Back to Journals » Infection and Drug Resistance » Volume 14
Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in Hospitalized Patients in Eastern Heilongjiang Province, China
Authors Yang X, Zhao J , Wang Y, Wu J, Wang X, Wang Y, Zhang Y , Li H
Received 22 February 2021
Accepted for publication 14 April 2021
Published 28 April 2021 Volume 2021:14 Pages 1635—1643
DOI https://doi.org/10.2147/IDR.S307856
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xueying Yang,1 Jing Zhao,1 Yong Wang,1 Jian Wu,2 Xiaohong Wang,1 Yuchao Wang,1 Yiru Zhang,1 Huiling Li1
1Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, 154002, Heilongjiang, People’s Republic of China; 2Department of Biochemistry, Jiamusi Maternal and Child Health Hospital, Jiamusi, 154002, Heilongjiang, People’s Republic of China
Correspondence: Huiling Li
Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, 154002, Heilongjiang, People’s Republic of China
Tel +86 15694549052
Email [email protected]
Background: Recently, owing to antibiotic resistance, the incidence of methicillin-resistant Staphylococcus aureus (MRSA) colonization among intensive care unit (ICU) patients has increased rapidly. So far, there are few studies on active screening of MRSA. The purpose of the current study was to verify the effectiveness of active screening and analyze the molecular epidemiological characteristics of MRSA in the region.
Methods: We collected 30 samples of the MRSA strains from a tertiary hospital in the Eastern Heilongjiang Province. Among them, 7 were retrieved through nasal vestibular swabs at the emergency ICU and 23 were obtained from clinical specimens. Additionally, relevant patient medical information was examined retrospectively and molecular epidemiology and risk factor analysis for MRSA were performed.
Results: Molecular epidemiology studies revealed that all strains of bacteria carried the mecA resistance gene. The Panton Valentine leukocidin (PVL), for instance, was detected at a rate of 13.33% (4/30). The Staphylococcus aureus protein A (spa) types, found amongst our samples, were mainly t324, t437, t034, etc., and we discovered a new spa type t19702. We also revealed 3 types of SCCmec, namely, SCCmec type II, SCCmec type IVa, and SCCmec type V, with the most prevalent clonotypes being ST72 and ST59. In addition, we also found 7 new ST types, namely, ST6567, ST6568, ST6569, ST6570, ST6571, ST6572, and ST6573. Using risk factor analysis, we also demonstrated that long, invasive procedures used in the ICU, such as tracheal intubation and ventilator usage, along with patients with cerebral infarction and other embolism are more susceptible to developing MRSA colonization and further infections.
Conclusion: We recommend the infection control department within hospitals to actively screen for MRSA and perform risk factor analysis in order to establish accurate preventive measures for controlling MRSA spread.
Keywords: methicillin-resistant Staphylococcus aureus, MRSA, active screening, mecA, molecular epidemiology, risk factors, Panton Valentine leukocidin
Introduction
In recent years, susceptible strains of Staphylococcus aureus, exposed to antibiotics, have become resistant.1 As a result, methicillin-resistant Staphylococcus aureus (MRSA) has become a common pathogen causing infections in hospitalized patients.2 It has also been implicated in surgical site infections (SSI).3 MRSA strains produce Panton Valentine leukocidin, an exotoxin that can infect skin and soft tissues.4 Around the world, many different countries and regions including Europe, Africa, and China have conducted research on the molecular epidemiology of MRSA.5–7 Nevertheless, there is a lack of research on the epidemiological characteristics of MRSA in the Eastern part of Heilongjiang Province.
The principle drug resistance gene in MRSA strains is mecA, which is also known to exist in coagulase-negative Staphylococci (CoNS).8 In addition, MRSA carrying the mecC gene has also been reported.9 The common colonization sites of MRSA are the nasal vestibule, pharynx, gastrointestinal tract, and axilla. Over the years, the MRSA colonization rate, producing infections, has dramatically increased among different populations, the most susceptible being children and elderly >85 years of age.10,11 As such, active screening of MRSA strains has become both necessary and urgent. In this study, we demonstrated that the active screening of nasal vestibular swabs in ICU patients can successfully achieve early detection, early intervention, and early treatment of hospitalized patients. This protocol can, therefore, effectively prevent the extensive spread of MRSA strains among hospitalized patients.
