Back to Journals » Infection and Drug Resistance » Volume 10
Molecular evaluation of colistin-resistant gene expression changes in Acinetobacter baumannii with real-time polymerase chain reaction
Authors Sepahvand S, Davarpanah MA, Roudgari A, Bahador A, Karbasizade V, Kargar Jahromi Z
Received 5 May 2017
Accepted for publication 24 July 2017
Published 27 November 2017 Volume 2017:10 Pages 455—462
DOI https://doi.org/10.2147/IDR.S141196
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Shahriar Sepahvand,1 Mohammad Ali Davarpanah,2 Amir Roudgari,3 Abbas Bahador,4 Vajihe Karbasizade,5 Zahra Kargar Jahromi6
1Department of Microbiology, Falavarjan Branch, Islamic Azad University, Isfahan, Iran; 2Shiraz HIV/AIDS Research Center, Institute of Health, Shiraz University of Medical Sciences, Shiraz, Iran; 3Shiraz Trauma Center, Shiraz University of Medical Sciences, Shiraz, Iran; 4Department of Microbiology, Faculty of Medicine, Tehran University of Medical Sciences, Tehran, Iran; 5Department of Microbiology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran; 6Zoonoses Research Center, Jahrom University of Medical Sciences, Jahrom, Iran
Background: Acinetobacter baumannii is an important human pathogen which has recently gained increased attention due to the occurrence of drug-resistant nosocomial infections in patients suffering from immune system disorders, and those in hospital intensive care units. The aim of this research was to identify and isolate A. baumannii strains resistant to colistin, determine antibiotic resistance pattern of this bacteria, investigate the presence of colistin-resistant genes, and finally assess the effect of expression changes in pmrA and pmrB genes resistant to A. baumannii against colistin via real-time polymerase chain reaction.
Methods: The samples were initially purified and isolated using biochemical tests and Microgen kit. Later, the resistance pattern evaluation of validated samples to different antibiotics and colistin was carried out using two methods viz., disc diffusion and E-test. This was followed by the assessment of genes resistant to colistin via polymerase chain reaction besides gene expression changes via real-time polymerase chain reaction.
Results: The results of this study indicated that eleven strains of A. baumannii isolated from Shahid Rajaee Trauma Hospital were resistant to colistin. However, in the resistance pattern evaluation of A. baumannii isolated from Ali Asghar Hospital, all the strains were sensitive to colistin. In the evaluation of genes resistant to pmrA and pmrB, most of the strains resistant to colistin were carriers of these genes. Besides, in the expression assessment of these genes, it was demonstrated that expression of pmrA in the strains resistant to colistin significantly increased in relation to sensitive strains, but the expression of pmrB increased at a lower rate in the strains resistant to colistin as compared to the sensitive strains.
Conclusion: Thus, it can be safely mentioned that increased expression of pmrA was due to the resistance of A. baumannii to colistin.
Keywords: Acinetobacter baumannii, colistin, real-time PCR, gene expression, PCR
Introduction
The microorganisms causing nosocomial infections bring about several issues from the viewpoint of treatment failure as well as patients’ mortality, this is particularly due to their antibiotic-resistant property. Among these bacteria, Acinetobacter baumannii is an important human pathogen which has recently gained much importance.1 A. baumannii is a Gram negative non-fermentative, anaerobic coccobacilli which is widely dispersed in the hospital environment. In addition, it is an important opportunistic pathogen and is responsible for various nosocomial infections.2 This pathogen has been considered among the six most dangerous nosocomial microorganisms by the Infectious Diseases Society of America (IDSA).3 A. baumannii generally infects intensive care unit (ICU) patients including those who have suffered stroke, injuries, burns, and need mechanical ventilation.4 One of the most interesting peculiarities of A. baumannii is that it can easily attain resistance toward different antibiotics. A. baumannii strains have shown resistance to most antibiotics so far.5 The factor which has led to fortification of this resistant system is the unnatural inherited abilities of A. baumannii during long-term viability in nosocomial environments, thereby causing nosocomial expansion of this bacteria.6
Today, this bacteria is resistant to all antibiotics ; thus, some drugs have been excluded from treatment of A. baumannii infections such as penicillins, cephalosporins, aminoglycosides, quinolones, and tetracyclines.7 The last treatment line against this bacteria is colistin which today, unfortunately has been globally reported to be resistant. Colistin via Colistinus subspecies (Bacillus polymixa) is synthesized non-ribosomally. The colistin resistance mechanisms of this bacteria are different, and include: 1) colistin resistance mediated by complete loss of lipopolysaccharides (LPS),8 2) the insertion sequence ISAball is involved in colistin resistance and loss of LPS,9 3) LPS-deficient A. baumannii shows altered signaling through host TLRs and increased susceptibility to the host antimicrobial peptide LL-37,10 4) gene expression changes in the two-component system of pmrAB, in which resistance to colistin is influenced by changes in the expression of pmrA and pmrB genes.11 In this research, we studied the relationship between the resistance of A. baumannii to colistin and gene expression changes in pmrA and pmrB.
