030.2021.01364
35. Ainoda Y, Aoki K, Ishii Y, et al. Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae ST258 isolated from a Japanese patient without a history of foreign travel - a new public health concern in Japan: a case report. BMC Infect Dis. 2019;19(1):20. doi:10.1186/s12879-018-3649-9
36. Alghoribi MF, Binkhamis K, Alswaji AA, et al. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: a new public health concern in Saudi Arabia. J Infect Public Health. 2020;13(4):647–650. doi:10.1016/j.jiph.2020.01.003
37. Fontana C, Favaro M, Campogiani L, et al. Ceftazidime/avibactam-resistant Klebsiella pneumoniae subsp. pneumoniae isolates in a tertiary Italian hospital: identification of a new mutation of the carbapenemase type 3 (KPC-3) gene conferring ceftazidime/avibactam resistance. Microorganisms. 2021;9(11):2356. doi:10.3390/microorganisms9112356
38. Liao Q, Deng J, Feng Y, et al. Emergence of ceftazidime-avibactam resistance due to a novel blaKPC-2 mutation during treatment of carbapenem-resistant Klebsiella pneumoniae infections. J Infect Public Health. 2022;15(5):545–549. doi:10.1016/j.jiph.2022.04.002
39. Mataseje LF, Boyd DA, Fuller J, et al. Characterization of OXA-48-like carbapenemase producers in Canada, 2011-14. J Antimicrob Chemother. 2018;73(3):626–633. doi:10.1093/jac/dkx462
40. Bakthavatchalam YD, Routray A, Mane A, et al. In vitro activity of ceftazidime-avibactam and its comparators against carbapenem resistant Enterobacterales collected across India: results from ATLAS surveillance 2018 to 2019. Diagn Microbiol Infect Dis. 2022;103(1):115652. doi:10.1016/j.diagmicrobio.2022.115652
41. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124.e1–124.e4. doi:10.1016/j.cmi.2019.08.020
42. Yu W, Xiong L, Luo Q, et al. In vitro activity comparison of ceftazidime-avibactam and aztreonam-avibactam against bloodstream infections with carbapenem-resistant organisms in China. Front Cell Infect Microbiol. 2021;25(11):780365. doi:10.3389/fcimb.2021.780365
43. Cui X, Shan B, Zhang X, et al. Reduced ceftazidime-avibactam susceptibility in KPC-producing Klebsiella pneumoniae from patients without ceftazidime-avibactam use history - a multicenter study in China. Front Microbiol. 2020;23(11):1365. doi:10.3389/fmicb.2020.01365
44. Humphries RM, Hemarajata P. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother. 2017;61(6):e00537–17. doi:10.1128/AAC.00537-17
45. Castanheira M, Doyle TB, Hubler C, Sader HS, Mendes RE. Ceftazidime-avibactam activity against a challenge set of carbapenem-resistant Enterobacterales: ompk36 L3 alterations and β-lactamases with ceftazidime hydrolytic activity lead to elevated MIC values. Int J Antimicrob Agents. 2020;56(1):106011. doi:10.1016/j.ijantimicag.2020.106011
46. Shen Z, Ding B, Ye M, et al. High ceftazidime hydrolysis activity and porin OmpK35 deficiency contribute to the decreased susceptibility to ceftazidime/avibactam in KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother. 2017;72(7):1930–1936.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.