Materials and Methods
Strain Source and Ethical Statement
Seven MRSA strains were obtained from active screening of the nasal vestibular swabs and 23 MRSA strains were collected during clinical work. The Staphylococcus aureus strain ATCC25923 was employed as quality control. For patients who actively screened, we obtained oral consent from the patients. The study protocol was approved by the Ethics Committee of Jiamusi University Clinical Medical College (Approval number 0326).
Active Screening Methods
We utilized a bilateral collection of nasal vestibular swabs. The specific protocol of sample retrieval was as follows: a sterile cotton swab was moistened with sterile sodium chloride before slowly inserting it into the patient’s nasal vestibule, followed by gentle rotation of the swab for 5 seconds to collect as much of the nasal specimen as possible. The same swab was used to collect specimens from both nostrils. The retrieved specimen was immediately inoculated in a special medium like Mannitol Salt Agar. Staphylococcus aureus formed a yellow-colored colony on Mannitol Salt Agar.12
DNA Extraction
We used the boiling procedure of bacterial DNA extraction. In brief, 5 hemolytic colonies, cultured overnight on the blood agar, were selected and mixed with 1 mL of sterile sodium chloride, followed by centrifugation, removal of supernatant, addition of 100μL of DNA extract to the pellet, gentle mixing of the pellet in the DNA extract, and heating at 100°C for 10 min. Subsequently, the sample was centrifuged again at 13000 rpm for 1 min and the supernatant, carrying the bacterial DNA, was maintained at −20°C until needed.
Antimicrobial Susceptibility Test (AST)
The antibiotic minimum inhibitory concentrations (MICs) of all MRSA strains were detected using the Siemens Walk Away 40 plus drug susceptibility machine. The MIC results were further confirmed using the disk diffusion method. All drug susceptibility data were based on the criteria developed by the Clinical and Laboratory Standard Institutes (CLSI-2020). The Staphylococcus aureus strain ATCC25923 was employed as the control.
Polymerase Chain Reaction (PCR) to Detect the Resistance Gene and Virulence Gene
The drug resistance gene mecA and the virulence gene PVL of MRSA strains were identified using PCR. The primer sequences used for the mecA gene are as follows: MecA1 (5ʹ-GTA GAA ATG ACT GAA CGT CCG ATA A-3ʹ) and MecA2 (5ʹ-CCA ATT CCA CAT TGT TTC GGT CTA A-3ʹ), as described previously.13 Alternately, the PVL gene was identified using previously reported primers,14 specifically, luk-PV-1 (5ʹ-ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A-3ʹ) and luk-PV-2 (5ʹ-GCA TCA AST GTA TTG GAT AGC AAA AGC-3ʹ). The sequencing results were confirmed using GenBank database BLAST search.
Spa Typing and SCCmec Typing
The Staphylococcus aureus spa gene X region was amplified using ordinary PCR. The primer sequences of the spa gene used were 1095F (5ʹ-AGACGATCCTTCGGTGAGC-3ʹ) and 1517R (5ʹ-GCTTTTGCAATGTCATTTACTG-3ʹ), as described previously.15 Subsequently, the sequencing results were compared and analyzed using the spa dedicated website (https://www.spaserver.ridom.de/). Unlike traditional PCR procedure, the current SCCmec typing used a multiplex PCR method, involving nine pairs of primers and with strict reaction parameters requirements. The sequence and reaction parameters were set, according to published literature.16 Lastly, SCCmec amplified products were analyzed using 2% agarose gel electrophoresis.
Homology Analysis
When the drug susceptibility spectrum of pathogens from the same department is the same or similar, the homology is highly suspected, and then the MLST genotyping method is used to confirm. In order to better reflect the molecular epidemiological characteristics of the region, we have done homology analysis of all 30 MRSA strains.7 housekeeping genes of Staphylococcus aureus were selectively amplified using PCR, sequenced, and compared to the PubMLST database (http://mlst.zoo.ox.ac.uk) to obtain ST typing. The primers used were based on published literature.17
Statistical Analysis
MRSA risk factor analysis used SPSS 22.0 for binary logistic regression analysis. Significance threshold was set at p-value of ≤0.05.