Methods
This study was approved by the Medical Ethics Committee at Shiraz University of Medical Sciences, and all patients provided written informed consent. This cross-sectional study was carried out for a period of 16 months on 100 clinical samples collected from a variety of infections of patients hospitalized in six different wards of ICU of two hospitals in Shiraz city (Shahid Rajaei Trauma Hospital and Ali Asghar Hospital), including internal units, neurology, surgery, recovery, and screening.
Isolation and identification of microorganism
Various clinical samples were cultured in different media, e.g., blood agar, chocolate agar, and MacConkey agar. Then, the cultured plates were incubated at 37°C for 24–48 h. After Gram staining, the catalase, oxidase, triple sugar iron agar, and motility biochemical tests were carried out. After that, clinical strains of A. baumannii were identified using Microgen kits (Novacyt, Vélizy-Villacoublay, France).
Microgen GN-A
The kit contains of lysine, ornithine, H2S, glucose, mannitol, xylose, ONPG, indole, urease, VP, citrate, and TDA. The kit works as follows. A single colony from the culture medium was used to prepare a bacterial suspension in 3 mL saline according to McFarland 0.5 turbidity standard. The plastic strip over the kit was removed and two or three drops of the suspension (100 μL) was added to the wells in each strip. Oil was dripped into the wells which were then covered by the plastic strip. The strips were then incubated at 37°C for 24 h, and the necessary reagents were added to the wells. The results were obtained based on comparison of color formed in the wells with the color guide, and to find the diagnosis code, they were inserted in a report form, and ultimately the 4-digit code was entered in Microgen specific software.
Antibiotic sensitivity assessment
For antibiotic sensitivity assessment, disc diffusion and E-test methods were used in accordance with CLSI standards.
Antibiotic sensitivity test by disc diffusion method
After bacterial inoculation in tryptic soy broth medium and incubation at 37˚C for 2–6 h and concentration preparation, the McFarland dilution and culturing on Mueller Hinton agar (MHA) was carried out. Later, ampicillin, Augmentin, aztreonam, Cefotaxime, ceftazidime, ticarcillin, cefixime, meropenem, imipenem, amikacin, imipenem-ethylenediaminetetraacetic, tigecycline, and colistin from Mast Company (Bootle, UK) were placed on the MHA plate and the results were studied after 18 h incubation at 37˚C.
Antibiotic sensitivity test using E-test method
Antibiotic sensitivity test was performed using the E-test method and according to CLSI standards. quantitative tests for ampicillin, Augmentin, aztreonam, Cefotaxime, ceftazidime, ticarcillin, cefixime, meropenem, imipenem, amikacin, imipenem-ethylenediaminetetraacetic, tigecycline, and colistin were performed by E-test (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s instructions. In certain cases, the minimum inhibitory concentration (MIC) was performed using microdilution and was based on CLSI standards. Escherichia coli ATCC 25922 was used as a pen-sensitive for all tests.
Polymerase chain reaction (PCR)
At this stage, using the gene or sequence specific primers (Table 1) in the PCR reaction, the presence of pmrA, pmrB, and 16S rRNA genes was assessed in the samples. The reactions were carried out in 25 µL volume containing PCR 1× buffer, 0.5 µL dNTP mixture, 0.4 µL DNA polymerase Taq, and 0.75 µL MgCl2, 1 µL of starters, 2 µL of DNA, and 17 µL of double deionized water. After the preparation of the Master Mix and covering it with mineral oil, the process was conducted in a Thermo-cycler system at the the temperatures listed in Table 2. Later, the PCR products were analyzed on ethidium bromide containing 1% agarose gel.
  | Table 1 Sequences of primers used |
  | Table 2 Polymerase chain reaction thermal steps |
Real-time PCR
For real-time PCR, a One Step kit from Kapa Biosystems, Inc. (Wilmington, MA, USA) was used. Kits include KAPA RT Mix, dUTP, and KAPA SYBR FAST qPCR Master Mix, which contains KAPA SYBR FAST DNA Polymerase, reaction buffer, dNTPs, SYBR Green I dye, and MgCl2 at a final concentration of 2.5 mM. 16S rRNA was used as the housekeeping gene. Housekeeping genes are used as a control to compare gene expression.