Results
Antimicrobial Susceptibility
MRSA strains drug susceptibility was evaluated using the Siemens Walk Away 40 plus drug susceptibility machine. Based on our results, all examined MRSA strains contained multi-drug resistance (Table 1). In fact, the resistance rate of amoxicillin-clavulanic acid, ampicillin-sulbactam, penicillin, ceftriaxone, cefoxitin, and oxacillin was 100%, whereas the resistance rate of clindamycin and erythromycin was 83.3%, levofloxacin was 10%, and ciprofloxacin was 16.7%. We also demonstrated that among the effective drugs for MRSA strains were gentamicin, vancomycin, and linezolid. Our specimens did not contain any Vancomycin-resistant Staphylococcus aureus.
 |
Table 1 Antimicrobial Susceptibility, Molecular Characteristics, and Outcomes of MRSA |
MRSA Resistance Gene and Virulence Gene
All 30 MRSA strains examined contained the mecA gene, whereas only 4 MRSA (13.3%) carried the virulence gene PVL. For further confirmation, both mecA and PVL genes were sequenced and compared to the GenBank database, namely to the mecA gene (GenBank accession number AB236888) and the PVL genes (GenBank accession numbers X72700 and AB006796) respectively. Our results showed that none of the 7 actively screened MRSA strains carried the PVL gene.
Spa Typing and SCCmec Typing
We further amplified the variable region of the spa gene, using PCR, and found polymorphism or fragments with varying sizes. The product was about 300 bp. Upon sequencing and comparing to the GenBank database, we found multiple spa types; mainly t324, followed by t437 and t034, along with smaller amounts of t9353, t2465, t8347, t664, t091, etc. We also discovered a new spa type among the actively screened strains, namely t19702 (Table 2). We, next, performed SCCmec typing using multiplex PCR, which, unlike traditional PCR, requires multiple pairs of primers and has strict reaction parameters requirements. The sequencing results were further confirmed with GenBank database BLAST search. We discovered a total of 3 SCCmec typings, namely, SCCmec type II, SCCmec type IVa and SCCmec type V (GenBank accession numbers D86934, AB063172 and AB12121), of which the latter two types were prevalent among community-related MRSA. Internal standard gene MecA147 (GenBank accession number X52593) was used as the control.
 |
Table 2 The Characteristics and Typing of the X District of MRSA Spa Gene Repeats |
Homology Analysis
Among the 7 actively screened MRSA strains, the most common type was ST59, followed by ST72, ST7, ST398, and ST2690. Among other MRSA strains, ST861 was also found. We, also, discovered 7 new ST types, namely, ST6567, ST6568, ST6569, ST6570, ST6571, ST6572, and ST6573. The strain sequences were further confirmed with MLST sequence typing reference database (http://mlst.zoo.ox.ac.uk). The genetic relationship between the strains is summarized in Figure 1. This figure is uploaded separately as an attached file.
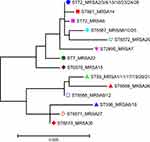 |
Figure 1 Phylogenetic tree of 30 methicillin-resistant Staphylococcus aureus isolates |
Risk Factor Analysis for MRSA Colonization and Infection
We performed binary logistic regression analysis on 30 experimental MRSA strains and 52 methicillin-sensitive Staphylococcus aureus (MSSA) strains. Based on our results, ICU hospitalization history, cerebral infarction and other embolism, tracheal intubation, ventilator usage, and other invasive procedures provide a high risk for MRSA colonization (Table 3).
 |
Table 3 Risk Factor Analysis of MRSA Colonization |
Discussion
The principle antibiotic resistance pathway used by MRSA involves the mecA gene, which produces a low-affinity penicillin-binding protein, namely PBP2a, thereby leading to resistance. Simultaneously, mecA belongs to the SCCmec mobile genetic element. Therefore, the drug resistance gene mecA can be transferred horizontally with SCCmec, conferring antibiotic resistance to other Staphylococcus aureus strains. As a result, the MRSA strains, in community and hospital environments, have become highly resistant to antibiotics.18 Unfortunately, there are reports that the resistance of Staphylococcus aureus to methicillin has increased, even in people not exposed to hospital risk factors.19
MRSA strains are mainly separated into hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA), based on its existence location and strain characteristics. HA-MRSA can usually be found in patients after a 48 h hospital stay, whereas, CA-MRSA can potentially be present in patients within 48 hours of outpatient or hospitalization visitation. HA-MRSA strains usually carry SCCmec type I, SCCmec type II, and SCCmec type III, whereas, CA-MRSA carried SCCmec type IV or SCCmec type V. However, multiple studies have demonstrated that the CA-MRSA and HA-MRSA strains can coexist within communities and hospital environments. For example, in Brazil, both SCCmec types IVa and V were detected in the HA-MRSA isolates.20,21
This article is the first published report on the active screening and molecular epidemiological research of MRSA in hospitalized patients in Eastern Heilongjiang Province. The active MRSA screening was performed via nasal vestibular swabs from ICU patients. Using the active screening method, we isolated 7 MRSA strains. We further extracted 23 MRSA strains from clinical specimens. The mecA resistance gene was discovered in all examined strains, and only 4 strains (4/30; 13.33%) contained the PVL gene, which is similar to a report from a dental hospital in Egypt.22 Interestingly, studies show that SCCmec carries the resistance gene, and the resistance gene can spread horizontally among different staphylococcal species.23 Using multiplex PCR, we isolated three forms of SCCmec, namely, SCCmec type II, SCCmec type IVa, and SCCmec type V, of which SCCmec type IVa was the most abundant. This finding is in accordance with the SCCmec classification in Hainan and with a study by Alkharsah et al24,25. We further used PCR to amplify the variable region of the spa gene in Staphylococcus aureus. Upon GenBank database comparison and analysis of the sequencing results, a total of 14 spa types were found, of which the main types were t324, t437, and t034. In addition, there were t9353, t2465, t8347, t664, and so on, among our samples. Notably, we also found a new classification, termed t19702. There is no general consensus on the ST typing of MRSA. The main types of MRSA strains in our hospital were ST72, ST59, ST398, ST861, and other ST types. Among them, we discovered 7 new ST types, namely, ST6567, ST6568, ST6569, ST6570, ST6571, ST6572, and ST6573. Our finding is consistent with a study conducted in Iran, who also reported MRSA ST typing, including ST1465, ST861, ST889 and ST772.26 In Eastern Heilongjiang Province, the most important MRSA clones were ST59-SCCmecIV-t437 and ST72-SCCmecIV-t324. In Shenzhen, the main MRSA clone was ST59-SCCmecIV-t437.27 In Northwest China, the most common clone was ST239-SCCmecIII-t030.28 Based on published reports, the ST398 typing is a zoonotic clonotype.29
MRSA infection can infect the skin and soft tissues, induce bacteremia, and endocarditis, and significantly increase the risk of morbidity and mortality.30 A combination of daptomycin and ceftaroline has been reported to lower fatality risk of MRSA bacteremia patients.31 In this study, a binary logistic regression analysis was performed to assess the risk factors of MRSA. Based on our analysis, cerebral infarction, embolism in other areas, ICU hospitalization, and invasive procedures like tracheal intubation and ventilator usage provided the main risk factors for MRSA colonization. Therefore, we recommend physicians to remain vigilant and provide timely intervention measures to high-risk patients, in order to prevent MRSA infection. The colonization rate of MRSA in ICU patients in mainland China is remarkably high, especially in patients whose nasal vestibular swabs test positive for MRSA.32 There are also reports of MRSA strains extracted from the tracheal intubation of ICU patients.33 Other researchers have suggested that the ICU admission history may be a risk factor for MRSA infection.34 Therefore, active monitoring of MRSA in ICU patients is crucial and must be considered. Additionally, there are emerging evidences of MRSA surges in non-hospital related environments. One study revealed that chronic skin diseases and invasive procedures, conducted in nursing homes, can elevate MRSA infection risk.35 In addition, patients with weakened immunity and diabetes are prone to MRSA infection.36,37 Moreover, medical staff have been reported to carry MRSA, increasing the potential for transmission in both hospital and non-hospital environments.38 There are also reports of livestock-related MRSA (LA-MRSA) colonization, suggesting a possible spread of MRSA strains between humans and animals.39,40 Special care needs to be given in all of these special cases to maintain MRSA infections to a minimum. In future research, we plan to screen ICU medical staff for MRSA, which has special significance for the prevention and control of MRSA in hospitals.