RNA extraction
RNA extraction was carried out with bacterial RNA isolation kit Trizol Max (Thermo Fisher Scientific, Waltham, MA, USA). After spinning, bacterial pellet was dissolved in 200 µL hastening factor (pre-prepared in the kit). It should be noted that the bacteria have already been lysed when the bacterial pellet is incubated at 95°C. Afterwards the bacterial pellet was lysed in 1 mL Trizol. Then, the lysed bacterial pellet was incubated at room temperature for 5 min in Trizol. Then chloroform was added and it was shaken for 15 s and incubated at room temperature for 2–3 min. The phases were separated with centrifugation at 12,000× g for 15 min. In this stage, 90% of the blue phase (transparent color) was transferred to the fresh Eppendorf tubes containing 500 µL of isopropanol for RNA precipitation. RNA was sedimented with the mixture. After that, the tubes were incubated at room temperature for 10 min. Then, centrifugation was carried out at the speed of 12,000× g for 10 min. The total RNA pellets were washed once with 70% ethanol. They were also centrifuged for 5 min at 7,500× g. RNA sediment in water without nuclease was again dissolved and its concentration was determined in an ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific).
Primers’ preparation
To perform real-time PCR, the specific primers related to pmrA, pmrB, and 16S rRNA genes, listed in Table 1, were used. The specificity of the primers was checked using primer blast software at NCBI. Using diethyl pyrocarbonate water, the primers reached the desired volume, and a dilution of one tenth of each primer was ultimately prepared. The non-diluted primers were kept at -70°C and the diluted primer was kept at -20°C.
Real-time PCR reaction stages
The real-time PCR reaction was conducted in the ABI Thermocycler System (Thermo Fisher Scientific) and analyzed using the ∆∆ Ct method. Initially, the samples containing RNA were removed from the freezer while they were on ice. Even the Master Mix was removed from the freezer, and was placed in its specific tube holder and kept on ice. Then, the amount of material needed for the real-time PCR reaction was calculated and ultimately 20 μL of the real-time PCR mixture was obtained. The material required for testing was poured into the real-time PCR micro tubes as shown in Table 3. The tubes were kept in the real-time PCR system according to the related cycles listed in Table 4.
  | Table 3 Real-time polymerase chain reaction materials Abbreviations: FP, forward primer; RP, reverse primer. |
  | Table 4 Real-time polymerase chain reaction Abbreviation: RT, reverse transcription. |
Eventually, Ct related to pmrA, pmrB, and housekeeping genes was obtained from the flow charts of the real-time PCR system. The raw data were extracted in Ct form from the system. The analysis of Cts was carried out by SDS software. Additionally, the rate of gene expression was measured using the Pfaffl method.
Results
After the elimination of duplicated isolates as well as the isolates that were clinically unrelated, a total of 100 A. baumannii strains were reviewed and identified via microscopic and macroscopic methods and using Microgen GN-A Kit. In terms of frequency (Table 5), the strains were from 41 patients (41%) in internal care units, 33 patients (33%) in the central ward, 13 patients (13%) in surgical ward, eight patients (8%) in neurology, and five (5%) in recovery and screening. In this study, A. baumannii strains were isolated from four different clinical specimens including urinary tract infections, wound specimens, respiratory system infections, and blood circulation (Figure 1).
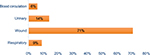  | Figure 1 The distribution of clinical samples. |
  | Table 5 The hospitals supplying the strains |
Antibiotic resistance
In this study, all A. baumannii isolates were resistant to ampicillin, Augmentin, aztreonam, Cefotaxime, ceftazidime, ticarcillin, cefixime, meropenem, tetracycline, and ceftriaxone. According to the results obtained from the study, imipenem and amikacin antibiotics were found to have low efficiency in treating the strains of A. baumannii. However, imipenem-ethylenediaminetetraacetic, colistin, tigecycline, and rifampin antibiotics were the most effective treatments, respectively. Since colistin is the last alternative to treat A. baumannii which is multidrug-resistant, resistance of A. baumannii to this antibiotic is considered treatment failure. In the current study, although eleven strains resistant to colistin were isolated in Shahid Rajaei Trauma Hospital, no strain resistant to colistin was found in Ali Asghar Hospital. As this antibiotic is the last line of treatment, even low resistance to this antibiotic results in failure in the treatment of A. baumannii. The antibiotic resistance pattern, using the disc diffusion method, in the Shahid Rajaei Trauma Hospital is shown in Figure 2, and in Ali Asghar Hospital is demonstrated in Figure 3. The results of E-test in the 100 isolated strains are depicted in Table 6.