Conclusions
In summary, we report here, for the first time, the epidemiological characteristics of MRSA in Eastern Heilongjiang Province and recommend measures for active ICU screening of MRSA. Active screening proved to be an effective preventive measure for MRSA infection. The main MRSA clones in this area, according to our study, were ST72 and ST59. With the use of active ICU patient screening and risk factors analysis of MRSA colonization, clinical intervention can be performed on drug-resistant bacteria to prevent infection. This is of great significance in the improvement of patient survival and in preventing further spread of this drug-resistant bacteria.
Abbreviations
MRSA, Methicillin-resistant Staphylococcus aureus; MSSA, Methicillin-sensitive Staphylococcus aureus; CoNS, Coagulase-negative Staphylococci; ICU, Intensive Care Unit; CLSI, Clinical and Laboratory Standard Institutes; PVL, Panton Valentine leukocidin; Spa, Staphylococcus aureus protein A; SCCmec, Staphylococcal cassette chromosome mec; MICs, Minimum inhibitory concentrations; AST, Antimicrobial susceptibility test; PCR, Polymerase chain reaction; MLST, Multilocus sequence typing.
Data Sharing Statement
Any undisclosed data from this study is available upon request.
Ethics Approval and Consent to Participate
All protocols were agreed upon by the Ethics Committee of Jiamusi University Clinical Medical College (0326). Patient informed consent was waived as the samples were collected from existing specimens retrieved during routine procedures and did not require further involvement from patients. For patients who actively screened, we obtained oral consent from the patients.
Consent for Publication
Not applicable.
Acknowledgments
We thank the First Affiliated Hospital of Jiamusi University for their assistance in our study. We are, also, grateful to the curator team at the University of Wuerzburg, Germany for assigning novel profiles at http://mlst.zoo.ox.ac.uk. We thank Professor Xiaoli Zhang from Yongchuan Hospital of Chongqing Medical University for her guidance on our experimental research.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, drafting and revising the article, gave final approval of the version to be published, have agreed on the journal to which the article will be submitted and agreed to be accountable for all aspects of the work. The final manuscript was reviewed and approved by all authors.
Funding
The Heilongjiang Provincial Medicine and Health Research Project(2020-352) funded this research.
Disclosure
The authors have no competing interests.
References
1. Goering RV, Swartzendruber EA, Obradovich AE, Tickler IA, Tenover FC. Emergence of oxacillin resistance in stealth methicillin-resistant Staphylococcus aureus due to mecA sequence instability. Antimicrob Agents Chemother. 2019;63:e00558–19.
2. Gajdács M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics (Basel). 2019;8:52. doi:10.3390/antibiotics8020052
3. Sun Y, Wang H, Tang Y, et al. Incidence and risk factors for surgical site infection after open reduction and internal fixation of ankle fracture: a retrospective multicenter study. Medicine (Baltimore). 2018;97:e9901. doi:10.1097/MD.0000000000009901
4. Hanawa T, Shimoda-Komatsu Y, Araki K, et al. Skin and soft tissue infections caused by different genotypes of PVL-Positive community-acquired methicillin-resistant Staphylococcus aureus strains. Jpn J Infect Dis. 2020;73:72–75. doi:10.7883/yoken.JJID.2019.162
5. Hetem DJ, Derde LP, Empel J, et al.; MOSAR WP3 Study Group. Molecular epidemiology of MRSA in 13 ICUs from eight European countries. J Antimicrob Chemother. 2016;71:45–52. doi:10.1093/jac/dkv298
6. Singh-Moodley A, Lowe M, Mogokotleng R, Perovic O. Diversity of SCCmec elements and spa types in South African Staphylococcus aureus mecA-positive blood culture isolates. BMC Infect Dis. 2020;20:816. doi:10.1186/s12879-020-05547-w
7. Kong H, Yu F, Zhang W, Li X, Wang H. Molecular epidemiology and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus strains in a tertiary hospital in China. Front Microbiol. 2017;8:838. doi:10.3389/fmicb.2017.00838
8. Shrestha LB, Bhattarai NR, Rai K, Khanal B. Antibiotic resistance and mecA gene characterization of coagulase-negative Staphylococci isolated from clinical samples in Nepal. Infect Drug Resist. 2020;13:3163–3169. doi:10.2147/IDR.S274163
9. Khan AA, Ali A, Tharmalingam N, Mylonakis E, Zahra R. First report of mecC gene in clinical methicillin resistant S. aureus (MRSA) from tertiary care hospital Islamabad, Pakistan. J Infect Public Health. 2020;13:1501–1507. doi:10.1016/j.jiph.2020.05.017
10. Tuta KE, Okesola AO, Umeokonkwo CD. The prevalence and risk factors associated with nasal methicillin-resistant Staphylococcus aureus colonization among children in a tertiary hospital in Nigeria. Ethiop J Health Sci. 2019;29:487–494. doi:10.4314/ejhs.v29i4.10
11. Kasela M, Grzegorczyk A, Korona-Głowniak I, Ossowski M, Nowakowicz-Dębek B, Malm A. Transmission and long-term colonization patterns of Staphylococcus aureus in a nursing home. Int J Environ Res Public Health. 2020;17:8073. doi:10.3390/ijerph17218073
12. Gajdács M, Urbán E. Epidemiology and resistance trends of Staphylococcus aureus isolated from vaginal samples: a 10-year retrospective study in Hungary. Acta Dermatovenerol Alp Pannonica Adriat. 2019;28:143–147.
13. Zhang K, Sparling J, Chow BL, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol. 2004;42:4947–4955. doi:10.1128/JCM.42.11.4947-4955.2004
14. Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi:10.1086/313461
15. Harmsen D, Claus H, Witte W, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–5448. doi:10.1128/JCM.41.12.5442-5448.2003
16. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi:10.1128/JCM.43.10.5026-5033.2005
17. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi:10.1128/JCM.38.3.1008-1015.2000
18. Gittens-St Hilaire MV, Chase E, Alleyne D. Prevalence, molecular characteristics and antimicrobial susceptibility patterns of MRSA in hospitalized and nonhospitalized patients in Barbados. New Microbes New Infect. 2020;35:100659. doi:10.1016/j.nmni.2020.100659
19. Rodriguez F, Salinas C, Fernandez S, et al. Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) clones from Paraguayan children. J Infect Dev Ctries. 2020;14:290–297. doi:10.3855/jidc.12108
20. Kateete DP, Bwanga F, Seni J, et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob Resist Infect Control. 2019;8:94. doi:10.1186/s13756-019-0551-1
21. de Carvalho SP, de Almeida JB, Andrade YMFS, et al. Molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from hospital and community environments in northeastern Brazil. Braz J Infect Dis. 2019;23:134–138. doi:10.1016/j.bjid.2019.04.005
22. Khairalla AS, Wasfi R, Ashour HM. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment. Sci Rep. 2017;7:7390. doi:10.1038/s41598-017-07713-8
23. Smyth DS, Wong A, Robinson DA. Cross-species spread of SCCmec IV subtypes in staphylococci. Infect Genet Evol. 2011;11:446–453. doi:10.1016/j.meegid.2010.12.005