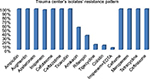  | Figure 2 Antibiotic resistance pattern in the Shahid Rajaei Trauma Hospital. |
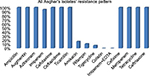  | Figure 3 Resistance pattern in Ali Asghar Hospital. |
PCR
The results obtained from the assessments of pmrA, pmrB, and 16SrRNA genes indicated that all the strains contained 16S rRNA gene. Most of the strains were carriers of pmrA and pmrB genes. This analysis was conducted from the combination of strains in both hospitals (Figure 4).
  | Figure 4 Column M (50 bp DNA ladder); column 1, negative control (distilled water); column 2, positive control; columns 3 to 9, pmrB gene; column 10, 16S rRNA; columns 11 to 13, pmrA gene. |
Real-time PCR
Using real-time PCR, the obtained results and pmrA, pmrB, and 16S rRNA gene replication curve indicated that the replication was conducted well and with a suitable operation and without any parasite (Figure 5).
  | Figure 5 Replication curve. |
Due to the lack of specification of fluorescent SYBR Green color, in order to assure the production of specific parts and the absence of non-specific bands and secondary structures besides dual primer in the real-time PCR products, the melt curve was traced. The results of melt curve for the applied starters showed that starters operated in a specific manner and lacked any type of non-specific parts. The fold change in the prmA and pmrB genes was in line with the obtained results 2 - ∆∆Ct-PmrA was equivalent to 249.68 and 2 - ∆∆Ct-PmrB was equivalent to 0.309145. The fold change in pmrA gene exhibited increased expression of this gene in the samples resistant to colistin in relation to the samples sensitive to colistin. The expression increase in pmrA gene results in the resistance of A. baumannii against colistin. On the other hand, the fold change in the pmrB gene in the samples resistant to colistin was quite insignificant in proportion to the samples sensitive in comparison to pmrA gene. Therefore, a significant increase in the expression of pmrA gene in relation to pmrB gene was observed which had an insignificant increase (Figures 6 and 7).
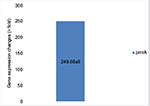  | Figure 6 Gene expression changes in pmrA genes. |
  | Figure 7 Gene expression changes in pmrB genes. |
Discussion
Today, numerous microorganisms lead to different nosocomial infections in hospitals, especially in ICUs. IDSA has recently reported a list of dangerous, infectious nosocomial microorganisms including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter “ESKAPE”. It is noteworthy that one of the microorganisms considered in the list is A. baumannii. This bacteria was underscored in the past, but today it is highly considered as one of the most important nosocomial pathogens due to its survival in different hospital sections, especially in ICU, and its fast resistance against different antibiotics. A. baumannii has developed resistance to most antibiotics wherein few antibiotics have been sidelined in the first and second line treatment of this bacterium.7
Today, the last effective treatment option for A. baumannii is colistin, but unfortunately different reports worldwide are indicative of the increase in the resistance of this pathogen against colistin. In our study from two Shiraz Hospitals (Shahid Rajaee Trauma Hospital and Ali Asghar), the resistance of this pathogen against colistin was demonstrated as follows: in Shahid Rajaee Trauma Hospital there were eleven strains resistant to colistin, but in Ali Asghar Hospital, no resistant strain was found, this is indicative in the ICU of Shahid Rajaee Trauma Hospital of Shiraz. Due to the large number of patients having been in accidents in this section, the control of infection is difficult and the transmission of this pathogen could be via the personnel of this hospital, from one individual to another. Like several other studies conducted on this organism, the highest isolates of our study were from the wound samples of the individuals hospitalized in the ICU. The patients have a lack of infection-prone immunity to this bacterium, thus, the few isolates resistant to colistin in our study were from individuals with a weak immune system. From the viewpoint that no new drug is on the way that could be an alternative therapy against A. baumannii resistant to colistin, the cognition of the resistance mechanisms of this bacterium against colistin is an essential matter for the delivery of novel therapeutic approaches and infection control. In the studies carried out recently on the resistance mechanisms of this bacterium, several mechanisms have been considered. One of the mechanisms of A. baumannii resistance against colistin is the expression changes of pmrA and pmrB gene, which is why, in our study this resistance mechanism has been covered. Initially, the number of strains resistant to colistin were identified via Microgen Kit. Later, we assessed the presence of pmrA and pmrB genes via the PCR technique which showed that they were carrying the highest sensitive and resistant strains of these genes.