24. Li X, Huang T, Xu K, Li C, Li Y. Molecular characteristics and virulence gene profiles of Staphylococcus aureus isolates in Hainan, China. BMC Infect Dis. 2019;19:873. doi:10.1186/s12879-019-4547-5
25. Alkharsah KR, Rehman S, Alkhamis F, Alnimr A, Diab A, Al-Ali AK. Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann Clin Microbiol Antimicrob. 2018;17:7. doi:10.1186/s12941-018-0260-2
26. Firoozeh F, Omidi M, Saffari M, Sedaghat H, Zibaei M. Molecular analysis of methicillin-resistant Staphylococcus aureus isolates from four teaching hospitals in Iran: the emergence of novel MRSA clones. Antimicrob Resist Infect Control. 2020;9:112. doi:10.1186/s13756-020-00777-8
27. Qin Y, Wen F, Zheng Y, Zhao R, Hu Q, Zhang R. Antimicrobial resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from child patients of high-risk wards in Shenzhen, China. Jpn J Infect Dis. 2017;70:479–484. doi:10.7883/yoken.JJID.2016.328
28. Yuan W, Liu J, Zhan Y, et al. Molecular typing revealed the emergence of pvl-positive sequence type 22 methicillin-susceptible Staphylococcus aureus in Urumqi, Northwestern China. Infect Drug Resist. 2019;12:1719–1728. doi:10.2147/IDR.S202906
29. Pirolo M, Visaggio D, Gioffrè A, et al. Unidirectional animal-to-human transmission of methicillin-resistant Staphylococcus aureus ST398 in pig farming; evidence from a surveillance study in southern Italy. Antimicrob Resist Infect Control. 2019;8:187. doi:10.1186/s13756-019-0650-z
30. Leibler JH, León C, Cardoso LJP, et al. Prevalence and risk factors for MRSA nasal colonization among persons experiencing homelessness in Boston, MA. J Med Microbiol. 2017;66:1183–1188. doi:10.1099/jmm.0.000552
31. Geriak M, Haddad F, Rizvi K, et al. Clinical data on daptomycin plus ceftaroline versus standard of care monotherapy in the treatment of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob Agents Chemother. 2019;63:e02483–18. doi:10.1128/AAC.02483-18
32. Qiao F, Huang W, Cai L, Zong Z, Yin W. Methicillin-resistant Staphylococcus aureus nasal colonization and infection in an intensive care unit of a university hospital in China. J Int Med Res. 2018;46:3698–3708. doi:10.1177/0300060518777812
33. Cabrera R, Fernández-Barat L, Motos A, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus clinical strains from the endotracheal tubes of patients with nosocomial pneumonia. Antimicrob Resist Infect Control. 2020;9:43. doi:10.1186/s13756-020-0679-z
34. Huang L, Zhang R, Hu Y, et al. Epidemiology and risk factors of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci infections in Zhejiang China from 2015 to 2017. Antimicrob Resist Infect Control. 2019;8:90. doi:10.1186/s13756-019-0539-x
35. Peters C, Dulon M, Kleinmüller O, Nienhaus A, Schablon A. MRSA prevalence and risk factors among health personnel and residents in nursing homes in Hamburg, Germany - A Cross-Sectional Study. PLoS One. 2017;12:e0169425. doi:10.1371/journal.pone.0169425
36. Hsu YY, Wu D, Hung CC, et al. Methicillin-resistant Staphylococcus aureus nasal colonization among HIV-infected patients in Taiwan: prevalence, molecular characteristics and associated factors with nasal carriage. BMC Infect Dis. 2020;20:254. doi:10.1186/s12879-020-04979-8
37. Kananizadeh P, Ohadian Moghadam S, Sadeghi Y, Rahimi Foroushani A, Adibi H, Pourmand MR. Molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) isolated from diabetic foot infection. Iran J Pathol. 2019;14:329–337. doi:10.30699/IJP.2019.101092.2035
38. Buenaventura-Alcazaren FA, Dela TA, Ong-Lim A, Destura RV. Prevalence and molecular characteristics of MRSA nasal carriage among hospital care workers in a tertiary hospital in the Philippines. J Microbiol Immunol Infect. 2020;53:739–745. doi:10.1016/j.jmii.2018.12.016
39. Pirolo M, Gioffrè A, Visaggio D, et al. Prevalence, molecular epidemiology, and antimicrobial resistance of methicillin-resistant Staphylococcus aureus from swine in southern Italy. BMC Microbiol. 2019;19:51. doi:10.1186/s12866-019-1422-x
40. Asanin J, Misic D, Aksentijevic K, et al. Genetic profiling and comparison of human and animal methicillin-resistant Staphylococcus aureus (MRSA) isolates from Serbia. Antibiotics (Basel). 2019;8:26. doi:10.3390/antibiotics8010026
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