Furthermore, the expression changes of these genes (pmrA and pmrB) were measured and compared in the sample sensitive to colistin, as well as the samples resistant to colistin. Adams et al, in 2009, carried out assessments in the expression of pmrA and pmrB genes.12 They reported that pmrA had an increased expression in the samples resistant to colistin. The results obtained in the present study showed a significant increase of pmrA gene expression in the samples resistant to colistin, in relation to the samples sensitive to colistin. Besides, in the investigations of pmrB gene expression changes, given the achieved results, it was shown that expression of this gene in the resistant samples was very insignificant in relation to the sensitive samples. Expression increase of pmrA and pmrB genes in the samples resistant to colistin were attributed to LPS loss; colistin has a positive load, thus, influences the LPS part of lipid A, having a negative load, which alters membrane permeability in the samples resistant to colistin in relation to the samples sensitive to colistin. Nevertheless, expression in pmrA and pmrB genes increases, especially in the pmrA gene, where the increase is significant, causing LPS loss which, in turn, results in the lack of membrane permeability alteration and colistin inefficiency on A. baumannii membrane followed by resistance of this organism to colistin. In the study of Alejandro et al, it was stated that in the bacteria resistant to colistin, the gene expression of pmrA increases; however, in the sensitive samples, the expression of this gene is not altered.11 Our findings indicate that what has importance in the resistance of A. baumannii against colistin, is the expression ratio of pmrA and pmrB genes in the samples resistant to colistin. The expression of pmrA gene is dominant in relation to the expression rate of pmrB gene. This domination is the reason for resistance against colistin. Likewise, in the samples sensitive to colistin, in general, the expression of none of these genes is in dominant form. In a bulk of previous research conducted in this regard, the expression rate of pmrA gene in the resistant samples was increased, even in our research this issue is very clearly shown, wherein the expression of pmrA gene in relation to pmrB gene was significantly increased. Therefore, taking into account these results, pmrA gene can be considered responsible for the resistance of A. baumannii against colistin.
Conclusion
During the resistance of A. baumannii against colistin, the expression of pmrA gene is increased, thus, this increased expression has a meaningful relationship with the resistance of this bacteria against colistin. Nevertheless, these results are indicative of the significance and effect of this gene against the advancement of A. baumannii resistance against colistin. Therefore, given the results of this research, pmrA gene can be introduced as a resistant molecular index against colistin. The suppression of this gene can be considered as a perspective for future research on this bacterium.
Acknowledgments
The authors express their gratitude to the staff of ICU and laboratory of Shahid Rajai Hospital and Ali Asghar Hospital in Shiraz for their cooperation.
Disclosure
The authors report no conflicts of interest in this work.
References
Curtis LT. Prevention of hospital-acquired infections: review of non-pharmacological interventions. J Hosp Infect. 2008;69(3):204–219. | ||
Wang H, Guo P, Sun H, et al. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob Agents Chemother. 2007;51(11):4022–4028. | ||
Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An Update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. | ||
Walsh SE, Maillard JY, Russell AD, Catrenich CE, Charbonneau DL, Bartolo RG. Development of bacterial resistance to several biocides and effects on antibiotic susceptibility. J Hosp Infect. 2003;55(2):98–107. | ||
Wroblewska MM, Towner KJ, Marchel H, Luczak M. Emergence and spread of carbapenem-resistant strains of Acinetobacter baumannii in a tertiary-care hospital in Poland. Clin Microbiol Infectm. 2007;13(5):490–496. | ||
Karah N, Sundsfjord A, Towner K, Samuelsen O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist Updat. 2012;15(4):237–247. | ||
Valencia R, Arroyo LA, Conde M, et al. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol. 2009;30(3):257–263. | ||
Moffatt JH, Harper M, Harrison P, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54(12):4971–4977. | ||
Moffatt JH, Harper M, Adler B, Nation RL, Li J, Boyce JD. Insertion Sequence ISAba11 Is Involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob Agents Chemother. 2011;55(6):3022–3024. | ||
Moffatt JH, Harper M, Mansell A, et al. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun. 2013;81(3):684–689. | ||
Beceiro A, Llobet E, Aranda J, et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother. 2011;55(7):3370–3379. | ||
Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;53(9):3628–3634. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

